Abstract
Background
Childhood Central Nervous System Primitive Neuro-Ectodermal brain Tumours (CNS-PNETs) are highly aggressive brain tumours for which molecular features and best therapeutic strategy remains unknown. We interrogated a large cohort of these rare tumours in order to identify molecular markers that will enhance clinical management of CNS-PNET.
Methods
Transcriptional and copy number profiles from primary hemispheric CNS-PNETs were examined using clustering, gene and pathways enrichment analyses to discover tumour sub-groups and group-specific molecular markers. Immuno-histochemical and/or gene expression analyses were used to validate and examine the clinical significance of novel sub-group markers in 123 primary CNS-PNETs.
Findings
Three molecular sub-groups of CNS-PNETs distinguished by primitive neural (Group 1), oligo-neural (Group 2) and mesenchymal lineage (Group 3) gene expression signature were identified. Tumour sub-groups exhibited differential expression of cell lineage markers, LIN28 and OLIG2, and correlated with distinct demographics, survival and metastatic incidence. Group 1 tumours affected primarily younger females; male: female ratios were respectively 0.61 (median age 2.9 years; 95% CI: 2.4–5.2; p≤ 0.005), 1.25 (median age 7.9 years; 95% CI: 6–9.7) and 1.63 (median age 5.9 years; 95% CI: 4.9–7.8) for group 1, 2 and 3 patients. Overall outcome was poorest in group 1 patients which had a median survival of 0.8 years (95% CI: 0.47–1.2; p=0.019) as compared to 1.8 years (95% CI: 1.4–2.3) and 4.3 years; (95% CI: 0.82–7.8) respectively for group 2 and 3 patients. Group 3 tumours had the highest incidence of metastases at diagnosis; M0: M+ ratio were respectively 0.9 and 3.9 for group 3, versus group 1 and 2 tumours combined (p=0.037).
Interpretation
LIN28 and OLIG2 represent highly promising, novel diagnostic and prognostic molecular markers for CNS PNET that warrants further evaluation in prospective clinical trials.
Introduction
Brain tumours are the most common paediatric solid neoplasms1 and a leading cause of childhood cancer related morbidity and mortality2. Embryonal tumours comprise the largest group of malignant paediatric brain tumours and include medulloblastoma, atypical rhabdoid teratoid tumour (ATRT) and central nervous system primitive neuro-ectodermal tumours (CNS PNETs). Despite histologic resemblance to medulloblastoma, patients with CNS-PNETs fare poorly even with intensified therapy designed for patients with metastatic medulloblastoma3,4. In contrast to medulloblastoma where substantial progress has been made in molecular understanding5,6 and clinical outcomes7, the molecular and cellular make-up of CNS-PNET remain largely unknown8 and tumour treatments are frequently ineffective. To advance CNS-PNET therapeutics, it will be important to delineate the cellular and molecular pathogenesis of CNS-PNET in order to better inform diagnosis, prognosis and tumour specific treatment design.
CNS-PNETs are predominantly hemispheric tumours comprising ~3–5% of all paediatric brain tumours. They are histologically heterogeneous with variable neuronal, ependymal or glial differentiation9 and can be challenging to diagnose by routine histo-pathology10. In contrast to ATRT11 and medulloblastoma, where significant advances have been made in specific diagnostic tools and molecular-based tumour classification, working classification for CNS-PNET remains in flux thus presenting challenges for both therapeutic and molecular studies. In recent studies our research groups identified a distinctly aggressive sub-group of CNS-PNETs with frequent amplification of an oncogenic miRNA cluster (C19MC)12,13. However, the molecular make-up of the majority of CNS-PNET remains unknown. Although genomic studies to date reveal substantial heterogeneity in DNA copy number profiles of CNS-PNETs8,12,14, the significance of these findings in relation to clinical phenotypes remains unclear. Similarly, gene expression studies of small cohorts to date12,15 have yielded limited insights into the clinical diversity of CNS-PNETs.
Recognizing the challenges and limitations of prior studies, we undertook a multi-institutional, international collaboration to enable concerted molecular analyses of a substantial numbers of primary CNS-PNETs. In this study we integrated gene expression, copy number and immuno-histochemical analyses to characterize 123 primary hemispheric CNS-PNETs and established that CNS-PNET comprise three molecular subgroups. Importantly, we demonstrate that expression of cell lineage markers, LIN28 and OLIG2, identifies sub-groups of CNS-PNET with distinct demographics, survival and incidence of metastases. Our study is the first to provide biological markers for clinically relevant subgroups of CNS PNET and establishes an important foundation for prospective therapeutic studies.
Methods and Materials
Clinical cohort and tumour materials
Tumour samples and patient clinical information were collected with consent as per protocols approved by the Hospital Research Ethics Board at participating institution (Supplemental Table 1) including 6 Children's Cancer and Leukemia Group (CCLG) registered centers in the UK, and from the Cooperative Human Tissue Network (CHTN, Columbus, Ohio USA). CNS-PNET tissue microarrays used in this study were constructed at the Hospital for Sick Children12, University of Nottingham14 and Institute of Cancer Research in Sutton, UK. All collected tumour samples were re-reviewed blindly (CEH), and tested for loss of INI1 immuno-reactivity or INI1 alterations by sequencing or MLPA analyses to rule out misdiagnosed ATRT. Only hemispheric tumours diagnosed as CNS-PNET according to the 2007 WHO CNS tumour classification criteria9 and without alterations of INI1 were included. For clinical correlative analyses, only tumours with complete clinical information were included; patient and tumour information are listed in Supplemental Table 2.
Gene expression and DNA copy number profiles
DNA and RNA were extracted using standard methods from 51 and 85 primary CNS-PNET samples respectively and analysed using Illumina Omni 2.5M SNP and Illumina HT-12 v4 gene expression arrays (http://www.illumina.com) to generate gene expression and DNA copy number profiles. DNA and RNA hybridisations were performed at The Centre of Applied Genomics Facility (TCAG), Hospital for Sick Children, Toronto (http://www.tcag.ca/), according to the manufacturer's protocol. For clinical correlative analyses, only 59 of the 85 tumours with copy number profiles with complete clinical information were included. Details of molecular analyses performed on individual tumour samples are shown in Supplemental Table 1; all data are deposited in the Wellcome Trust, European Genome-phenome Archive (www.EBI.AC.UK; Accession number:EGAS00000000116).
