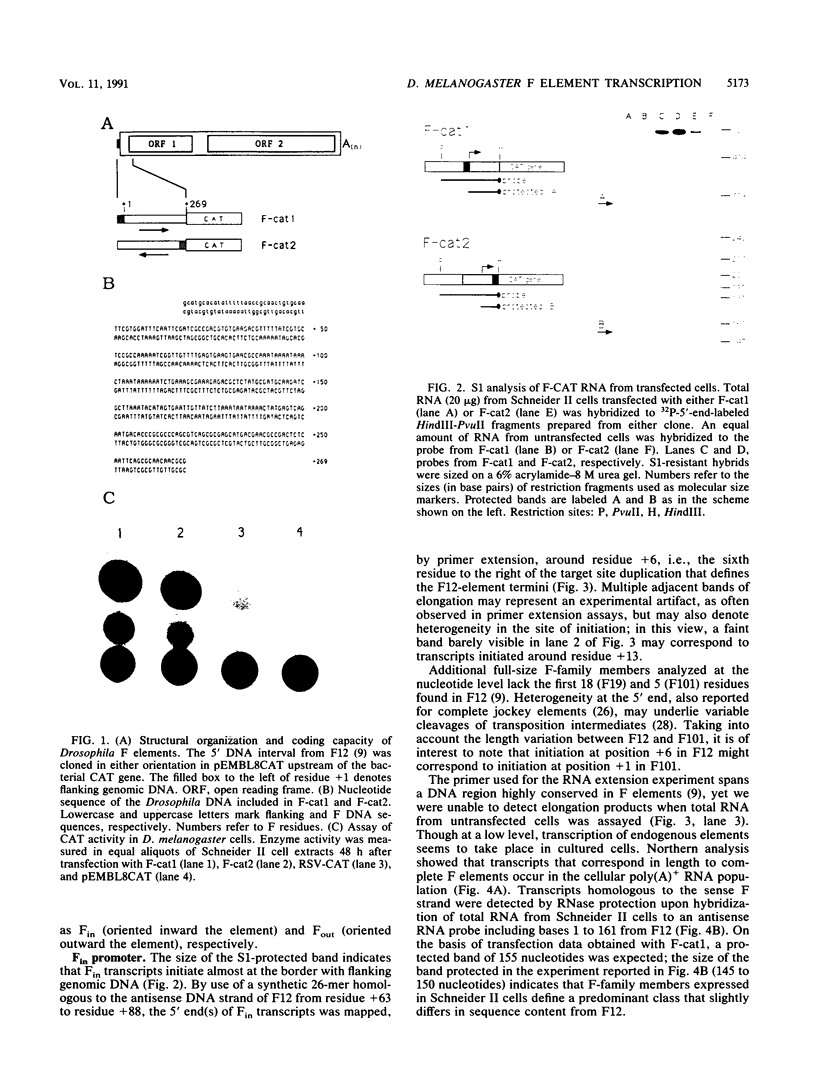
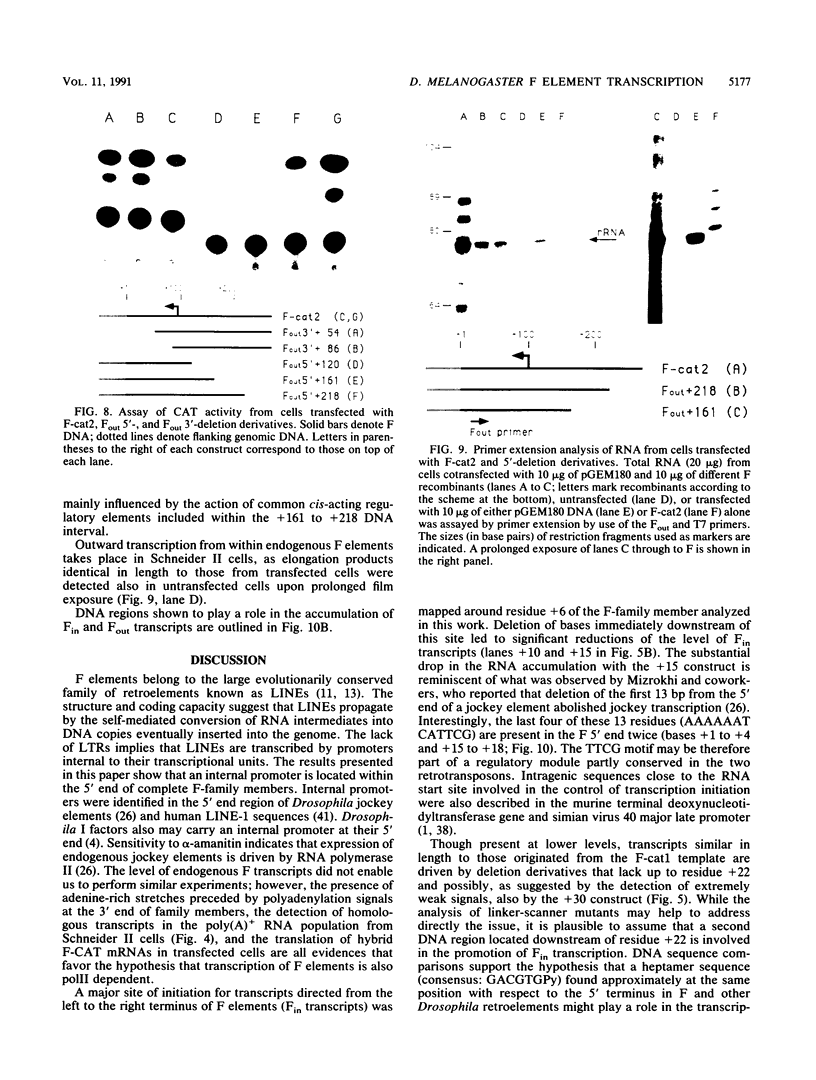
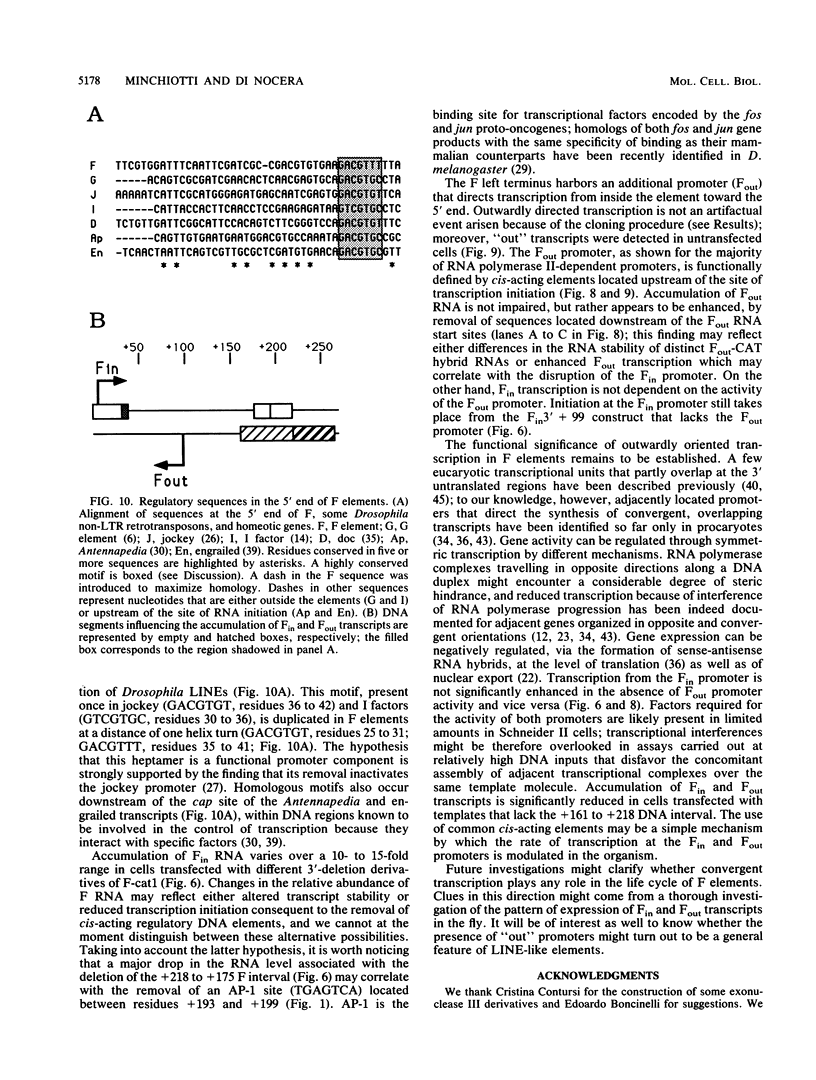
Abstract
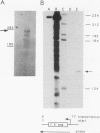
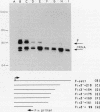
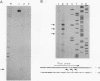
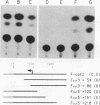
Drosophila melanogaster F elements are mobile, oligo(A)-terminated DNA sequences that likely propagate by the retrotranscription of RNA intermediates. Plasmids bearing DNA segments from the left-hand region of a full-length F element fused to the CAT gene were used as templates for transient expression assays in Drosophila Schneider II cultured cells. Protein and RNA analyses led to the identification of two promoters, Fin and Fout, that transcribe in opposite orientations. The Fin promoter drives the synthesis of transcripts that initiate around residue +6 and are directed toward the element. Fin, that probably controls the formation of F transposition RNA intermediates and gene products, is internal to the transcribed region. Sequences important for accumulation of Fin transcripts are included within the +1 to +30 interval; an additional regulatory element may coincide with a heptamer located downstream of this region also found in the 5' end regions of F-like Drosophila retrotransposons. Analysis of the template activity of 3' deletion derivatives indicates that the level of accumulation of Fin RNA is also dependent upon the presence of sequences located within the +175 to +218 interval. The Fout promoter drives transcription in the opposite orientation with respect to Fin. Fout transcripts initiate at nearby sites within the +92 to +102 interval. Sequences downstream of these multiple RNA start sites are not required for the activity of the Fout promoter. Deletions knocking out the Fin promoter do not impair Fout transcription; conversely, initiation at the Fin promoter still takes place in templates that lack the Fout promoter. At a low level, both promoters are active in cultured cells.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayer D. E., Dynan W. S. Simian virus 40 major late promoter: a novel tripartite structure that includes intragenic sequences. Mol Cell Biol. 1988 May;8(5):2021–2033. doi: 10.1128/mcb.8.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A., Paro R., Sang H. M., Pelisson A., Finnegan D. J. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984 Aug;38(1):153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- Chaboissier M. C., Busseau I., Prosser J., Finnegan D. J., Bucheton A. Identification of a potential RNA intermediate for transposition of the LINE-like element I factor in Drosophila melanogaster. EMBO J. 1990 Nov;9(11):3557–3563. doi: 10.1002/j.1460-2075.1990.tb07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey S. N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986 Jan 24;14(2):623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Casari G. Related polypeptides are encoded by Drosophila F elements, I factors, and mammalian L1 sequences. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5843–5847. doi: 10.1073/pnas.84.16.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P. Close relationship between non-viral retroposons in Drosophila melanogaster. Nucleic Acids Res. 1988 May 11;16(9):4041–4052. doi: 10.1093/nar/16.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Dawid I. B. Transient expression of genes introduced into cultured cells of Drosophila. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Digan M. E., Dawid I. B. A family of oligo-adenylate-terminated transposable sequences in Drosophila melanogaster. J Mol Biol. 1983 Aug 25;168(4):715–727. doi: 10.1016/s0022-2836(83)80071-0. [DOI] [PubMed] [Google Scholar]
- Di Nocera P. P., Graziani F., Lavorgna G. Genomic and structural organization of Drosophila melanogaster G elements. Nucleic Acids Res. 1986 Jan 24;14(2):675–691. doi: 10.1093/nar/14.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 1989 Feb;3(2):185–197. doi: 10.1101/gad.3.2.185. [DOI] [PubMed] [Google Scholar]
- Fanning T. G., Singer M. F. LINE-1: a mammalian transposable element. Biochim Biophys Acta. 1987 Dec 8;910(3):203–212. doi: 10.1016/0167-4781(87)90112-6. [DOI] [PubMed] [Google Scholar]
- Fawcett D. H., Lister C. K., Kellett E., Finnegan D. J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986 Dec 26;47(6):1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J., Fawcett D. H. Transposable elements in Drosophila melanogaster. Oxf Surv Eukaryot Genes. 1986;3:1–62. [PubMed] [Google Scholar]
