SUMMARY
Assembly factors (AFs) prevent premature translation initiation on small (40S) ribosomal subunit assembly intermediates by blocking ligand binding. However, it is unclear how AFs are displaced from maturing 40S ribosomes, if or how maturing subunits are assessed for fidelity, and what prevents premature translation initiation once AFs dissociate. Here we show that maturation involves a translation-like cycle whereby the translation factor eIF5B, a GTPase, promotes joining of large (60S) subunits with pre-40S subunits to give 80S-like complexes, which are subsequently disassembled by the termination factor Rli1, an ATPase. The AFs Tsr1 and Rio2 block the mRNA channel and initiator tRNA binding site, and therefore 80S-like ribosomes lack mRNA or initiator tRNA. After Tsr1 and Rio2 dissociate from 80S-like complexes Rli1-directed displacement of 60S subunits allows for translation initiation. This cycle thus provides a functional test of 60S subunit binding and the GTPase site before ribosomes enter the translating pool.
INTRODUCTION
Ribosome biosynthesis involves the transcription of rRNA precursors, their processing into mature rRNAs, the binding of ribosomal proteins, and the folding of rRNAs. In eukaryotes, this process is facilitated by a large machinery of mostly essential assembly factors (AFs) and occurs in two phases: an early nucleolar/nuclear phase followed by cytoplasmic maturation steps (Karbstein, 2011; Strunk and Karbstein, 2009). For assembly of the small (40S) subunit, ribosomal proteins (Rps) from the “body” and the “platform” regions of the subunit bind during transcription (Ferreira-Cerca et al., 2005; Karbstein, 2011). Most of the remaining proteins, which bind to the “head” region, bind prior to the export of the small subunit to the cytoplasm to produce a 40S assembly intermediate that contains all Rps except Rps10 and Rps26, and which thus substantially resembles mature 40S subunits (Strunk et al., 2011).
In addition to pre-18S rRNA (20S rRNA) and most Rps, the cytoplasmic pre-40S precursor is stably bound to seven AFs: the nuclease Nob1, which produces the mature 18S rRNA 3’-end, and its regulator Pno1; the methylase Dim1; the kinase Rio2; the export adaptor Ltv1, and its binding partner Enp1; and the GTPase-like Tsr1. These AFs co-purify with pre-40S subunits over two columns and remain bound for days (Strunk et al., 2011). As this assembly intermediate resides in the cytoplasm, where mRNAs, tRNAs and translation factors are abundant, premature translation initiation may present a problem as incompletely assembled ribosomes could be prone to translational errors and stalled ribosomes and their bound mRNAs are rapidly degraded (Cole et al., 2009; Soudet et al., 2010).
A major function of AFs bound to late cytoplasmic 40S assembly intermediates is to prevent premature translation initiation (Strunk et al., 2011). AFs sterically block the binding of translation initiation factors to the 40S subunit and prevent mRNA recruitment. Additionally, Tsr1 impedes the spontaneous joining of 40S and 60S subunits, and the decoding site is deformed. While the positioning of these AFs explains how premature translation initiation is blocked, their presence raises new questions: How is dissociation of assembly factors promoted without leading to translation initiation in the process? Does this conversion of inactive assembly intermediates to mature 40S ribosomes require any checkpoints to assure that subunits are functional? Furthermore, purified cytoplasmic 40S ribosome assembly intermediates are stable for days, while maturation, and thus dissociation of AFs, takes only minutes in vivo (Granneman et al., 2005; Kos and Tollervey, 2010). Taken together these data suggest that extrinsic factors are required to promote dissociation of AFs.
Taking advantage of the sequential dissociation of AFs from pre-40S subunits, and using yeast strains deleted or depleted for factors promoting progress in this dissociation cascade, we have dissected individual late steps in 40S ribosome maturation. These data reveal the surprising finding that 40S maturation involves a translation-like cycle, whereby mature large (60S) subunits join pre-40S subunits in an eIF5B-dependent manner. However, the resulting 80S-like ribosomes are not translation initiation intermediates as they do not contain mRNA or initiator tRNA. Thus, mRNA recruitment and translation initiation require dissociation of 60S subunits mediated by the termination factor Rli1.
Collectively, these findings define this translation-like cycle as a final quality control step in which major functions of the maturing small subunit, such as binding to 60S subunits and binding to translation factors are inspected.
RESULTS
Fap7 Depletion Leads to Accumulation of pre-40S Ribosomes in 80S-like Ribosomes
Pre-40S ribosomes that accumulate in the cytoplasm contain seven stably bound assembly factors (AFs), which are strategically positioned to prevent the premature initiation of translation on pre-40S subunits (Strunk et al., 2011). When purified from cells these pre-40S subunits are stable, suggesting that dissociation of AFs is extrinsically controlled, and perhaps even requires energy input in the form of ATP or GTP. However, how this is accomplished and how this occurs without triggering premature translation initiation is unknown.