PCR, Fluorescence in-situ hybridization and immuno-histochemical studies
For gene specific qRT-PCR validation of array data, 10ng cDNA synthesized from 1ug of RNA (TaqMan® Reverse Transcription Kit, Applied Biosystems) was amplified using specific TaqMan probes/primer sets (Supplemental Table 3) and mRNA expression levels were determined relative to actin using the ΔCt method. All RT-PCR assays were performed in triplicate. Immuno-histochemical analyses on tumour tissue microarray or FFPE tumour slides were performed by the Pathology Research Program laboratory (http://www.uhnres.utoronto.ca). All tissue sections were treated with heat-induced epitope retrieval and blocked for endogenous peroxidase and biotin. Antibodies used in this study included: anti-LIN28 (Cell Signalling Technology, Boston, USA), OLIG2, (Immuno-Biological Laboratories, Minneapolis, USA), GFAP (DAKO, Burlington, CA) and SYNAPTOPHYSIN (Millipore, Massachusetts). Antibody reactions were visualized using a Biogenix detection kit (BioGenix Laboratories, San Ramon, USA). Immuno-reactivity for LIN28, GFAP and SYNAPTOPHYSIN were scored manually based on intensity (1=low, 2=mod, 3=high) and distribution of stains (1=≤10%, 2=10–50%, 3>50%). OLIG2 immuno-stains were quantified using the Aperio Scanscope (Aperio, Vista CA, USA) system and the ImageScope software nuclear IHC algorithm. For tumours on TMA, IHC values was determined based on average staining score of at least 2 tissue cores, while tumours with FFPE slides were scored based on the extent of staining in relation to the entire tumour section. Normal testicular tissue (human and murine) and Oligodendroglioma tumour tissue were used respectively as positive controls for LIN28 and OLIG2 immuno-stains; samples processed in parallel without primary antibodies served as negative controls. All IHC stains were scored blindly by DP and TC, and reviewed by AH and CEH. FISH was performed on FFPE TMA or individual slides using established protocols. Test MYCN (2p24) and p16 (9p21) specific PlatinumBright550 probe with corresponding LAF (2q11) and 9q21 PlatinumBright495 control probes (Kreatech, Stretton Scientific, Stretton, UK) were used.
Informatics and Statistical Analyses
CNS-PNETs were classified into molecular sub-groups by unsupervised hierarchical clustering (HCL), Non-negative Matrix Factorization (NMF)16 and principle component (PCA) analyses of genes with the highest co-efficient of variation using the Partek Genomics Suite (Partek, St Louis, MO), version 6.5. Genes enriched within tumour sub-groups were determined using a supervised t-test adjusted for multiple hypotheses testing using the FDR method. Ingenuity pathway analyses were performed on supervised gene sets to identify canonical signalling pathways in each tumour sub-group. To determine regions of copy number gains and losses, inferred copy number data was generated using the Illumina Genome studio software and was imported into Partek for CNV partitioning/segmentation analyses using a SNP window of 150. Significance of Copy Number Alterations (CNAs) in tumour sub-groups was then determined using Fisher's exact test. Log rank analysis using the Kaplan Meier method and Chi-square analyses was used respectively to compare survival time and proportion of survivors across tumour sub-groups, while ANOVA was used to determine significance of tumour sub-group in relation to age. To analyse the significance of molecular sub-groups in relation to gender and metastatic status at diagnosis, features in an individual molecular sub-group was compared to a pooled cohort of the other two molecular sub-groups using Fisher's exact test. Adjustment for multiple testing was not performed as patients with complete information available for each clinical parameter varied. All statistical analyses were performed using the SPSS software package, version 19.
Role of funding source
The sponsors of the study only provided financial support and had no role in collection, analyses or interpretation of data or writing of this manuscript. DP, SM, RG and AH had access to the raw data. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Results
Gene expression profiles segregate CNS-PNET into 3 distinct molecular sub-groups
We performed multiple unsupervised analyses on expression profiles of 51 primary hemispheric CNS-PNET. HCL and NMF clustering using 200–1000 genes consistently identified 3 distinct molecular sub-groups of CNS-PNET with NMF analyses indicating strongest cophenetic co-efficient at k=3 (Figure 1A; Supplemental Figure 1A, B). Principle Component Analyses showed group 1 tumours, which have frequent C19MC locus amplification, segregated distinctly while group 2 and 3 tumours showed greater proximity and some overlap (Supplemental Figure 1C).
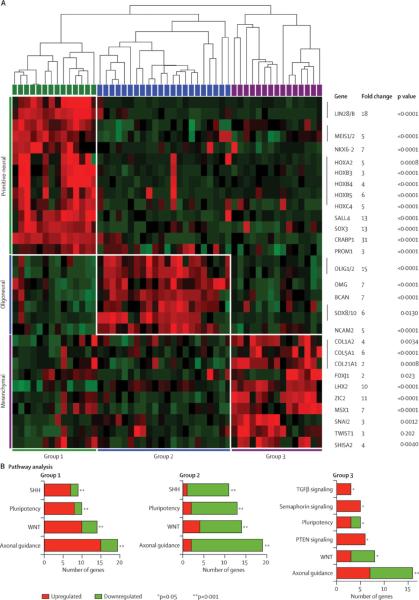
Figure 1. Molecular sub-groups of CNS-PNET exhibit distinct cell lineage and signalling signatures.
A. Unsupervised cluster analyses were performed on human HT-12v4 expression array (Illumina) data from 51 primary CNS-PNET samples to identify the most stable tumour grouping with a minimal gene set (Supplemental Figure 1). Heat map shows the most highly expressed cell lineage genes in each sub-group identified using a supervised t-test adjusted for multiple testing (FDR≤0.05), relative to a hierarchical cluster map of all tumours. Magnitude and significance of cell lineage genes most significantly up-regulated in each tumour sub-groups are denoted respectively by fold change and p-values.