- Franzè A., Archidiacono N., Rocchi M., Marino M., Grimaldi G. Isolation and expression analysis of a human zinc finger gene (ZNF41) located on the short arm of the X chromosome. Genomics. 1991 Apr;9(4):728–736. doi: 10.1016/0888-7543(91)90367-n. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Di Nocera P. P. Multiple repeated units in Drosophila melanogaster ribosomal DNA spacer stimulate rRNA precursor transcription. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5502–5506. doi: 10.1073/pnas.85.15.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Kuhara S., Takenaka O., Sakaki Y. L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature. 1986 Jun 5;321(6070):625–628. doi: 10.1038/321625a0. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Jakubczak J. L., Xiong Y., Eickbush T. H. Type I (R1) and type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J Mol Biol. 1990 Mar 5;212(1):37–52. doi: 10.1016/0022-2836(90)90303-4. [DOI] [PubMed] [Google Scholar]
- Kim S. K., Wold B. J. Stable reduction of thymidine kinase activity in cells expressing high levels of anti-sense RNA. Cell. 1985 Aug;42(1):129–138. doi: 10.1016/s0092-8674(85)80108-2. [DOI] [PubMed] [Google Scholar]
- Lloubes R., Baty D., Lazdunski C. The promoters of the genes for colicin production, release and immunity in the ColA plasmid: effects of convergent transcription and Lex A protein. Nucleic Acids Res. 1986 Mar 25;14(6):2621–2636. doi: 10.1093/nar/14.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckow B., Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987 Jul 10;15(13):5490–5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrokhi L. J., Georgieva S. G., Ilyin Y. V. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988 Aug 26;54(5):685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- Mizrokhi L. J., Mazo A. M. Evidence for horizontal transmission of the mobile element jockey between distant Drosophila species. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9216–9220. doi: 10.1073/pnas.87.23.9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossie K. G., Young M. W., Varmus H. E. Extrachromosomal DNA forms of copia-like transposable elements, F elements and middle repetitive DNA sequences in Drosophila melanogaster. Variation in cultured cells and embryos. J Mol Biol. 1985 Mar 5;182(1):31–43. doi: 10.1016/0022-2836(85)90025-7. [DOI] [PubMed] [Google Scholar]
- Perkins K. K., Dailey G. M., Tjian R. In vitro analysis of the Antennapedia P2 promoter: identification of a new Drosophila transcription factor. Genes Dev. 1988 Dec;2(12A):1615–1626. doi: 10.1101/gad.2.12a.1615. [DOI] [PubMed] [Google Scholar]