We reasoned that AFs were likely to dissociate sequentially and not all together, and that depletion of a protein required for AF removal would lead to accumulation of pre-40S subunits containing only a subset of the seven stably bound AFs: those whose dissociation occurs after or dependent upon the depleted protein. The binding of AFs to pre-40S subunits can be conveniently monitored by sucrose gradient sedimentation followed by Western blotting for AFs.
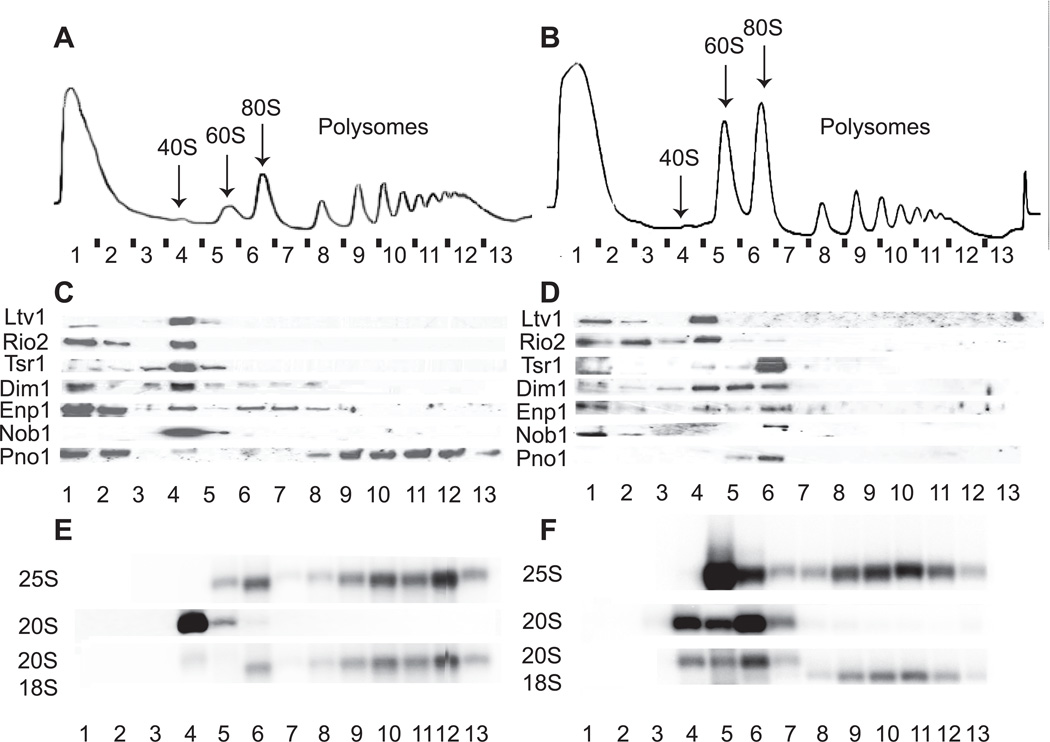
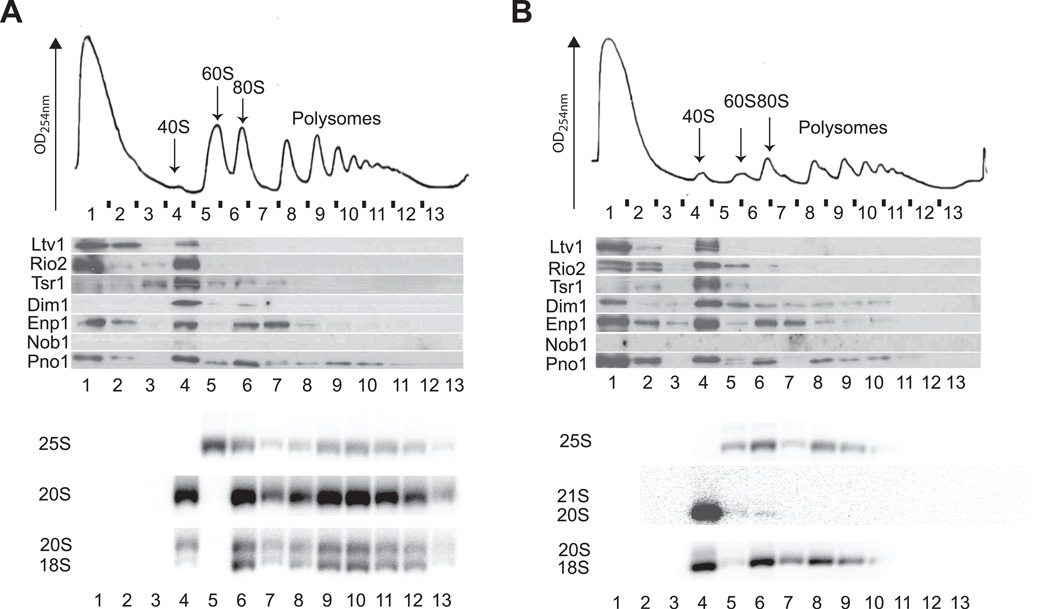
Several proteins that are required for the maturation of cytoplasmic pre-40S subunits are not stably associated with these subunits. One of these is Fap7, which contains an ATPase motif (Granneman et al., 2005) and has ATPase activity (BSS & KK, unpublished data). To assess if Fap7 is required for displacement of AFs from late pre-40S subunits, extracts were prepared from yeast depleted of Fap7 and analyzed by sucrose gradient centrifugation. Surprisingly, depletion of Fap7 triggers the accumulation of 80S ribosomes (Figure 1B), a highly unusual phenotype for cells depleted for 40S assembly factors (Strunk et al., 2011, and data not shown). Interestingly, immunoblot analyses demonstrate that in the absence of Fap7, nearly all AFs, except for Ltv1 and Rio2, are found within these 80S-like ribosomes (Figure 1D). Moreover, these 80S-like complexes comprise 60S subunits and pre-40S subunits, as they contain 25S rRNA and 20S pre-rRNA (Figure 1F), and lack hallmarks of ~90S early nucleolar precursors, such as pre-18S rRNA intermediates (21/22/23S or 35S) (Figure 1F and data not shown). Additionally, no evidence for 25.5S rRNA, the precursor for 25S rRNA, was found in these 80S-like ribosomes (data not shown). These data are consistent with the previously reported accumulation of 20S rRNA in the cytoplasm upon Fap7 depletion (Granneman et al., 2005), and indicate that the substrates for Fap7 during 40S maturation are not free pre-40S ribosomes, but instead 80S-like complexes, which contain large ribosomal subunits in addition to pre-40S subunits. Thus, 40S maturation occurs within 80S-like ribosomes. Furthermore, the data also strongly suggest that Fap7 acts after Rio2 and Ltv1 