B. Signalling pathways enriched in each tumour sub-group were determined by Ingenuity Pathways Analyses of group-specific gene sets derived from supervised analyses. Most significantly altered canonical pathways determined from analyses of 343, 276 and 325 genes respectively in groups 1, 2 and 3 are represented in relation to tumour sub-group. Proportion of up or down regulated genes within each category are respectively indicated in red and green.
To define genes or pathways that characterize each CNS-PNET sub-group, we performed supervised analyses of each sub-group relative to the others, and examined the most highly differentially expressed gene sets between sub-groups for gene and pathway enrichment. The 3 sub-groups showed significant differences in neural lineage and differentiation genes (Figure 1A). Expression profiles of group 1 were most significantly enriched for genes associated with embryonic or neural stem cells. Notably, LIN28 and CRABP117 which are implicated in stem cell pluripotency, were amongst the top over-expressed genes with nearly 20–30 fold greater expression in group 1 as compared to group 2 and 3 CNS-PNET. In group 2 tumours, OLIG1/2, SOX10 and BCAN, which are markers of oligo-neural differentiation18, were the most highly up-regulated genes, while group 3 tumours demonstrated more limited expression of neural differentiation genes, but showed significant up-regulation of epithelial and mesenchymal differentiation genes including COL1A, COL5A, FOXJ119 and MSX120.
Pathway enrichment analyses also indicated significant differences in the signalling gene profiles of each tumour sub-group (Figure 1B). Consistent with differential enrichment of lineage related genes in tumour sub-groups, we observed significant differences in expression of axonal guidance genes amongst the CNS-PNET sub-groups. Genes involved in WNT and SHH signalling were most significantly up- and down regulated respectively in group 1 and 2 tumours, while TGF-β and PTEN signalling pathway genes were specifically up-regulated in group 3 tumours.
Expression of cell lineage markers, LIN28 and OLIG2, distinguish molecular sub-groups of CNS-PNET
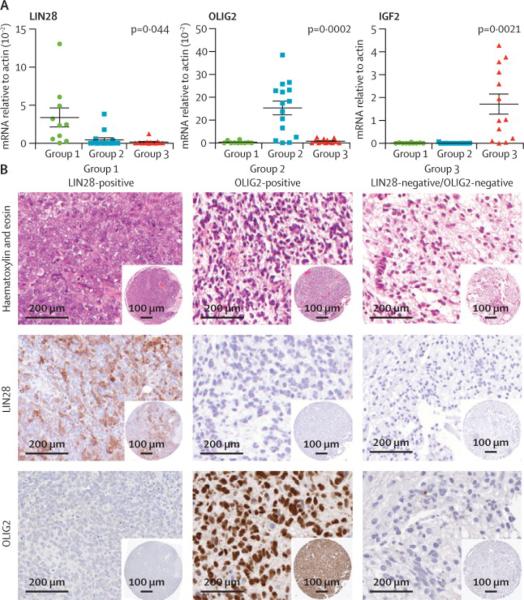
In order to determine the clinical significance of the molecular sub-groups, we sought markers of each sub-group that could be examined by IHC on a larger cohort of clinically well characterized tumours with only FFPE materials available for study. We performed qRT-PCR analyses to validate group-specific gene clusters identified by supervised analyses and examined expression levels of individual genes across groups using t-tests in order to identify the most robust, up-regulated loci that can distinguish tumour sub-groups. These analyses revealed that LIN28, OLIG2 and IGF2 genes were highly differentially expressed in CNS-PNET groups 1, 2 and 3 tumours respectively (Figure 2A, Supplemental Figure 2A, B). IGF2 protein expression could not be reliably scored on tumour samples (Supplemental Figure 3), however IHC analyses for LIN28 and OLIG2 was robust and correlated well with gene expression levels determined by arrays and qRT-PCR analyses. IHC analyses on a test cohort of 22 tumours indicated that extent of cytoplasmic LIN28 and nuclear OLIG2 immuno-staining also correlated with tumour sub-group assignment based on gene expression profiles (Supplemental Figure 4A, B). LIN28 and OLIG2 expression were characteristically high and low respectively in Group 1 tumours, while Group 2 tumours exhibited high OLIG2 and minimal LIN28 immuno-positivity. LIN28 and OLIG2 protein expression was low or absent in Group 3 tumours (Figure 2B).
Figure 2. Cell lineage markers, LIN28 and OLIG2, identify molecular sub-groups of CNS-PNET.
A. Enrichment of specific lineage genes in CNS-PNET sub-groups was confirmed by qRT-PCR analyses of 51 primary CNS-PNET profiled by gene expression arrays (Supplemental Figure 2). Meanexpression levels of LIN28, OLIG2 and IGF2 (n=3 replicas), which were most highly enriched respectively in group 1, 2 and 3 CNS-PNETs, are represented with SEM (horizontal bars). Transcript levels within CNS-PNET sub-groups 1, 2 and 3 are indicated by green, blue and purple bars or spheres respectively.
B. Distinct expression patterns of LIN28 and OLIG2 in molecular sub-groups of CNS-PNET was validated and further characterized by immuno-histochemical analyses in a larger cohort of CNS-PNET (n=72) (Supplemental Figure 2). Characteristic LIN28 and OLIG2 immuno-stains (10× magnification) in CNS-PNET sub-groups are shown in relation to a hematoxylin and eosin (H&E) stain. Corresponding tissue microarray core is shown at low magnification in inset
We performed LIN28 and OLIG2 IHC analyses on an additional 72 primary CNS-PNETs with only FFPE samples available for analyses; 15 tumours had inconclusive IHC analyses (Supplemental Table 4). In sum, 108 primary CNS-PNETs could be assigned to molecular sub-groups based on gene expression and/or IHC analyses of LIN28 or OLIG2 protein expression. Group 1, 2 and 3 tumours respectively comprised 27%, 33% and 40% of tumours analysed (Supplemental Figure 4C). Expression of markers for primitive neural, glial (Nestin, GFAP) or neuronal (synaptophysin) differentiation which are conventionally used in histo-pathologic diagnosis of CNS-PNET, was also performed on all tumours. Group 1 tumours with high LIN28 expression generally also expressed high levels of Nestin, but exhibited limited to no GFAP expression. GFAP and synaptophysin expression varied substantially between each of the tumour groups and did not consistently correlate with LIN28 or OLIG2 expression (data not shown). Notably, qRT-PCR/expression analyses indicated that expression of other neuronal differentiation genes also do not significantly differ amongst the molecular sub-groups of CNS-PNETs (Supplemental Figure 5A, B). These findings collectively highlight the limitations of conventional markers to capture the molecular diversity of CNS-PNETs.
