Summary
Extended longevity is often correlated with increased resistance against various stressors. Insulin/IGF-1-like signaling (IIS) is known to have a conserved role in aging and cellular mechanisms against stress. In C. elegans, genetic studies suggest that heat-shock transcription factor HSF-1 is required for IIS to modulate longevity. Here we report that the activity of HSF-1 is regulated by IIS. This regulation might occur at an early step of HSF-1 activation via two HSF-1 regulators, DDL-1 and DDL-2. Inhibition of DDL-1/2 increases longevity and thermotolerance in an hsf-1 dependent manner. Furthermore, biochemical analyses suggest that DDL-1/2 negatively regulates HSF-1 activity by forming a protein complex with HSF-1. The formation of this complex (DHIC) is affected by the phosphorylation status of DDL-1. Both the formation of DHIC and the phosphorylation of DDL-1 are controlled by IIS. Therefore, DDL-1/2 may serve as the link between IIS and HSF-1 pathway.
Introduction
Studies in a variety of organisms have revealed that extended longevity is often correlated with increased resistance against the deleterious effects of environmental and physiological stresses, including heat-shock and oxidizing conditions (Lithgow and Walker, 2002). In the nematode C. elegans, alterations in a number of independent pathways, including the insulin/IGF-1-like signaling (IIS) pathway, are known to increase longevity and stress resistance (Kenyon, 2010). For example, loss-of-function mutations affecting the insulin/IGF-1-like receptor DAF-2 have been shown to increase longevity and resistance to heat stress (Kenyon et al., 1993; Lithgow et al., 1995).
The IIS pathway also regulates other physiological processes independent of longevity in worms, such as fat production, reproduction, and dauer formation (Kimura et al., 1997). The FOXO transcription factor DAF-16, which is negatively regulated by the IIS pathway, is required for most known phenotypes that are associated with reduced IIS (Kenyon, 2010). DAF-16 controls the expression of a diverse set of downstream antioxidant, metabolic, chaperone, antimicrobial, and other genes that act in a cumulative way to influence longevity and stress response (Murphy et al., 2003). While many genes controlled by the IIS pathway require DAF-16 for their expression, several co-regulators of DAF-16, such as HSF-1, SMK-1, and HCF-1, have recently been found to function collaboratively with DAF-16 to regulate different subsets of target genes independently in response to different environmental stressors (Hsu et al., 2003; Li et al., 2008; Wolff et al., 2006). It appears that, among these co-regulators of DAF-16, only HSF-1 is required for IIS to up-regulate the transcription of genes involved in the heat-shock response.
The heat-shock response is a fundamental and critical cellular defensive mechanism against heat stress that results in the rapid expression of a group of proteins known as heat-shock proteins (HSPs). HSPs function as molecular chaperones, playing a variety of roles including assisting in protein folding, targeting damaged proteins for degradation and other responses associated with the protection of cell from damage (Hartl, 1996; Jolly and Morimoto, 2000). The induction of the heat-shock response is mediated by heat-shock transcription factors (HSFs). While invertebrate animals such as nematodes typically have only one HSF (Clos et al., 1990; Garigan et al., 2002), four distinct but related HSF isoforms have been found in vertebrates (HSF1~4) (Pirkkala et al., 2001). In response to stress, HSF acquires DNA-binding activity to the heat-shock elements (HSE) located in the promoters of hsp genes, thereby mediating their transcription. The activation of vertebrate HSF1 appears to be a multistep process that includes oligomerization, post-translational modifications, nuclear localization, and acquisition of DNA-binding activity (Sarge et al., 1993). However, not all of these steps are essential for the activation of HSF in invertebrate species (Sorger et al., 1987).
A decrease in the effectiveness of the heat-shock response has been associated with aging and is partly responsible for the age-related increase in mortality (Finkel and Holbrook, 2000). This age-related attenuation of the heat-shock response is often related to a decrease in the capacity of cells to produce HSPs (Soti and Csermely, 2000). Several studies have directly implicated heat-shock response genes, including HSF, in the regulation of longevity. First, it has been reported that the expression of genes encoding small heat-shock proteins (sHSPs) is increased in Drosophila lines selected for increased longevity (Kurapati et al., 2000), as well as in C. elegans daf-2 mutants (Murphy et al., 2003). Moreover, mild heat stress in C. elegans results in a small but significant extension of lifespan (Lithgow et al., 1995). Similarly, mild heat stress in Drosophila causes a period of decreased mortality rate (Khazaeli et al., 1997). It also has been found that over-expression of hsf-1 lead to increased longevity, while inhibition of HSF-1 activity by RNAi shortens the animal’s lifespan (Hsu et al., 2003).
Recent studies have demonstrated that HSF-1 and its downstream targets may act in concert with the IIS pathway to regulate longevity in C. elegans. It has been reported that HSF-1, similar to DAF-16, is required for daf-2 mutations to extend lifespan (Hsu et al., 2003). It is also known that the expression of a subset of heat-shock response genes is increased in daf-2 mutants under unstressed conditions and is at least partially required for the lifespan phenotypes of daf-2 mutants (Hsu et al., 2003). Together, these observations imply that the IIS pathway might, at least in part, influence longevity by regulating heat-shock response.
Here, we show that both the DNA-binding and the transcriptional activity of HSF-1 are directly regulated by IIS and that this regulation likely occurs at an early step of HSF-1 activation. We show that the proteins DDL-1 and DDL-2, previously implicated in lifespan extension, modulate HSF-1 activity by forming an inhibitory heterocomplex with HSF-1, and that formation of this complex is regulated by IIS. Our findings suggest that these HSF-1 regulators may link insulin/IGF-1 signalling and the cellular response to heat stress.
Results
The activation of HSF-1 is a multistep process in C. elegans
Heat-shock transcription factor (HSF-1) is a key transcriptional regulator of the cellular response to various proteotoxic stresses, including heat, in C. elegans. The mammalian HSF1 is constitutively present in cells and is activated when cells encounter proteotoxic stresses. It has been well-documented in other animal models that activation of HSF1 appears to be a multistep process, involving oligomerization, inducible post-translational modification, nuclear/subnuclear localization, acquisition of DNA-binding activity, and acquisition of transcriptional activity. All of these steps are shown to be tightly controlled [reviewed in (Morimoto, 1998; Voellmy, 2004)]. However, given the important role that C. elegans HSF-1 might play in determining DAF-2 longevity, the activation and regulation of HSF-1 have not been thoroughly investigated. Thus, we first examined how C. elegans HSF-1 responds to heat stress.
To examine whether oligomerization of HSF-1 also occurs in C. elegans, worm whole cell extracts (WCE) were incubated with cross-linking reagent EGS before being analyzed by western blotting with anti-human HSF1 antibody. We found that worm HSF-1 forms both dimers and trimers upon heat-shock (Fig. S1A). We then asked whether HSF-1 is post-translationally modified in response to heat-shock. Results from our western blotting analysis indicate that post-translational modifications (PTM) on HSF-1 proteins occur in a time-dependent manner upon heat-shock (Fig. S1B). This change may represent a shift from a non-modified to a modified form of HSF-1 or a shift between two different post-translationally modified forms of HSF-1 in response to heat-shock. Further analysis with alkaline phosphatase (CIP) suggests that phosphorylation(s) may be responsible for the majority of the PTM observed in HSF-1 (Fig. S1C). There are at least 12 phosphorylation sites, both constitutive and inducible, that have been identified on human HSF1 (Guettouche et al., 2005). The effects of phosphorylation on HSF-1 activity, however, can be either positive or negative.