have already dissociated from pre-40S subunits, as these two AFs are not found in 80S-like ribosomes that accumulate upon depletion of Fap7 (Figure 1D). Notably, yeast expressing the Fap7 mutant D82AH84A, which is in its predicted Walker B motif, show an identical phenotype (Figure S1A), indicating that ATP hydrolysis is required for progression of the 40S maturation pathway as previously suggested (Granneman et al., 2005).
Figure 1. Depletion of Fap7 Leads to Accumulation of 40S Assembly Factors in 80S-Like Ribosomes.
10–50% sucrose gradients from cell lysates from Rio2TAP (A,C,E) or Rio2TAP, Gal1::Fap7 (B,D,F) cells grown in glucose for 16h.
(A,B) Absorbance profiles at 254 nm.
(C,D) Western blots for assembly factors.
(E,F) Northern blots for rRNAs and precursors. Exposure times were 4–6h, 2h and 1–2 h for 20S, 18S and 25S rRNAs, respectively.
See also Figure S1.
Fap7 Binds 80S Ribosomes
To test if Fap7 binds to 80S-like ribosomes during small subunit maturation and to confirm that 80S-like ribosomes are formed as an on-pathway intermediate rather than as a dead-end product that manifests only in the absence of functional Fap7, we tested if Fap7 binds to 80S ribosomes. Fap7 only transiently interacts with pre-ribosomes (Granneman et al., 2005). Therefore, to stabilize possible interactions between Fap7 and its substrate, formaldehyde crosslinking was performed as previously described (Valasek et al., 2007). Proteins associated with Fap7 were purified via TAP-purification from a strain expressing TAP-tagged Fap7 and analyzed by mass-spectrometry after reversal of the crosslink. Notably, the majority of proteins that co-purify with Fap7 are ribosomal proteins from both subunits (Table 1), strongly supporting the notion that 80S-like ribosomes are bona-fide substrates for Fap7. Mass spectrometry also identified some ribosome-associated chaperones, which might co-purify via their association with 60S subunits. These findings are consistent with previous gradient centrifugation experiments (Granneman et al., 2005). These experiments showed most of Fap7 to be free, consistent with transient binding interactions, while a small fraction of Fap7 was associated with 80S ribosomes (Granneman et al., 2005).
Table 1.