CNS-PNET sub-groups have distinct DNA copy number patterns
Studies in a number of other brain tumours indicate CNAs may frequently drive gene expression signatures5,6, and thus may also represent important sub-group specific markers. We performed ultra-high resolution copy number analyses of 85 CNS-PNETs using the Illumina Omni Quad arrays which interrogate 2.5 million SNPs. With the exception of the C19MC miRNA amplicon we previously identified12, there were few other recurrent high level copy number gains or amplification. Focal MYCN and CDK4 amplification was detected in isolated tumours. Deletions centred on CDKN2A/2B were the most frequent CNA observed (10 of 85 tumours) (Supplemental Figure 6). To determine whether there were characteristic CNAs within the CNS-PNET sub-groups, we analysed the copy number patterns of a sub-set of 59 tumours which could be sub-grouped based on LIN28 and OLIG2 gene or protein expression (Supplemental Figure 7). Copy number analyses showed that in addition to chr19q13.41 amplification and chr2 gains, group1 tumours had frequent gains of chr3. Group 2 tumours CNA profiles were characterized by more frequent gains of chr 8p (p=0.027), 13 (p=0.009) and 20 (p=0.039) compared to group 1 and 2 tumours. Notably, only group 2 and 3, but not group 1 tumours exhibited frequent chr 9p loss centred on the CDKN2A/2B locus. In addition, group 3 tumours showed frequent loss of chr14 (p=0.009). Thus CNS-PNET sub-groups correlate with distinct gene expression as well as genomic profiles.
Molecular sub-groups identify CNS-PNETs with distinct clinical phenotypes
In order to determine the clinical significance of CNS-PNET molecular sub-groups, we examined whether sub-groups differed in patient characteristics and outcome. Of 108 patients for which tumour sub-grouping could be established, demographic data on gender, age, survival time and tumour stage were respectively available for 107, 100, 58 and 66 cases (Table 1, 2; Supplemental Table 2).
Table 1.
Clinical and Molecular Features of CNS-PNET
| Group 1 | Group 2 | Group 3 | Total available for analysis | p Value | ¥Comparison | |
|---|---|---|---|---|---|---|
| Total Number | 29 | 36 | 43 | 108 | ||
| Variable | Number Available for Analyses | |||||
| Gender | 29 | 36 | 42 | 107 | ||
| male | 11 | 20 | 26 | |||
| female | 18 | 16 | 16 | |||
| male:female ratio | 0.62 | 1.25 | 1.63 | □ p=0.043 | Grp 1 vs [2&3] | |
| Age at Diagnosis | 26 | 32 | 42 | 100 | ||
| Median Age (yrs) | 2.9 | 7.9 | 5.9 | ° p=0.005 | ||
| 95% Confidence Interval | 2.4–5.2 | 6–9.7 | 4.9–7.8 | |||
| Age ≤ 4 years | 20 | 9 | 18 | 47 | ||
| Age > 4 years | 6 | 23 | 24 | 53 | ||
| ≤ 4:>4 years ratio | 3.33 | 0.39 | 0.75 | ~p=0.001 | ||
| Metastasis Status | 19 | 20 | 19 | 58 | ||
| M0 | 14 | 17 | 9 | |||
| M+ | 5 | 3 | 10 | |||
| M0:M+ ratio | 2.80 | 5.67 | 0.90 | □ p=0.037 | Grp 3 vs [1&2] | |
| Status | 26 | 26 | 34 | 86 | ||
| Dead | 21 | 20 | 20 | |||
| Alive | 5 | 6 | 14 | |||
| Dead:Alive ratio | 4.2 | 3.33 | 1.43 | ~p=0.13 | ||
| Survival Time | 20 | 23 | 23 | 66 | ||
| Median Survival (yrs) | 0.8 | 1.8 | 4.3 | *p=0.019 | ||
| 95% Confidence Interval | 0.47–1.2 | 1.4–2.3 | 0.82–7.8 | |||
Pearson Chi-Square
Fisher's Exact Test
Log Rank (Mantel-Cox) test
Feature in one individual group was compared to a pooled cohort of the other two groups
note: some patients not included in analyses due to lack of specific clinical data - details of all patients in cohort are shown in Supplemental Table 1
Table 2.