- Perkins K. K., Dailey G. M., Tjian R. Novel Jun- and Fos-related proteins in Drosophila are functionally homologous to enhancer factor AP-1. EMBO J. 1988 Dec 20;7(13):4265–4273. doi: 10.1002/j.1460-2075.1988.tb03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A. C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J. L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988 Jun;7(6):1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priimägi A. F., Mizrokhi L. J., Ilyin Y. V. The Drosophila mobile element jockey belongs to LINEs and contains coding sequences homologous to some retroviral proteins. Gene. 1988 Oct 30;70(2):253–262. doi: 10.1016/0378-1119(88)90197-7. [DOI] [PubMed] [Google Scholar]
- Saha S., Haggård-Ljungquist E., Nordström K. The cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. EMBO J. 1987 Oct;6(10):3191–3199. doi: 10.1002/j.1460-2075.1987.tb02631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuwly S., Kuroiwa A., Gehring W. J. Molecular analysis of the dominant homeotic Antennapedia phenotype. EMBO J. 1987 Jan;6(1):201–206. doi: 10.1002/j.1460-2075.1987.tb04739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Translational control of IS10 transposition. Cell. 1983 Sep;34(2):683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Baltimore D. The "initiator" as a transcription control element. Cell. 1989 Apr 7;57(1):103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Soeller W. C., Poole S. J., Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988 Jan;2(1):68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Spencer C. A., Gietz R. D., Hodgetts R. B. Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature. 1986 Jul 17;322(6076):279–281. doi: 10.1038/322279a0. [DOI] [PubMed] [Google Scholar]
- Swergold G. D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990 Dec;10(12):6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. F., Murray N. E. Convergent transcription in bacteriophage lambda: interference with gene expression. J Mol Biol. 1979 Sep 15;133(2):249–266. doi: 10.1016/0022-2836(79)90533-3. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Williams T., Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3' ends. Nature. 1986 Jul 17;322(6076):275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. The site-specific ribosomal DNA insertion element R1Bm belongs to a class of non-long-terminal-repeat retrotransposons. Mol Cell Biol. 1988 Jan;8(1):114–123. doi: 10.1128/mcb.8.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]