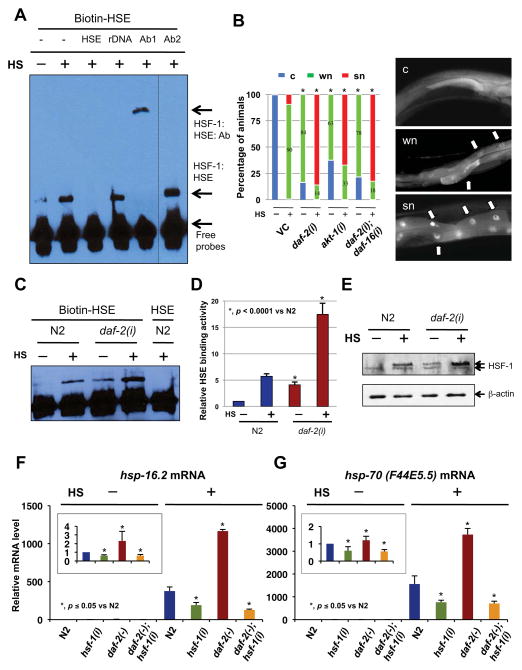
One of the most important steps of HSF activation is for HSFs to acquire DNA binding activity to the heat-shock element (HSE), located in the upstream regulatory region of its target genes. Thus, we examined whether heat-shock leads to an induction of HSF-1 DNA-binding activity in worms, using an electrophoretic mobility shift assay (EMSA) developed in our lab for C. elegans HSF-1. As observed in other systems upon stimulation, there is an increased level of HSF-1 that can bind to the biotin-labeled HSE probes (Fig. 1A). This binding can be out-competed by unlabeled HSE containing oligonucleotides, but not by randomly synthesized oligonucleotides, suggesting a specific interaction between HSF-1 and HSE probes. Moreover, activated HSF-1 and HSE form a larger complex with the anti-HSF1 polyclonal antibodies, but not with the anti-GFP antibodies (Fig. 1A).
Figure 1. Inactivation of daf-2 positively regulates HSF-1 activity and heat-shock response.
(A) DNA-binding activity of C. elegans HSF-1 in response to 90 min of heat-shock at 37°C (HS), measured by electrophoretic mobility shift assay. Nuclear extracts were incubated with biotin-labeled HSE probes (Biotin-HSE). The specificity of the DNA binding was determined by competition with 200x unlabeled HSE probes (HSE), randomly synthesized DNA (rDNA), anti-HSF-1 antibodies (Ab1), or anti-GFP antibodies (Ab2) before being applied to gel eletrophoresis. (B) Nuclear accumulation of HSF-1 in response to IIS inactivation. EQ73 animals (hsf-1::gfp) grown on vector control (VC) or different RNAi bacteria were unstressed or heat-shocked for 30 min (HS) before being classified into three groups according to the nuclear/cytosolic (n/c) ratio of GFP intensity in the intestinal cells (right panels). “c”, “wn”, and “sn” are animals with n/c ratio < 1.2, 1.2~2.0, and >2.0, respectively. The mean of three independent experiments were pooled and shown (left panel). *, p < 0.0001 vs VC under same conditions (chi2-test). n ≥ 300. (C–D) The DNA-binding activity of HSF-1 in daf-2(e1370) mutants in response to 90 min of heat-shock (HS). The result of a representative experiment is shown in (C). The mean ± SD of three independent experiments (mean ± SD), normalized to the control (N2 with unlabeled HSE), is presented in (D). (E) N2 or daf-2(RNAi) animals were unstressed or heat-shocked for 90 min (HS). Worm whole cell extracts (WCE) of these animals were subjected to immunoblotting analysis using anti-HSF-1 (top) or anti-β-actin (bottom) antibodies. Detail quantification in Fig. S1D. (F–G) Relative abundance of (F) hsp-16.2 and (G) hsp-70 (F44E5.5) mRNA in wild types (N2) or daf-2(e1370) mutants fed with control or hsf-1 RNAi bacteria. The inset shows the mRNA level under unstressed conditions (without 90 min HS). Data were combined from at least three experiments, and the mean ± SD of each treatment are shown.
While the formation of active oligomers to acquire DNA binding activity is conserved across species, there has been some controversy regarding the subcellular localization of HSF1 under unstressed conditions in different metazoan systems. Some studies have suggested that HSF1 is predominantly cytoplasmic prior to heat-shock and nuclear after stress (Sarge et al., 1993; Sistonen et al., 1994), whereas others have suggested that HSF1 is always nuclear (Mercier et al., 1999). Using a transgenic line that expresses gfp-tagged hsf-1 under the control of its own endogenous promoter, HSF-1-GFP was observed in intestinal cells, body wall muscle cells, hypodermal cells as well as many neurons in the head and tail. The GFP signal can be reduced upon treatment with hsf-1 RNAi. The HSF-1-GFP is evenly distributed between the nucleus and cytoplasm before heat-shock in intestinal cells (Fig. 1B). In response to 30 min of heat-shock, an accumulation of HSF-1-GFP was observed in the intestinal nuclei (Fig. 1B). Over-expression of the same hsf::gfp construct rescued the lifespan shortening effect of hsf-1(sy441) mutations (Hajdu-Cronin et al., 2004) (Fig. S1F) and produced a significant lifespan extension on its own in a wild-type background (Fig. S1G), suggesting that the tagged HSF is fully functional. Together, our findings suggest that the activation of HSF-1 in C. elegans is also a multistep process and that these assays can be very useful tools to assess HSF-1 activity.
DAF-2 insulin/IGF-1-like signaling (IIS) inhibits HSF-1 activity
If IIS pathway influences longevity, at least in part, by directly modulating HSF-1 activity and heat-shock response, one would expect that a reduction in DAF-2 activity should promote the activation of HSF-1 and raise its activity. To test this idea, we first investigated how reduction in DAF-2 activity affects HSF-1 nuclear translocation and found that there is a significantly higher level of HSF-1 localized in the nucleus of daf-2(RNAi) animals compared to control before or after heat-shock (Fig. 1B), indicating a elevated HSF-1 activation. This effect appears to be DAF-16 independent, as simultaneous knockdown of both daf-2 and daf-16 did not reverse the phenotype (Fig. 1B). Similarly, inactivation of another IIS component AKT-1 by RNAi resulted in an increased HSF-1 nuclear localization, although the effect was smaller (Fig. 1B).
We next examined how reduction in DAF-2 activity affects the DNA binding activity of HSF-1. As predicted, we found that the DNA binding activity of HSF-1 is increased 4-fold in unstressed daf-2(RNAi) animals, and more than 17-fold in heat-shocked daf-2(RNAi) animals as compared to the unstressed wild-type (N2) controls (Fig. 1C-D), suggesting that the basal level of HSF-1 activity might be higher in daf-2(RNAi) animals, which may allow for a stronger response to heat stress. Presumably, the increase in DNA binding activity of HSF-1 in response to heat stress or daf-2 knockdown may result from a nuclear accumulation of activated HSF-1 (i.e. the oligomerized and post-translationally modified HSF-1). Indeed, results from western blotting analysis indicate that there is an elevated level of post-translationally modified HSF-1 in daf-2 mutants under both unstressed and stressed condition (Fig. 1E, S1D), suggesting the presence of an increased amount of activated HSF-1 in daf-2 mutants. It is worth noting that the amount of total HSF-1 was also increased in response to heat stress or daf-2 knockdown, while the level of unmodified HSF-1 remained largely unaltered (Fig. S1D). However, this increase in total HSF-1 protein is unlikely to be regulated transcriptionally, as the mRNA level of hsf-1 does not appear to be different in daf-2(RNAi) animals compared to the control (Fig. S3A). In fact, mRNA level of hsf-1 is slightly reduced after heat-shock (Fig. S3A), implying a potential negative feedback regulation at the transcriptional level following the initial activation upon heat-shock. One possible explanation is that the modified and DNA-bound HSF-1 proteins are less accessible for degradation.