Clinical and Molecular Features of CNS-PNET by Age
| Group 1 | Group 2 | Group 3 | Total available for analysis | p Value | ¥Comparison | |
|---|---|---|---|---|---|---|
| Age ≤ 4 years | 20 | 9 | 18 | 47 | ||
| Variable | Number Available for Analyses | |||||
| Metastasis Status | 15 | 6 | 4 | 25 | ||
| M0 | 11 | 5 | 4 | |||
| M+ | 4 | 1 | 0 | |||
| M0:M+ ratio | 2.75 | 5 | 4 | ~p=0.48 | ||
| Status | 20 | 7 | 13 | 40 | ||
| Dead | 16 | 6 | 10 | |||
| Alive | 4 | 1 | 3 | |||
| Dead: Alive ratio | 4 | 6 | 3.33 | ~p=0.90 | ||
| Survival Time | 15 | 6 | 6 | 27 | ||
| Median Survival (yrs) | 1 | 0.75 | 2.7 | *p=0.70 | ||
| Confidence Interval | 0.7–1.3 | 0–4.8 | 1.9–3.5 | |||
| Group 1 | Group 2 | Group 3 | Total available for analysis | p Value | ¥Comparison | |
|---|---|---|---|---|---|---|
| Age > 4 years | 6 | 23 | 24 | 53 | ||
| Variable | Number Available for Analyses | |||||
| Metastasis Status | 4 | 14 | 15 | 33 | ||
| M0 | 3 | 12 | 5 | |||
| M+ | 1 | 2 | 10 | |||
| M0:M+ ratio | 3 | 6 | 0.50 | ~p=0.033 | Grp 3 vs [1&2] | |
| 6 | 0.50 | □p=0.014 | Grp 2 vs 3 | |||
| Status | 6 | 19 | 21 | 46 | ||
| Dead | 5 | 14 | 10 | |||
| Alive | 1 | 5 | 11 | |||
| Dead: Alive ratio | 5 | 2.8 | 0.91 | ~p=0.13 | ||
| 2.8 | 0.91 | □p=0.087 | Grp 2 vs 3 | |||
Pearson Chi-Square
ANOVA
Fisher's Exact Test
Log Rank (Mantel-Cox) test
Feature in one individual group was compared to a pooled cohort of the other two groups
note: some patients not included in analyses due to lack of specific clinical data - details of all patients in cohort are shown in Supplemental Table 1
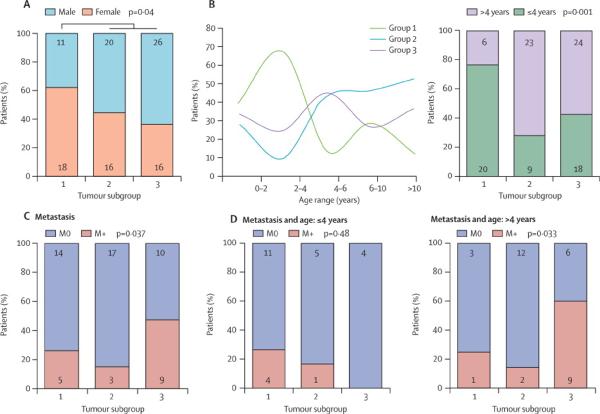
Gender and age distribution differed between the 3 molecular CNS-PNET sub-groups. Females comprised a significantly greater proportion of patients in group 1 as compared to group 2 and 3 combined (n=107, p=0.043). Male: female ratio in group 1, 2 and 3 CNS-PNETs were respectively 0.62 (11 males:18 females), 1.25 (20 males:16 females) and 1.63 (26 males: 16 females) (Figure 3A, Table 1). Group 1 and 2 patients exhibited bimodal age distributions with peak incidence at opposite age spectra, while group 3 patients had a single peak between 4–8 years. Overall group 1 patients were significantly younger (median age 2.9 yrs; 95% CI: 2.4–5.2, p=0.005) as compared to group 2 (median age 7.9; 95% CI: 6–9.7) and 3 (median age 5.9; 95% CI:4.9–7.8) patients (Table 1). Of 100 patients for which age information was known, 47 were ≤4 years, however young patients were significantly over-represented in group 1 as compared to group 2 and 3 (p=0.001); 77% (20/26) of group 1 as compared to 28% (9/32) of group 2 and 43% (18/42)of 3 patients were ≤4 years of age at diagnosis (Figure 3B, Table 1).
Figure 3. Age and gender distribution in molecular sub-groups of CNS-PNET.
Demographic information available on 108 primaries CNS-PNET (Table 1, 2 and supplemental Table 1) was examined to determine tumour sub-group specific correlation with: A- Gender and, B- Age at diagnosis. Significance was determined using ANOVA (gender) and Chi-square analyses (age). Number of patients in each category is indicated within the bar graphs.
Molecular sub-groups of CNS-PNET also exhibited significant differences in incidence of tumour metastases. Group 3 tumours demonstrated the highest incidence of disseminated disease at diagnosis: 10/19 group 3 patients were metastastic (M+) at diagnosis while a combined total of only 8/39 group1 and 2 patients were M+ at diagnosis (p=0.037) (Figure 4A). Proportion of localized M0 to M+ tumours in groups 1, 2 and 3 were respectively 2.8, 5.67 and 0.9 (Table 1). Interestingly, although metastatic disease is reported to be more frequent in younger children with embryonal brain tumours, analyses performed with stratification for age ≤ or > 4 years showed the incidence of tumour metastases was significantly different amongst CNS-PNET subgroups diagnosed in older children (Figure 4B, Table 2). A majority (10/15; 67%) of group 3 patients > 4 yrs of age at diagnosis had metastatic presentation as compared to only 1/4 (25%) of group 1 and 2/14 (14%) group 2 patients. Proportion of M0 to M+ tumours, which were respectively 3, 6 and 0.5 for group 1, 2 and 3 CNS-PNETs in children > 4 years, differed significantly in comparisons of group 3 to a combined cohort of group 1 and 2 patients (p=0.033) as well as to group 2 patients alone (0.014) (Figure 4B, Table 2).
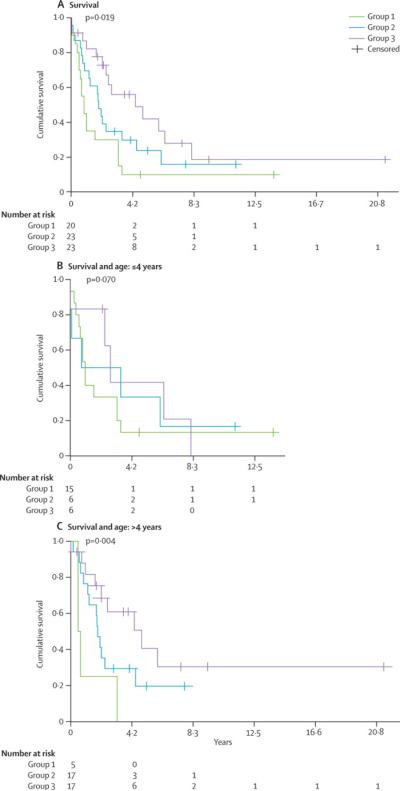
Figure 4. Molecular sub-groups of CNS-PNET exhibit distinct clinical phenotypes.