Finally, to examine whether increased DNA binding activity of HSF-1 results in an increase in its transcriptional activity, we measured the mRNA level of four known targets of HSF-1 by qRT-PCR in daf-2(RNAi) animals before and after heat stress. The mRNA levels of these genes, including hsp-16.2, sip-1, and two different hsp-70s, are all increased in daf-2(RNAi) animals under both stressed and unstressed conditions (Fig. 1F-G, S3C–D). The up-regulation of heat-shock protein expression observed in daf-2(RNAi) animals can be suppressed by hsf-1 RNAi treatments (Fig. 1F–G, S3C–D), suggesting that the observed changes in the expression is the result of alteration in HSF-1 activity. Together, our findings suggest that the HSF-1 activity is negatively controlled by the IIS pathway, and that multiple steps of HSF-1 activation are affected by inhibiting IIS under both stressed and unstressed conditions.
hsf-1 is required for ddl-1, ddl-2, and hsb-1 to influence longevity
Our findings, together with previous studies, strongly suggest that HSF-1 activity is regulated by IIS. To further understand this regulation and determine whether it is direct, we next attempted to elucidate the mechanism underlying this regulation. We focused on two ddl (daf-16-dependent longevity) genes identified from a previous genome-wide RNAi screen for longevity genes (Hansen et al., 2005).
ddl-1 and ddl-2 both encode evolutionarily conserved proteins. DDL-1 is homologous to human coiled-coil domain-containing protein 53 (CCDC53), whereas DDL-2 is the worm homolog of human WASH2 (Wiskott-Aldrich syndrome protein and SCAR homolog) protein. WASH2 and CCDC53 are both components of the WASH complex, which is involved in the actin polymerization (Derivery et al., 2009). CCDC53 has also been reported to potentially interact with the heat-shock factor binding protein-1 (HSBP1) (Rual et al., 2005), a known negative regulator of HSF-1 (Satyal et al., 1998). Results from yeast two-hybrid experiments also suggested that worm DDL-1 may interact with both HSB-1 and DDL-2 (Li et al., 2004). The inhibitory activity of HSB-1 is achieved via protein-protein interactions between HSB-1 and the oligomerization motif of HSF-1 (Satyal et al., 1998). We therefore hypothesized that DDL-1/2 (i.e. DDL-1 and DDL-2) and HSB-1 may regulate the activity of HSF-1 via the formation of an inhibitory complex.
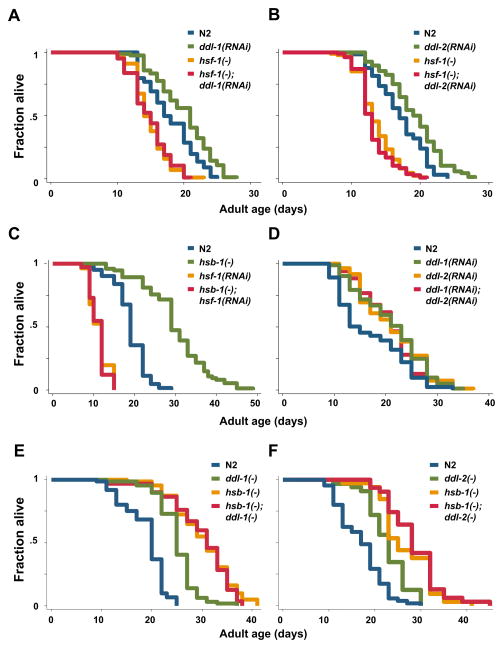
To test this hypothesis, we first asked whether hsf-1 is required for ddl-1 and ddl-2 to influence longevity. The lifespans of hsf-1(sy441) mutants, which have been reported to exhibit reduced heat-shock response and shortened lifespan (Hajdu-Cronin et al., 2004), grown on ddl-1 or ddl-2 RNAi bacteria were measured. We found that RNAi treatment of ddl-1 or ddl-2 failed to produce any significant lifespan extension on hsf-1(sy441) mutants (Fig. 2A–B; Table 1), while reducing expression of ddl-1 or ddl-2 by RNAi extends lifespan by 12–24% in our hands (Fig. 2A–B; Table 1, S1). This finding suggests that hsf-1 is required for the extended lifespan observed in ddl-1 or ddl-2 RNAi treated animals. Consistent with the idea that DDL-1/2 may influence longevity by altering HSF-1 activity, animals grown on ddl-1 or ddl-2 RNAi bacteria exhibit increased resistance to both heat and oxidative stresses (Fig. S2A–B). Although reducing ddl-1/2 expression extends wild-type lifespan, over-expression of ddl-1/2 is not sufficient to alter wild-type lifespan (Fig. S2C–E). Over-expression of ddl-1, however, does reverse the lifespan-extending phenotypes of ddl-1(ok2916) mutants (Fig. S2F). It is possible that the residual activity of HSF-1 is sufficient to ensure survival under experimental conditions (at 20°C, without heat stress).
Figure 2. A common hsf-1-dependent mechanism mediates the longevity effects of ddl-1, ddl-2 and hsb-1.
(A) Lifespan analysis of wild-type (N2) animals or hsf-1(sy441) mutants grown on empty vector control or ddl-1 RNAi bacteria at 20°C. (B) Lifespan analysis of N2 animals or hsf-1(sy441) mutants grown on control or ddl-2 RNAi bacteria. (C) Lifespan analysis of N2 animals or hsb-1(cg116) mutants grown on control or hsf-1 RNAi bacteria. (D) Lifespan analysis of N2 animals grown on control, ddl-1 RNAi, ddl-2 RNAi, or 1:1 mixture of ddl-1 and ddl-2 RNAi bacteria at 20°C. (E) Lifespan analysis of N2, ddl-1(ok2916), hsb-1(cg116), or ddl-1(ok2916);hsb-1(cg116) mutants. (F) Lifespan analysis of N2, ddl-2(ok3235), hsb-1(cg116), or ddl-2(ok3235);hsb-1(cg116) mutants. Statistical details are summarized in Table 1 and Table S1.
Table 1. Effects of ddl-1, ddl-2, and hsb-1 mutations on lifespan.
Adult mean lifespan ± SEM, in days. Lifespan experiments were carried out at 20°C. The 75th percentile is the age at which the fraction of animals alive reaches 0.25. ‘n’ shows the number of observed deaths relative to total number of animals started. The difference between these numbers represents the number of animals censored. Animals that exploded, bagged, or crawled off the plates were censored at the time of the event. The log-rank (Mantel-Cox) test was used for statistical analysis (p-Values). Data shown here represent one independent lifespan experiment. Please see Table S1 for additional repetitions of the experiments.
| Strain | Mean Life Span ± SEM (days) | 75th Percentile (days) | p Value | n |
|---|---|---|---|---|
| N2; control (i) | 17.2 ± 0.3 | 21 | - | 67/90 |
| N2; ddl-1(RNAi) (i) | 19.1 ± 0.3 | 22 | 0.0034a | 84/90 |
| hsf-1(sy441); control | 14.9 ± 0.1 | 16 | <0.0001a | 101/169 |
| hsf-1(sy441); ddl-1(RNAi) | 14.9 ± 0.3 | 17 | <0.0001a, <0.0001b, 0.74c | 96/170 |
| N2; control (ii) | 17.2 ± 0.2 | 20 | - | 63/82 |
| N2; ddl-2(RNAi) (ii) | 19.2 ± 0.2 | 22 | 0.0008a | 68/86 |
| hsf-1(sy441); control | 13.6 ± 0.2 | 15 | <0.0001a | 86/156 |
| hsf-1(sy441); ddl-2(RNAi) | 13.2 ± 0.1 | 14 | <0.0001a, <0.0001b, 0.26c | 107/156 |
| N2; control (iii) | 19.1 ± 0.2 | 22 | - | 62/72 |
| N2; hsf-1(RNAi) | 11.2 ± 0.2 | 12 | <0.0001a | 46/72 |
| hsb-1(cg116); control | 30.0 ± 0.4 | 35 | <0.0001a | 74/84 |
| hsb-1(cg116); hsf-1(RNAi) | 11.1 ± 0.2 | 12 | <0.0001a, 0.78b, <0.0001c | 33/84 |
| N2 (i) | 18.9 ± 0.2 | 22 | - | 60/72 |
| ddl-1(ok2916) | 25.1 ± 0.3 | 27 | <0.0001a | 65/72 |
| hsb-1(cg116) (i) | 30.3 ± 0.5 | 35 | <0.0001a | 62/72 |
| hsb-1(cg116); ddl-1(ok2916) | 29.9 ± 0.3 | 35 | <0.0001a, <0.0001d, 0.51e | 57/72 |
| N2 (ii) | 17.1 ± 0.4 | 21 | - | 52/72 |
| ddl-2(ok3235) | 22.6 ± 0.3 | 26 | <0.0001a | 32/72 |
| hsb-1(cg116) (ii) | 26.8 ± 0.4 | 32 | <0.0001a | 32/72 |
| hsb-1(cg116); ddl-2(ok3235) | 28.6 ± 0.4 | 32 | <0.0001a, <0.0026f, 0.32e | 31/72 |
| N2; control(RNAi) (iv) | 17.2 ± 0.6 | 23 | - | 82/90 |
| N2; ddl-1(RNAi) (iv) | 21.3 ± 0.5 | 25 | 0.0006a | 81/90 |
| N2; ddl-2(RNAi) (iv) | 21.4 ± 0.6 | 28 | 0.0001a | 81/90 |
| N2; ddl-1(RNAi); ddl-2(RNAi) | 20.9 ± 0.3 | 25 | 0.0072a, 0.34g, 0.26h | 86/90 |
p-Values calculated by pair-wise comparisons to N2 grown on vector control of the same experiment.