Clinical information available on 108 primaries CNS-PNET (Table 1, 2 and supplemental Table 1) was analysed to determine tumour sub-group specific correlation with A - Metastatic status at diagnosis, B- Age and metastatic status at diagnosis, C- Overall survival and D- Overall survival and age. Significance was determined using 2-sided Fisher's exact test (metastatic status at diagnosis) and log-rank (survival) analyses. Number of patients in each category is indicated within the bar graphs.
Log rank analysis of all tumour age groups showed overall survival for group 1 was significantly less than group 2 and 3 patients. Median survival for group 1, 2 and 3 were respectively 0.8 years (95% CI:0.47–1.2), 1.8 years (95% CI:1.4–2.3) and 4.3 years (CI: 0.82–7.8) p= 0.019 (Figure 4C, Table 1); with the exception of two longer term survivors, all group 3 patients were deceased within 4.2 years of diagnosis. As the majority of group 1 tumours arise in younger children who are often treated heterogeneously with radiation sparing therapeutic approaches due to concerns of neuro-cognitive damage3,21, we examined whether the poor prognostic association of LIN28 expression in group 1 tumours held true for older children who are conventionally prescribed intensified treatment regimens with higher dose cranio-spinal irradiation. As most infant brain tumour protocols enroll patients up to 3–4 years of age3,4,21, we stratified patients by age ≤ or >4 years, to remove age and potential treatment biases on survival. As shown in Figure 4D and Table 2, while overall survival for all young patients was similarly dismal, older children with LIN28 group 1 tumours fared significantly worse (median survival of 0.5 years, 95% CI: 0–1, p=0.004) than patients in group 1 (median survival of 1.8 years, 95% CI: 1.5–2.2) and 2 (median survival of 4.8 years, 95% CI:1.6–8). These findings indicate that immuno-positivity for LIN28 identifies a particularly high risk group of CNS-PNET across ages.
In summary, our findings demonstrate for the first time that differential expression of cell lineage markers, LIN28 and OLIG2, distinguishes 3 molecular sub-groups of CNS-PNET and identifies CNS-PNET sub-groups at very high risk of metastases and treatment failures.
Discussion
Therapeutic advances for childhood CNS-PNET have been difficult due to the rare incidence of CNS-PNET9, incomplete knowledge regarding the clinical and biological spectra of disease and lack of specific markers to aid histo-pathologic diagnoses8,10. In this study, we used integrated genomic analyses to examine a large cohort of primary childhood hemispheric CNS-PNET, and demonstrate that hemispheric CNS-PNETs comprises 3 molecular sub-groups with distinct lineage specific gene expression signature. Importantly, we show that immuno-histochemical analyses of two lineage markers, LIN28 and OLIG2, can distinguish molecular sub-groups of CNS-PNET with unique demographic and clinical features. Primitive neural group 1 tumours, with frequent C19MC amplification and high LIN28 expression, are distinctly aggressive tumours arising in younger children; oligo-neural group 2 tumours with high OLIG2 expression, arise in older children and are frequently localized while mesenchymal group 3 CNSPNET, which have limited LIN28 and OLIG2 expression, are associated with a high incidence of metastases and arise across ages. This study represents a first report of promising molecular markers for childhood CNS-PNET that can be applied to refine tumour diagnosis, classification and treatment risk stratification.
Comprehensive clinical and biological data on a substantial cohort of primary CNS-PNETs are lacking. Prior molecular studies have often included a spectrum of CNS-PNETs including rare variants, and tumours arising in different anatomical locations (pineoblastomas), as well as medulloblastoma8. As the biological relationship of CNS-PNET arising in different anatomical locations remains unclear, we restricted our current study to tumours arising in the cerebral hemispheres which comprises the majority of childhood CNS-PNET. By examining clinical features in relation to molecular sub-groups in a large cohort of primary tumours, we have been able to define distinct clinico-genetic sub-groups of CNS-PNETs arising in the cerebral hemispheres. Notably, our data shows both gender and age specific association with molecular sub-groups of hemispheric CNS-PNET. Although CNS-PNET are generally considered to be primarily a disease of younger children, our data shows that > 50% of CNS-PNETs arise in older children and that CNS-PNETs exhibit an age-dependent distribution of molecular sub-groups with group 1 and 2 tumours predominantly diagnosed respectively in children ≤ or > 4 years of age. The strong association of lineage specific gene expression signature and age with specific tumour sub-groups suggest molecular sub-groups of hemispheric CNS-PNET may derive from different precursor cell stage/type. Specifically, transcriptional signatures of group 1 and 2 tumours which are respectively enriched for CD133, CRABP1, LIN28 and, ASCL1 and OLIG1/2, suggest origin of group 1 and 2 tumours respectively from early neural or oligo-neural progenitors. The possible cellular origin of Group 3 CNS-PNETs which are enriched for mesenchymal differentiation genes including ZIC222 and LHX223, is less clear.
Studies of human brain tumours and brain tumour models demonstrate that cell lineage related gene expression signatures often correlate with and underlie clinical and biological heterogeneity in a spectrum of CNS tumours including malignant gliomas24, ependymoma25 and medulloblastoma26. Indeed, in addition to age and gender, we observed significant differences in survival and metastatic tendency between the three CNS-PNET sub-groups. We observed poorest survival in the primitive neural group 1/LIN28 expressing tumours, irrespective of age or metastatic status. Taken together with our prior observation which link C19MC amplification with a distinctly aggressive CNS-PNET phenotype12,13, our findings further highlight CNS-PNET with C19MC amplification and/or LIN28 expression as a unique clinico-pathologic entity and indicate LIN28 IHC as a new diagnostic tool for this distinct group of embryonal brain tumours.