Compared to N2 grown on the same RNAi bacteria.
Compared to the same mutants grown on vector control.
Compared to to ddl-1(ok2916) mutants.
Compared to hsb-1(cg116) mutants.
Compared to ddl-2(ok3235) mutants
Compared to N2 grown on the ddl-1 RNAi bacteria of the same experiment.
Compared to N2 grown on the ddl-2 RNAi bacteria of the same experiment.
Since HSB-1 is predicted to interact with both DDL-1 and HSF-1, we have also examined whether the loss of function mutation of hsb-1 increases lifespan. Indeed, the lifespan of hsb-1(cg116) mutants, which harbors a null mutation of hsb-1, is increased by up to 60% (Fig. 2C; Table 1). Moreover, hsf-1 is also required for the lifespan extension caused by hsb-1 mutations, as hsb-1(cg116); hsf-1(RNAi) animals’ lifespans are as short as hsf-1(RNAi) animals (Fig. 2C; Table 1).
Taken together, our findings suggest that ddl-1, ddl-2, and hsb-1 may share the same mechanism that involves hsf-1 to influence longevity. Results from our genetic epistasis analysis on ddl-1, ddl-2, and hsb-1 also support this model. First, inhibition of both ddl-1 and ddl-2 by RNAi does not lead to a larger lifespan extension compared to animals treated with ddl-1 or ddl-2 RNAi alone, suggesting that ddl-1 and ddl-2 may have common effectors and act in the same genetic pathway (Fig. 2D; Table 1). Secondly, we found that inhibition of ddl-1 does not further increase the lifespan of hsb-1(cg116) mutants, while it typically increases N2 lifespan by up to 33% (Fig. 2E; Table 1). We also tested the lifespan of hsb-1(cg116) mutants grown on ddl-1 RNAi and found no additive effect (data not shown). Similarly, the lifespan of hsb-1(cg116); ddl-2(ok3235) double mutants is not statistically different from hsb-1(cg116) single mutants (Fig. 2F; Table 1). Together, these findings imply that a common mechanism might mediate the lifespan phenotypes of ddl-1, ddl-2, and hsb-1 mutations.
DDL-1/2 negatively regulate HSF-1 activity
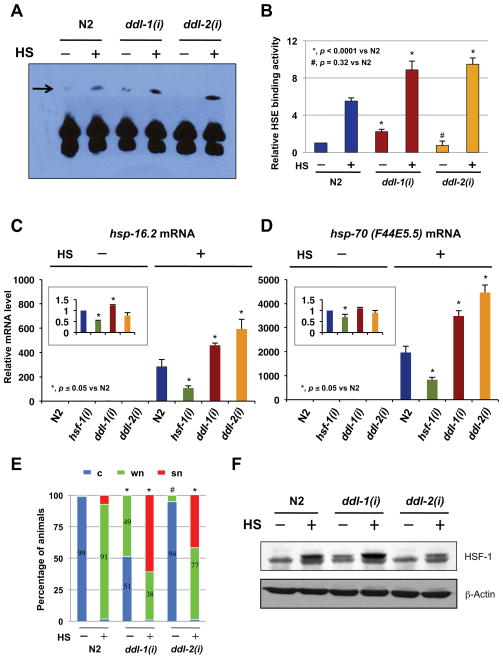
HSB-1 is known to control cellular response to heat stress by negatively regulating HSF-1 activity. Therefore, if ddl-1/2 and hsb-1 were to influence longevity through a common mechanism, the most likely common effectors would be HSF-1. To test whether DDL-1/2 exert their functions by negatively regulating HSF-1 activity, we measured the DNA-binding activity of HSF-1 in ddl-1(RNAi) and ddl-2(RNAi) animals. Inhibition of ddl-1 appears to increase DNA-binding activity of HSF-1 both before and after heat-shock (Fig. 3A-B). There is also a significant increase in DNA-binding activity of HSF-1 in ddl-2(RNAi) animals under stressed condition, whereas inhibiting ddl-2 under unstressed conditions produced no significant effect (Fig. 3A–B). We then asked whether increased HSF-1 DNA binding activity results in an increase in its transcriptional activity. A similar pattern was observed when we examined the effect of inhibiting ddl-1 or -2 on HSF-1 transcriptional activity by qRT-PCR. Inhibition of ddl-1 or -2 led to increases in mRNA transcription of all four hsp genes after heat-shock (Fig. 3C–D, S3E–F). However, under unstressed conditions, inhibition of ddl-1 or -2 did not significantly elevate the mRNA level of hsf-1 targets, except for the hsp-16.2 and sip-1 in ddl-1(RNAi) animals (Fig. 3C–D, S3E–F).
Figure 3. DDL-1 and DDL-2 negatively regulate HSF-1 activity.
(A-B) Lowering ddl-1 or ddl-2 expression increases HSF-1 DNA-binding activity. The DNA-binding activity of HSF-1 in N2, ddl-1(RNAi), or ddl-2(RNAi) animals with or without 90 min of heat-shock (HS). A representative experiment is shown in (A). Quantification of three independent experiments (mean ± SD) is presented in (B). (C–D) Relative abundance of (C) hsp-16.2, and (D) hsp-70 (F44E5.5) mRNA in N2, hsf-1(RNAi), ddl-1(RNAi), or ddl-2(RNAI) animals with or without heat-shock (90 min). The inset shows the mRNA level under unstressed conditions. The mean ± SD of three independent experiments were pooled and shown here. (E) Nuclear accumulation of HSF-1 in response to DDL-1/2 inactivation. EQ73 animals (hsf-1::gfp) grown on control, ddl-1, or ddl-2 RNAi bacteria were unstressed or heat-shocked for 30 min (HS). The result of three experiments were pooled and shown here. Data are mean, n ≥ 300 per RNAi treatment. *, p < 0.0001; #, p = 0.097 (chi2-test). (F) Worm extracts (WCE) prepared from N2, ddl-1(RNAi), or ddl-2(RNAi) animals with or without 90 min of HS were subjected to immunoblotting analysis using anti-HSF-1 (top) or anti-β-actin (bottom) antibodies. Detail quantification in Fig. S1D.
Presumably, the changes in HSF-1 activity observed in ddl mutants might be due to changes in the amount of nuclear localized and activated HSF-1. Indeed, we found that there is a significantly higher level of nuclear localized HSF-1 in ddl-1(RNAi) animals under both stressed and unstressed condition, and in ddl-2(RNAi) animals under stressed condition (Fig. 3E). We also found a increased level of post-translationally modified HSF-1 in ddl-1(RNAi) animals under stressed and unstressed condition (Fig. 3F, S1D). Curiously, the level of HSF-1 PTM is slightly reduced in ddl-2(RNAi) animals (Fig. 3F, S1D), suggesting that DDL-1 and DDL-2 may regulate HSF-1 activity via both overlapping and distinct mechanisms. Together, our findings strongly support the hypothesis that HSB-1, DDL-1 and DDL-2 might function as negative regulators of HSF-1 to influence longevity in worms.
DDL-1/2 form a heterocomplex with HSB-1 and HSF-1
Having established that DDL-1/2 inhibits HSF-1 activity, and thereby suppresses HSF-1-dependent gene expression, we next asked how DDL-1/2 controls HSF-1 activity. It has been shown that the activity of mammalian HSF1 can be negatively regulated by the interaction between HSF1 and its binding proteins (Zuo et al., 1995). Therefore, it is possible that DDL-1/2 may regulate HSF-1 activity by forming a heterocomplex with HSB-1 and HSF-1 to keep HSF-1 in its inactive form.