The overall survival for group 2 and 3 tumours, which more commonly presented in older children, did not differ significantly (p=0.089), but we observed a trend towards better survival for children > 4yrs with group 3 mesenchymal lineage tumours; 26% (5/19) of group 2 vs 52% (11/21) of group 3 patients were alive at last evaluation (Table 2). This is surprising, as group 3 tumours had the highest incidence of metastases at diagnosis; a clinical feature which is linked to poorer outcomes in medulloblastoma and other embryonal brain tumours. These observations which suggest greater sensitivity of group 3 tumours to medulloblastoma-type therapeutics, usually prescribed for older children with CNS-PNET, may reflect greater biological relatedness of group 3 tumours to medulloblastoma. Interestingly, although ~90% of group 2 tumours were localized at time of diagnosis, older group 2 patients trended towards poorer survival as compared to group 3 patients. Hence, high dose cranio-spinal radiation which is routinely prescribed for all older children with CNS-PNET may offer limited therapeutic benefit for most group 2 CNS-PNETs. These collective observations underscore the clinical heterogeneity of CNS-PNET arising in the cerebral hemisphere and suggest that group 2 and 3 CNS-PNET may require different therapeutic approaches tailored to their specific biology.
Our studies confirm the documented poor overall outcome of CNS-PNETs across age groups and highlight the need to seek novel therapeutics for this aggressive disease. Pathway enrichment analyses indicate that the non-canonical WNT pathway predominates in group 1 tumours and thus may represent an attractive pathway for therapeutic targeting. Group 1 tumours also showed significant up regulation of the SHH signalling pathway, suggesting that novel SHH pathway inhibitors currently in clinical trials27 may be attractive new therapeutics for this sub-group. Of note, CRABP1, a retinoid binding protein known to alter retinoic metabolism and confer ATRA resistance28, is very highly expressed in the LIN28/ group 1 CNS-PNET. Thus retinoic acid, which is being tested in current high risk medulloblastoma and CNS-PNET cooperative group clinical trials for older children, may be of limited therapeutic benefit in group 1 CNS-PNETs.
In contrast to group 3 tumours which showed up-regulation of multiple canonical pathways, the oligo-neural group 2 tumours exhibited down-regulation of both SHH and WNT signalling pathways. Although we observed higher expression of PDGFRA and ERBB3 in this sub-group (Supplemental Figure 5) pathway analyses did not indicate significant global enrichment of receptor tyrosine signalling pathways. However, these potential therapeutic pathways may emerge with studies of larger cohorts, and further delineation of CNS-PNET sub-groups. Consistent with the higher incidence of metastases observed in group 3 tumours, significant activation of semaphorin signalling genes was observed in this tumour group. In addition to activated TGF-β signalling, group 3 tumours exhibited up-regulation of PTEN signalling and IGF2 expression, thus making these pathways/genes attractive for potential sub-group specific therapeutics.
Our study demonstrates for the first time that hemispheric tumours diagnosed as CNS-PNET in children is comprised molecular sub-groups with distinct survival and metastatic characteristics that can be distinguished by immuno-stains for cell lineage markers, LIN28 and OLIG2. We anticipate these novel markers will help identify high risk group 1 tumours for novel therapies, and allow tailoring of chemo-radio-therapy for group 2 and 3 patients, which differ significantly in metastatic potential. Furthermore, as our study was restricted to hemispheric CNS-PNETs, it will be important to define the significance of these molecular groupings to non-hemispheric CNS-PNETs, such as pineoblastoma. Through an international collaborative effort, our study has enabled comprehensive new insights into the clinical and molecular spectra of a rare disease which to date has been poorly studied and understood. These first efforts provide an essential step towards improved working classification for CNS-PNET and development of specific models and therapeutic strategies for CNS-PNETs. Our report underscores the importance of concerted, collaborative efforts to study large retrospective tumour and patient cohorts in order to accelerate biological and ultimately therapeutic studies of rare tumours.
Research in Context
Current treatment strategies for CNS-PNET are largely designed based on close histologic similarities to medulloblastoma, although it is clear that medulloblastoma-type therapy does not have the same efficacy in CNS-PNETs. There is a need to tailor treatment for CNS-PNET to exploit their distinct biology, however, molecular studies of CNS-PNETs have been limited by rare disease incidence and lack of robust diagnostic markers. With the exception of two recent studies by our group, molecular studies of CNS-PNETs to date have been limited to small cohorts of CNS-PNETs from various anatomical sites and identified using varying histo-pathologic criteria. We performed integrated genomic analyses on 123 primary CNS-PNET restricted to the cerebral hemispheres which met the 2007 WHO CNS classification criteria for CNS-PNET and lacked alterations at the Rhabdoid tumour locus. In keeping with prior clinical observations, our study demonstrates that CNS-PNET comprises a heterogeneous spectrum of tumours and defines three molecular subtypes of CNS-PNETs with distinct survival and metastatic features. Our study, which provides the first molecular prognostic markers for CNS-PNET, represents a significant advance towards biology-driven therapeutic strategies for CNS-PNET.
Supplementary Material
ACKNOWLEDGEMENTS
Funding from the Canadian Institute of Health Research, grant no.102684, b.r.a.i.n.child (AH) and Samantha Dickson Brain Tumour Trust, grant no. 17/53 (RG), assistance of clinicians from the Children's Cancer Leukemia Group centres and CCLG Biological studies committee, neuro-pathology review by Keith Robson and James Lowe at CBTRC, statistical consultations from Derek Stephen (Statistical Support Unit, Hospital for Sick Children), and technical help from Jonathon Torchia are gratefully acknowledged.
This study was funded by the Canadian Institute of Health Research, Brainchild and the Samantha Dickson Brain Tumour Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
STATEMENT OF CONFLICT OF INTEREST: The authors declared no conflict of interest.
AUTHOR CONTRIBUTION: Concept and design: Annie Huang, Eric Bouffet, Richard G Grundy
Financial Support: Annie Huang, Richard G Grundy
Provision of study material or patients: Cynthia Hawkins, Richard G Grundy, Amar Gajjar, Stefan M Pfister, Eric Bouffet, Andrey Korshunov, Jennifer Chan, Lucie Lafay-Cousin, Charles Eberhart, Helen Toledano, Jason Fangusaro, Seung-Ki Kim, Young-Shin Ra, Timothy Van Meter, Ching C Lau, Scott L Pomeroy, Ho-Keung Ng, Chris Jones, Steve C Clifford, James T Hayden, Xing Fan, Karin M Muraszko, Hideo Nakamura and Annie Huang
Data analyses and interpretation: Daniel Picard, Annie Huang, Suzanne Miller, Cynthia Hawkins, Ping Zhao Hu
Manuscript writing: Daniel Picard, Annie Huang, Suzanne Miller.