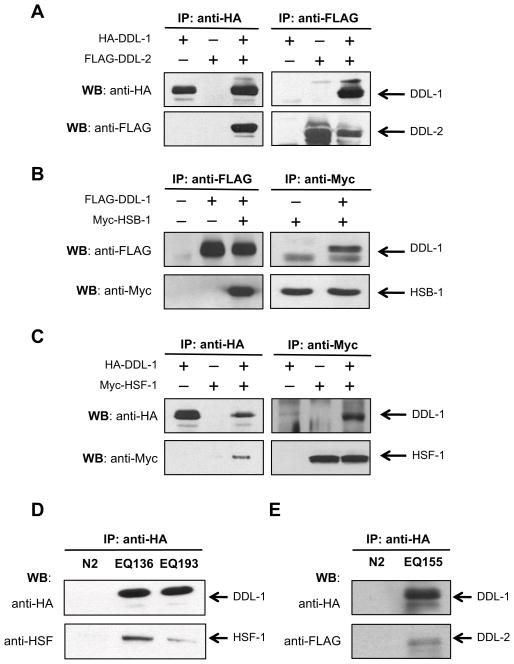
To test this hypothesis, we first examined the interactions between DDL-1 and DDL-2 in a cell culture system. Plasmids containing HA-tagged DDL-1 and/or FLAG-tagged DDL-2 were transfected into the 293T human renal epithelial cells. Co-immunoprecipitation (Co-IP) analyses were then performed to examine whether DDL-1 interacts with DDL-2. When cell extracts were immunoprecipitated with antibodies against HA-tag and blotted with antibodies against FLAG-tag, we were able to detect the FLAG-DDL-2 proteins (Fig. 4A). Similarly, HA-DDL-1 proteins can be pulled down together with DDL-2 using the anti-FLAG antibody (Fig. 4A). We then asked whether DDL-1 interacts with HSB-1. Our Co-IP results indicated that HSB-1 can be pulled down by an antibody against tagged DDL-1 and vice versa (Fig. 4B). These findings suggest that DDL-1 may physically interact with HSB-1 and DDL-2, or at least be present in a protein complex containing all three of them. Thus, if DDL-1 functions by forming an inhibitory complex of HSF-1, one may predict the presence of HSF-1 in this heterocomplex consisting of at least HSB-1, DDL-1, and DDL-2. Indeed, immunoprecipitation with antibodies against either tagged HSF-1 or DDL-1 successfully pulled down each other in 293T cells (Fig. 4C).
Figure 4. DDL-1 forms a protein complex with HSF-1, HSB-1, and DDL-2 in a cell culture model and in C. elegans.
(A) DDL-1 interacts with DDL-2 in 293T cells. Here and in (B-C), 293T cells were transfected with indicated combinations of pCMV-HA-DDL-1, pFLAG-CMV2-DDL-2, pFLAG-CMV2-DDL-1, pCMV-Myc-HSB-1, or pCMV-Myc-HSF-1 plasmids. Whole cell extracts were then immunoprecipitated (IP) and subsequently western-blotted (WB) using indicated antibodies. (B) DDL-1 interacts with HSB-1 in 293T cells. (C) DDL-1 may form a protein complex with HSF-1 in 293T cells. (D) DDL-1 may form a protein complex with HSF-1 in C. elegans. Worm whole cell extracts (WCE) were prepared from N2, EQ136 [HA-ddl-1 oe], or EQ193 [HA-ddl-1 oe; hsb-1(cg116)] animals. Samples were immunoprecipitated and subsequently western-blotted using indicated antibodies. (E) DDL-1 interacts with DDL-2 in C. elegans. WCE were prepared from N2 or EQ155 animals expressing both HA-DDL-1 and FLAG-DDL-2 proteins. Samples were immunoprecipitated and western-blotted using indicated antibodies.
To examine whether a similar protein complex is formed in C. elegans, we constructed a transgenic line over-expressing HA-ddl-1. Co-IP experiments were then carried out with protein samples isolated from these worms. Consistent with our observations in cell culture model, we were able to pull down endogenous worm HSF-1 together with DDL-1 using antibodies against HA-tagged DDL-1 (Fig. 4D). Similarly, interaction between DDL-1 and DDL-2 in worms is confirmed by Co-IP using a transgenic line over-expressing both HA-ddl-1 and Flag-ddl-2 (Fig. 4E). Co-IP experiments with transgenic animals in hsb-1(-) background indicate that the formation of this heterocomplex largely depends on the presence of HSB-1 (Fig. 4D). Together, our findings suggest that there is an evolutionarily conserved interaction between DDL-1 and its binding partners.
The formation of DDL-1 containing HSF-1 inhibitory complex (DHIC) is promoted by IIS
As we have demonstrated, the ability of HSF-1 to turn on transcription is negatively regulated by the IIS (Fig. 1). However, one major question that remains to be addressed is how DAF-2 controls the HSF-1 activity, which consequentially influences both longevity and cellular responses to heat. As a first step in exploring a molecular connection between the IIS pathway and HSF-1 activity, we investigated the impact of inhibiting IIS on the formation of DHIC (DDL-1 containing HSF-1 inhibitory complex). N2 and EQ136 animals (i.e. the HA-ddl-1 o.e. line) grown on control, daf-2, or akt-1 RNAi bacteria were harvested and subjected to Co-IP analysis. The proteins pulled down with anti-HA antibodies were then subjected to western blotting analysis using anti-HSF or anti-HA antibodies. We found that the amount of HSF-1 pulled down with DDL-1 is significantly reduced in EQ136 animals fed with daf-2 or akt-1 RNAi bacteria (Fig. 5A, S4A). The decreases in DDL-1-coprecipitated HSF-1 is likely due to an alteration on the formation of DHIC by IIS, since the mRNA levels of hsf-1, ddl-1, ddl-2 or hsb-1 are not altered (Fig. S3A, S4E) and the total protein level of HSF-1 is actually increased (Fig. 1E, S1D) in daf-2(RNAi) animals. This effect of IIS on the formation of DHIC appears to be DAF-16-independent, as daf-16 knockdown does not rescue the phenotype observed in daf-2 mutants (Fig. S4A). Intriguingly, we also found that heat stress does not affect the formation of DHIC (Fig. S4A), suggesting the presence of a separate DHIC-independent regulation on HSF-1 activity upon heat stress.
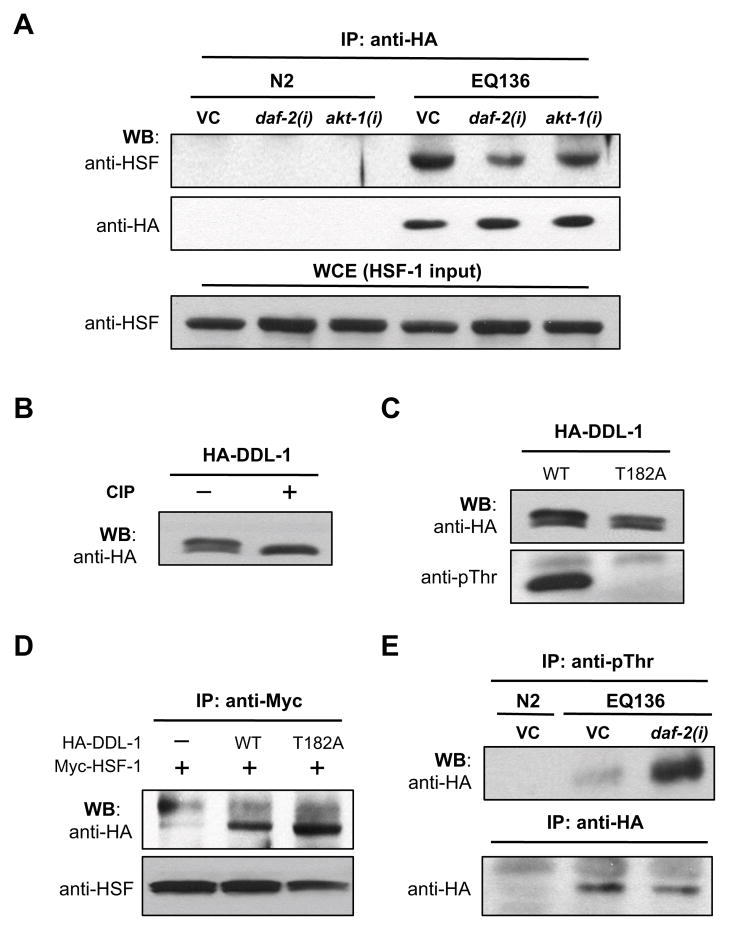
Figure 5. Both the formation of DHIC and the threonine phosphorylation of DDL-1 are regulated by IIS.
(A) The formation of DHIC is disrupted by IIS inactivation. Worm whole cell extracts (WCE) prepared from N2 or EQ136 [HA-ddl-1 oe] adult animals grown on control, daf-2, akt-1, or a 1:1 mixture of daf-2 and daf-16 RNAi bacteria were subjected to immunoprecipitation (IP) and western blotting analysis (WB) with indicated antibodies. The total HSF-1 input of each IP experiment was measured by blotting each WCE sample with anti-HSF1 antibodies. (B) Whole cell extracts prepared from 293T cells over-expressing HA-DDL-1 were treated with buffer or 1U/μg protein CIP (alkaline phosphatase) for 1 hr. Samples were then subjected to western blotting analysis (WB) with anti-HA antibodies. (C) Whole cell extracts prepared from 293T cells over-expressing HA-tagged wild-type or mutated (T182A) DDL-1 were subjected to western blotting analysis (WB) using anit-HA or anti-phosphothreonine antibodies. (D) 293T cells were transfected with indicated combinations of pCMV-driven HA-DDL-1(WT), HA-DDL-1(T182A) or Myc-HSF-1 plasmids. Whole cell extracts prepared from these cells were immunoprecipitated (IP) and subsequently western-blotted (WB) using indicated antibodies. (E) The level of threonine phosphorylated DDL-1 is elevated in daf-2 mutants. WCE prepared from N2 or EQ136 worms grown on control or daf-2 RNAi bacteria were immunoprecipitated and western-blotted using indicated antibodies. Shown here (A, D, and E) are the immunoblots of a representative experiment. Quantification of at least three experiments is shown in Fig. S4.
Phosphorylation of DDL-1 disrupts the formation of DHIC
Similar to HSF-1, results from our western blotting analysis has suggested that DDL-1 is also post-translationally modified (Fig. 5B). CIP treatment results indicate that phosphorylation accounts for a majority of PTM on DDL-1 (Fig. 5B). Western blotting analysis with anti-pThr antibodies suggest that at least one of the threonine residues on DDL-1 is phosphorylated (Fig. 5C). There are five putative phospho-threonine (pThr) sites on DDL-1, as predicted by PredPhospho (Kim et al., 2004). Two of which, Thr-171 and Thr-182, are located in close proximity to an evolutionarily conserved region that also contains a predicted pThr site (Thr-181) on human CCDC53. Thus, we constructed expression plasmids that encode DDL-1 with a threonine-to-alanine mutation at Thr-171 or Thr-182. Our results indicate that the Thr-182 to Ala (T182A) mutation completely eliminates the threonine phosphorylation on DDL-1 (Fig. 5C). Conversely, T171A mutation produced no detectable effect on the phosphorylation of DDL-1 (data not shown). This finding suggests that Thr-182 might be the only Thr residue that is phosphorylated on DDL-1. However, additional phosphorylations on non-Thr residues (e.g. Ser or Tyr) are likely to be present, since the PTM on DDL-1 is decreased but not abolished by T182A mutation (Fig. 5C).
Next, we asked whether the formation of DHIC is affected by the phosphorylation status of DDL-1. To address this, we carried out Co-IP experiments with either native or T182A DDL-1 in 293T cells. We found that the amount of DDL-1 that co-precipitated with HSF-1 is significantly higher when the Thr-182 residue of DDL-1 is mutated and not phosphorylated (Fig. 5D, S4C). Together, our findings suggest that the phosphorylation at Thr-182 of DDL-1 may promote DHIC dissociation and consequently the oligomerization and activation of HSF-1, although the kinase(s) that phosphorylates DDL-1 remains unknown.
Threonine phosphorylation of DDL-1 is regulated by IIS
How might IIS control DHIC formation? Having established that the threonine phosphorylation of DDL-1 negatively impacts DHIC formation, and thereby promotes HSF-1 activation, one possibility is that IIS may control the HSF-1 activity by modulating the phosphorylation status of DDL-1. To test this idea, we examined the level of threonine-phosphorylated DDL-1 in daf-2(RNAi) animals. Samples harvested from EQ136 animals (HA-ddl-1) fed with vector control or daf-2 RNAi bacteria were IP with anti-pThr antibodies and blotted with antibodies against HA tag. Consistent with the model, we found that inactivating DAF-2 elevates the ratio of threonine-phosphorylated DDL-1 to total DDL-1 by more than 5-fold (Fig. 5E, S4D).
Discussion
Although the roles of HSF-1 and heat-shock responsive genes in the regulation of stress responses and longevity have been previously described in C. elegans, how the HSF-1 activity is modulated at the molecular level in response to different environmental or even hormonal cues remains unknown. The work presented here provides an array of evidence for a possible mechanism underlying the regulation of HSF-1 activation by the insulin/IGF-1-like signaling (IIS), one of the major regulatory pathway for longevity. In this model, DDL-1/2 and HSB-1 negatively regulates HSF-1 activity by forming a protein complex with HSF-1 that consequently reduces the amount of HSF-1 susceptible to heat stress-induced activation. IIS controls HSF-1 activity, at least in part, by regulating the formation of this DDL-1 containing HSF-1 inhibitory complex (DHIC), possibly via modulating the threonine phosphorylation status of DDL-1 (Fig. 6).
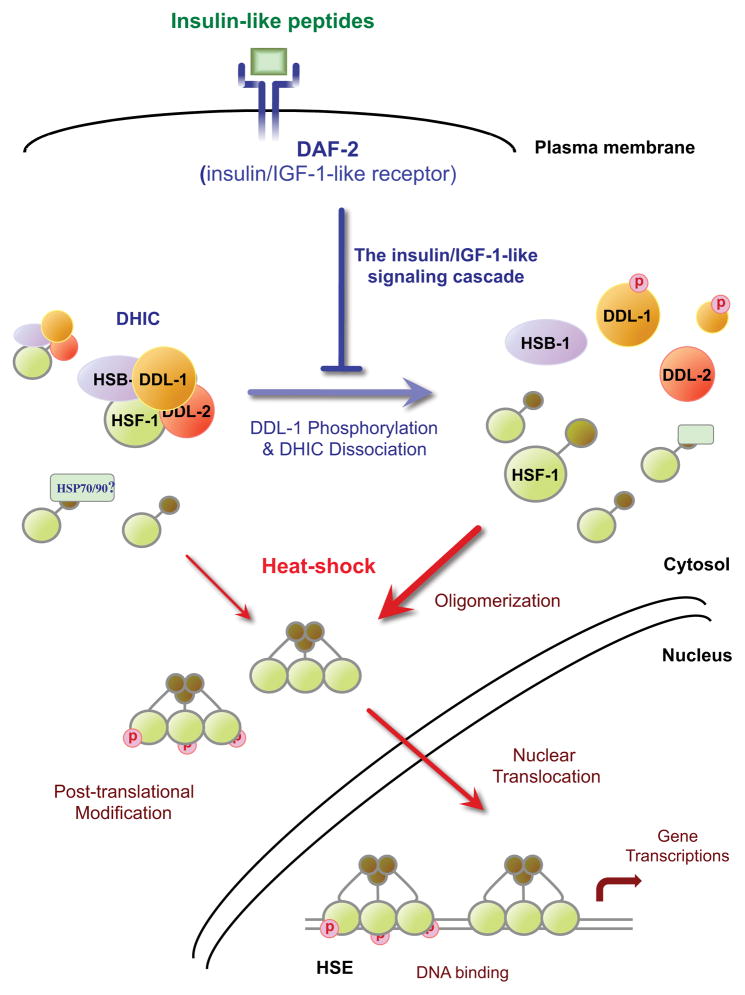
Figure 6. Model of HSF-1 activation regulated by IIS in C. elegans.