Final Approval of Manuscript: All authors
REFERENCES
- 1.Kohler BA, Ward E, McCarthy BJ, et al. Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. J Natl Cancer Inst. 2011;103:714–36. doi: 10.1093/jnci/djr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turner CD, Rey-Casserly C, Liptak CC, et al. Late effects of therapy for pediatric brain tumor survivors. J Child Neurol. 2009;24:1455–63. doi: 10.1177/0883073809341709. [DOI] [PubMed] [Google Scholar]
- 3.Timmermann B, Kortmann RD, Kuhl J, et al. Role of radiotherapy in supratentorial primitive neuroectodermal tumor in young children: results of the German HIT-SKK87 and HIT-SKK92 trials. J Clin Oncol. 2006;24:1554–60. doi: 10.1200/JCO.2005.04.8074. [DOI] [PubMed] [Google Scholar]
- 4.Pizer BL, Weston CL, Robinson KJ, et al. Analysis of patients with supratentorial primitive neuro-ectodermal tumours entered into the SIOP/UKCCSG PNET 3 study. Eur J Cancer. 2006;42:1120–8. doi: 10.1016/j.ejca.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 5.Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–14. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho YJ, Tsherniak A, Tamayo P, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–30. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gajjar A, Chintagumpala M, Ashley D, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7:813–20. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 8.Li MH, Bouffet E, Hawkins CE, et al. Molecular genetics of supratentorial primitive neuroectodermal tumors and pineoblastoma. Neurosurg Focus. 2005;19:E3. doi: 10.3171/foc.2005.19.5.4. [DOI] [PubMed] [Google Scholar]
- 9.Louis DN, Wiestler OD. WHO Classification of Tumours of the Central Nervous System. ed 4th IARC; Lyon: 2007. [Google Scholar]
- 10.Burger PC. Supratentorial primitive neuroectodermal tumor (sPNET) Brain Pathol. 2006;16:86. doi: 10.1111/j.1750-3639.2006.tb00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson EM, Sievert AJ, Gai X, et al. Genomic analysis using high-density single nucleotide polymorphism-based oligonucleotide arrays and multiplex ligation-dependent probe amplification provides a comprehensive analysis of INI1/SMARCB1 in malignant rhabdoid tumors. Clin Cancer Res. 2009;15:1923–30. doi: 10.1158/1078-0432.CCR-08-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Lee KF, Lu Y, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–46. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korshunov A, Remke M, Gessi M, et al. Focal genomic amplification at 19q13.42 comprises a powerful diagnostic marker for embryonal tumors with ependymoblastic rosettes. Acta Neuropathol. 2010;120:253–60. doi: 10.1007/s00401-010-0688-8. [DOI] [PubMed] [Google Scholar]
- 14.Miller S, Rogers HA, Lyon P, et al. Genome-wide molecular characterization of central nervous system primitive neuroectodermal tumor and pineoblastoma. Neuro Oncol. 2011;13:866–79. doi: 10.1093/neuonc/nor070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers HA, Miller S, Lowe J, et al. An investigation of WNT pathway activation and association with survival in central nervous system primitive neuroectodermal tumours (CNS PNET) Br J Cancer. 2009;100:1292–302. doi: 10.1038/sj.bjc.6604979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet JP, Tamayo P, Golub TR, et al. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skottman H, Mikkola M, Lundin K, et al. Gene expression signatures of seven individual human embryonic stem cell lines. Stem Cells. 2005;23:1343–56. doi: 10.1634/stemcells.2004-0341. [DOI] [PubMed] [Google Scholar]
- 18.Ligon KL, Fancy SP, Franklin RJ, et al. Olig gene function in CNS development and disease. Glia. 2006;54:1–10. doi: 10.1002/glia.20273. [DOI] [PubMed] [Google Scholar]
- 19.Jacquet BV, Muthusamy N, Sommerville LJ, et al. Specification of a Foxj1-dependent lineage in the forebrain is required for embryonic-to-postnatal transition of neurogenesis in the olfactory bulb. J Neurosci. 2011;31:9368–82. doi: 10.1523/JNEUROSCI.0171-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houzelstein D, Auda-Boucher G, Cheraud Y, et al. The homeobox gene Msx1 is expressed in a subset of somites, and in muscle progenitor cells migrating into the forelimb. Development. 1999;126:2689–701. doi: 10.1242/dev.126.12.2689. [DOI] [PubMed] [Google Scholar]
- 21.Fangusaro J, Finlay J, Sposto R, et al. Intensive chemotherapy followed by consolidative myeloablative chemotherapy with autologous hematopoietic cell rescue (AuHCR) in young children with newly diagnosed supratentorial primitive neuroectodermal tumors (sPNETs): report of the Head Start I and II experience. Pediatr Blood Cancer. 2008;50:312–8. doi: 10.1002/pbc.21307. [DOI] [PubMed] [Google Scholar]
- 22.Elms P, Siggers P, Napper D, et al. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev Biol. 2003;264:391–406. doi: 10.1016/j.ydbio.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Hoch RV, Rubenstein JL, Pleasure S. Genes and signaling events that establish regional patterning of the mammalian forebrain. Semin Cell Dev Biol. 2009;20:378–86. doi: 10.1016/j.semcdb.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 25.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–35. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Gibson P, Tong Y, Robinson G, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–9. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudin CM, Hann CL, Laterra J, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–8. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boylan JF, Gudas LJ. The level of CRABP-I expression influences the amounts and types of all-trans-retinoic acid metabolites in F9 teratocarcinoma stem cells. J Biol Chem. 1992;267:21486–91. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.