Upon heat stress stimulation, HSF-1 undergoes oligomerization, post-translational modification, and nuclear translocation in an undefined order before acquiring DNA-binding and transcriptional activity. The formation of a DDL-1 containing HSF-1 inhibitory complex (DHIC), consisting of HSF-1, HSB-1, DDL-1 and DDL-2, reduces the pool of HSF-1 susceptible to heat stress stimulation. Increased insulin/IGF-1-like signaling (IIS) promotes the formation of DHIC, while reducing DAF-2 activity promotes DDL-1 phosphorylation and disrupts DHIC formation and consequently increases HSF-1 activity under both stressed and unstressed conditions.
DDL-1 and DDL-2 as negative regulators of HSF-1
In mammalian cells, HSF1 activity may be negatively regulated by the intramolecular interaction between HSF1 and its binding proteins (Zuo et al., 1995). For example, mammalian HSF-1 forms inhibitory complexes with proteins such as Hsp90, Hsp70, and p23 (Voellmy, 2004). Similarly, we found that DDL-1/2 may also regulate HSF-1 activity by forming an inhibitory complex containing at least HSF-1, HSB-1 and DDL-1/2 (i.e. DHIC). In both cell culture and C. elegans models, DDL-1 appears to interact with, or at least co-exist in the same complex with HSF-1, DDL-2, and HSB-1 (Fig. 4A-E), while HSB-1 is required for the formation of this protein complex (Fig 4D).
Although our Co-IP results strongly suggest the presence of the DHIC in vivo, we cannot completely exclude the possibility that protein-protein interactions occurred after cell lysis. However, several lines of evidence suggest that this is unlikely to be the case. First, the protein-protein interaction observed between DDL-1 and HSF-1/HSB-1 can be disrupted by genetic manipulations, such as RNAi knockdown of daf-2, without affecting the overall protein levels of DDL-1 (Fig. 5A). This finding implies that the interaction observed is specific and likely occurs in vivo. Furthermore, human orthologs of DDL-1, DDL-2, and HSB-1 have been reported to either interact with each other or co-exist in the same protein complex (Derivery et al., 2009; Rual et al., 2005), suggesting that the formation of DHIC may be conserved across species. Finally, HSF-1 is expressed in almost all cell types as observed in our hsf-1::gfp transgenic lines, whereas HSB-1 is found to be expressed in a variety of tissues such as pharynx, intestine, muscles, and tail neurons (Hunt-Newbury et al., 2007). Similarly, strong DDL-1 expression is observed in the cytoplasm of many of the same tissues (e.g. pharynx, intestine, body wall muscle, and a subset of head and tail neurons, Fig. S5). The overlap in expression pattern among HSF-1, HSB-1 and DDL-1 support the idea that they do co-localize and interact with each other in vivo.
DDL-2, on the contrary, is mainly expressed in a subset of neurons, larval body wall muscles, and a small number of adult intestinal cells (Fig. S6). However, it is not clear whether the expression of DDL-2 in those neurons is indeed critical in regulating HSF-1 activity and longevity. Further studies are required to address these questions.
Direct regulation of HSF-1 activity by IIS
HSF-1 has been postulated to play a key role in mediating many of the beneficial health effects observed in long-lived IIS pathway mutants (Cohen et al., 2006; Hsu et al., 2003; Morley and Morimoto, 2004). It is also known that the up-regulation of certain HSF-1 targets is at least partially responsible for the lifespan extension observed in IIS pathway mutants (Hsu et al., 2003). However, it was not clear whether the activity of HSF-1 is under the direct control of IIS. In a recent study, McColl et al. reported that while the overall mRNA levels of several HSPs are significantly increased in IIS mutants, the DNA binding activity of HSF-1 and the stability of HSPs mRNA may not be altered by IIS (McColl et al., 2010). However, the EMSA experiments were done using a DNA probe containing four predicted HSF-1 binding motifs (HSE) without surrounding sequences found in endogenous hsp promoters. Using a different probe containing endogenous HSEs found in hsp16.2 promoter, our results indicate that the DNA binding activity of HSF-1 is up-regulated by IIS in a daf-16-independent manner (Fig. 1). Moreover, multiple steps of HSF-1 activation, including PTM and nuclear translocation, are also modulated by IIS (Fig. 1).
The most compelling evidence for a direct regulation of HSF-1 activity by IIS comes from our study on the effects of IIS inhibition on DHIC formation. First, we found that the formation of DHIC is largely diminished when IIS is reduced (Fig. 5A), suggesting that IIS may regulate HSF-1 activity by controlling the formation of DHIC. Furthermore, we found that the formation of DHIC is affected by the phosphorylation status of DDL-1, particularly at the Thr-182 residue, and that the level of theronine-phosphorylated DDL-1 is dramatically increased in daf-2(-) mutants (Fig. 5D–E). Although DAF-16 is known to act downstream of IIS to control the expression of several stress response genes, it appears that DAF-16 is not involved in this regulation (Fig. S4A).
While the phosphorylatin status of DDL-1 may play a key role in the regulation of DHIC formation and HSF-1 activity, the protein kinase(s) that phosphorylates DDL-1 remains unknown. The DDL-1 Thr-182 residue is predicted to be phosphorylated by a glycogen synthase kinase 3 (GSK-3)-like kinase by PredPhospho. However, RNAi knockdown of both GSK-3 isoforms in worms does not significantly affect the phosphorylation status of T182 (Fig. S4D). Similarly, inhibition of AKT-1, a kinase downstream of DAF-2, does not attenuate pT182 level. In fact, the level of pT182 is increased, as observed in daf-2 mutants (Fig. S4D), suggesting that DDL-1 is not a substrate of AKT-1. Further investigations are required to identify the kinase(s) responsible for the phosphorylation of DDL-1.
Multiple layers of regulations of HSF-1 activity
It is well known that an acute increase of HSF-1 activity can be induced by heat or other environmental stresses. Our study has shown that HSF-1 activity is also subjected to hormonal regulation, which occurs in a more chronic fashion. In addition to their temporal differences, the IIS appears to regulate HSF-1 activity via a molecular mechanism independent of the stress-induced activation of HSF-1, as heat stress does not affect the formation of DHIC (Fig. S4A). This also implies that the IIS pathway and DHIC may be important for controlling the amount of HSF-1 that is susceptible to heat stress stimulation, but is not required for the heat stress-induced HSF-1 activation. Therefore, IIS plays more of a modulatory role with respect to HSF-1 activity. This is different from the mechanism by which IIS regulates other transcription factors, such as DAF-16 and SKN-1, as reduced IIS leads to strong activations and dramatic increases in nuclear occupancy of these proteins.
Insulin and IGF-1 signaling are known to be involved in many key physiological processes, including metabolism, development, growth, reproduction, and aging. Recently, it has also been linked to the cellular stress responses. Similarly, while HSFs are best recognized as the master regulators of the heat-shock response and protein homeostasis, they also contribute to other physiological processes such as development and aging. Our findings presented here have provided a potential mechanism at the molecular level that links these two pathways together. Since most of the components of IIS and HSF-1 pathways found in worms are evolutionarily conserved, future studies aimed at better understanding the crosstalk between worm IIS and HSF pathways will shed light on the mechanisms by which the aging process is controlled across species.
Experimental Procedures
C. elegans Strains and Methods
Please see Supplemental Info for the list of strains used in this study. Animals were cultured on NGM plates seeded with E. coli at 15°C, using the standard method. All animals were cultured for at least two generations without starvation before the experiments were initiated.
Lifespan Analysis
Lifespan analysis was conducted at 20°C as previous ly described unless otherwise stated (Kenyon et al., 1993). RNAi treatments were carried out by adding synchronized eggs to plates seeded with the RNAi bacteria. Worms were moved to plates with fresh RNAi bacteria every two days until reproduction ceased. Worms were then moved to new plates every 5–7 days for the rest of the lifespan analysis. Viability of the worms was scored every 2–3 days.
Preparation of Worm Nuclear Extracts
Frozen worm pellets were homogenized in a Kontes Pellet Pestle® tissue grinder in the presence of an equal volume of 2X NPB buffer. Cells were pelleted (4000g, 5min, 4°C) and then homogenized 20 strokes with pestle A of the Dounce homogenizer. The suspension was then washed three times in NPB buffer containing 0.25% NP-40 and 0.1% Triton-X100. The nuclei were pelleted again and extracted with 4x volume of HEG buffer at 4°C for 45 min. The nuclear fraction was collected by centrifugation at 14,000g, 4°C for 15 min. Protein concentrations were determined by Bradford assay. Please see Supplemental Info for buffer recipes.
Electrophoretic Mobility Shift Assay (EMSA)
1 μg of worm nuclear extracts (NE) was incubated with 1μg/μL Poly (dI•dC) and 1 nM biotin-labeled oligonucleotide containing the HSE sequence for 15 min at room temperature in binding buffer (buffer recipes in Supplemental Info). The biotin-labeled oligonucleotides were synthesized based on the sequence covering the HSE in the promoter region of hsp-16.1 (detail sequence in Supplemental Info) Following native 3.5% polyacrylamide gel electrophoresis, HSF-1-HSE DNA complexes were visualized by LightShift® Chemiluminscent EMSA kit (Pierce).
HSF-1 nuclear localization assay
Day 2 adult animals carrying an integrated hsf-1::gfp array (EQ73) grown on a vector control (VC) or different RNAi bacteria were either unstressed or heat-shocked on 37°C heat block for 30 min. Fluorescence images of the animals were then taken and scored blindly for the nuclear accumulation of HSF-1::GFP protein in the intestinal cells (white arrows). At least 100 animals were scored per RNAi treatment per experiment. Worms were classified into separate groups according to the nuclear/cytosolic (n/c) ratio of GFP intensity in the intestinal cells.
Co-immunoprecipitation (Co-IP) in 293T cells
Twenty-four hours after 293T cells were transfected with various combinations of different plasmids containing HA-ddl-1, FLAG-ddl-1, FLAG-ddl-2, myc-hsb-1 or myc-hsf-1 cDNA, cells were washed with PBS and re-suspended in L-RIPA buffer (buffer recipes in Supplemental Info). The cell suspensions were then placed on ice for 10 min before being subjected to centrifugation at 14,000x g for 10 minutes at 4°C. The supernatants were then collected. The protein levels of whole cell extract (WCE) were quantified by Bradford assay. For each sample, 1,000 mg of total protein was used for the IP experiments. Anti-HA (Convance, #MMS101P), anti-FLAG (Sigma, #F3165) or anti-Myc (Cell Signaling, #2276) antibodies were added to WCE at 1:150, 1:300, and 1:500 dilutions, respectively. Five mg of anti-mouse rabbit polyclonal antibody was then added as bridge antibody. Reactions were incubated at 4°C with gentle shaking overnight. 30 ml of 50% Protein A-agarose beads (Sigma #P7786; pre-blocked by 10% BSA) were added into the solutions 4–5 hrs after the initiation of the incubation. The beads were washed three times with L-RIPA buffer supplemented with 50 mg/ml ABESF and 1 mM sodium orthovanadate, before being subjected to western blotting analysis.
Co-immunoprecipitation (Co-IP) in Worms
About 15,000 synchronized day 1 adult worms grown on either control or RNAi bacteria at 20°C were harvested by washing three times with cold M9 buffer and one more time with HB-high salt buffer (buffer recipes in Supplemental Info). Worm pellets were then resuspended in 3x volume of HB-high salt buffer supplemented with Protease Inhibitor Cocktail Complete Mini (Roche), 2.5 mM sodium pyrophosphate, 20 mM b-glycerolphosphate, and 1 mM sodium orthovanadate. The pellets were immediately frozen and stored in liquid nitrogen for future use. Frozen suspensions were thawed, homogenized with a Dounce homogenizer (30 strokes with a pestle B), and centrifuged at 14,000x g at 4 °C for 20 minutes. Supernatants (i.e. the worm WCE) were collected and total protein concentrations were quantified by Bradford assay. If necessary, cross-linking was done by incubating worm protein extracts with 1 mM EGS [ethylene glycol bis(succinimidyl succinate)] at 25°C for 30min. The cross-linking reactions were stopped by adding and incubating with 20mM Tris-HCl, pH 7.4 for an additional 30 min. For immunoprecipitation, 30 ml of anti-HA agarose beads (Sigma #A2095) were added to 1,500 mg of protein extract and incubated with gentle shaking at 4°C overnight. The beads were then washed 3 times with HB-high salt buffer supplemented with 50 mg/ml ABESF and 1 mM sodium orthovanadate before being subjected to western blotting analysis.
Western Blotting Analysis
The samples were subjected to SDS-PAGE and transferred to a PVDF membrane (Millipore). The transblotted membrane was washed three times with TBS containing 0.05% Tween 20 (TBST). After blocking with TBST containing 5% nonfat milk for 60 min, the membrane was incubated with the primary antibody indicated (e.g. anti-HSF1, Calbiochem, #385580) at 4°C for 12 h and washed three times with TBST. The membrane was then probed with HRP-conjugated secondary antibody for 1 h at room temperature and washed with TBST three times. Finally, the immunoblots were detected using a chemiluminescent substrate (Pierce) and visualized by autoradiography.
Supplementary Material
Acknowledgments
This work was supported by grant AG028516 from NIH/NIA and grant from the Ellison Medical Foundation to A.-L. Hsu. We thank the Caenorhabditis Genetic Center for providing the PS3551, CH116, VC2193, and RB2380 strains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Clos J, Westwood JT, Becker PB, Wilson S, Lambert K, Wu C. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell. 1990;63:1085–1097. doi: 10.1016/0092-8674(90)90511-c. [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Developmental cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Sternberg PW. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 2004;168:1937–1949. doi: 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92:1564–1572. doi: 10.1093/jnci/92.19.1564. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Khazaeli AA, Tatar M, Pletcher SD, Curtsinger JW. Heat-induced longevity extension in Drosophila. I. Heat treatment, mortality, and thermotolerance. J Gerontol A Biol Sci Med Sci. 1997;52:B48–52. doi: 10.1093/gerona/52a.1.b48. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee J, Oh B, Kimm K, Koh I. Prediction of phosphorylation sites using SVMs. Bioinformatics. 2004;20:3179–3184. doi: 10.1093/bioinformatics/bth382. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kurapati R, Passananti HB, Rose MR, Tower J. Increased hsp22 RNA levels in Drosophila lines genetically selected for increased longevity. J Gerontol A Biol Sci Med Sci. 2000;55:B552–559. doi: 10.1093/gerona/55.11.b552. [DOI] [PubMed] [Google Scholar]
- Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow GJ, Walker GA. Stress resistance as a determinate of C. elegans lifespan. Mech Ageing Dev. 2002;123:765–771. doi: 10.1016/s0047-6374(01)00422-5. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 2010;12:260–272. doi: 10.1016/j.cmet.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier PA, Winegarden NA, Westwood JT. Human heat shock factor 1 is predominantly a nuclear protein before and after heat stress. J Cell Sci. 1999;112(Pt 16):2765–2774. doi: 10.1242/jcs.112.16.2765. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 1998;12:1962–1974. doi: 10.1101/gad.12.13.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Morimoto RI. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol Cell Biol. 1994;14:2087–2099. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Lewis MJ, Pelham HR. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987;329:81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- Soti C, Csermely P. Molecular chaperones and the aging process. Biogerontology. 2000;1:225–233. doi: 10.1023/a:1010082129022. [DOI] [PubMed] [Google Scholar]
- Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9:122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Zuo J, Rungger D, Voellmy R. Multiple layers of regulation of human heat shock transcription factor 1. Mol Cell Biol. 1995;15:4319–4330. doi: 10.1128/mcb.15.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.