Proteins crosslinked to Fap7TAP
| # spectra | MW | function | ||
|---|---|---|---|---|
| Asc1 | 3 | 35 kDa | 40S protein | |
| Rps0 | 8 | 28 kDa | 40S protein | |
| Rps7 | 1 | 22 kDa | 40S protein | |
| 40S proteins | Rps12 | 2 | 15 kDa | 40S protein |
| Rps14 | 3 | 15 kDa | 40S protein | |
| Rps16 | 6 | 16 kDa | 40S protein | |
| Rps18 | 2 | 17 kDa | 40S protein | |
| Rps19 | 4 | 16 kDa | 40S protein | |
| Rps22 | 2 | 15 kDa | 40S protein | |
| Rps24 | 2 | 15 kDa | 40S protein | |
| Rpp0 | 7 | 34 kDa | 60S protein | |
| Rpl5 | 2 | 34 kDa | 60S protein | |
| 60S proteins | Rpl6 | 2 | 20 kDa | 60S protein |
| Rpl7 | 2 | 22 kDa | 60S protein | |
| Rpl11 | 2 | 20 kDa | 60S protein | |
| Rpl12 | 5 | 18 kDa | 60S protein | |
| Rpl14 | 1 | 15 kDa | 60S protein | |
| Rpl23 | 1 | 14 kDa | 60S protein | |
| Rpl26 | 3 | 14 kDa | 60S protein | |
| Dim1 | 7 | 36 kDa | 18S rRNA dimethylase | |
| Translation factors | eIF4A | 2 | 45 kDa | Translation initiation |
| eEF1α | 31 | 50 kDa | Translation elongation | |
| eEF1β | 2 | 23 kDa | Translation elongation | |
| eIF5A | 9 | 17 kDa | Translation elongation | |
| Tsa1 | 11 | 22 kDa | Ribosome-associated thioredoxin peroxidase | |
| Bmh2 | 11 | 31 kDa | 14-3-3 protein | |
| Ola1 | 6 | 44 kDa | GTPase | |
| Vma4 | 3 | 26 kDa | Vacuolar H+ATPase | |
| Sam1 | 3 | 42 kDa | SAM synthetase |
80S-like Ribosomes that Accumulate in the Absence of Fap7 Are Not Translation Initiation Intermediates
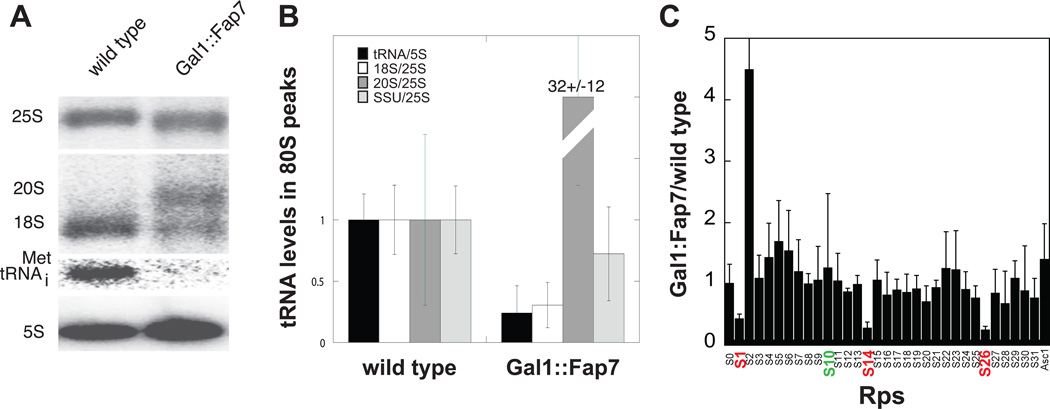
20S rRNA and Nob1 can be found in polysomes, suggesting that translation might initiate on pre-40S subunits containing Nob1 and perhaps other assembly factors (Soudet et al., 2010). To determine if the 80S-like ribosomes observed in yeast lacking Fap7 are intermediates in the translation initiation pathway, Northern blotting was performed to assess if initiator tRNA (Met-tRNAiMet) was present in these complexes. Notably, the levels of Met-tRNAiMet found in 80S-like ribosomes from yeast lacking Fap7 are strongly reduced relative to those associated with 80S ribosomes in wild type cells (Figure 2A&B, tRNA levels relative to 5S rRNA levels are 1 ± 0.21 and 0.24 ± 0.22 in wild type and Fap7 depleted cells, respectively). Furthermore, the levels of Met-tRNAiMet in the 80S-like complexes from Fap7-depleted yeast correlate with levels of remaining mature 40S subunits, but not with the highly increased levels of 20S rRNA (Figure 2B), indicating that the remaining Met-tRNAiMet is only associated with remaining mature 80S complexes (containing mature 40S and 60S subunits) in these cells. Therefore, the 80S-like ribosomes that accumulate in Fap7-depleted yeast do not contain initiator tRNA. Furthermore, mass spectrometry indicates that only the 80S peak from wild type cells, not the 80S-like peak from D82H84A cells contains eIF2, the initiation factor that delivers initiator tRNA to the ribosome (data not shown). These findings are consistent with the overlap of the binding site for Rio2 (Strunk et al., 2011) with that predicted for eIF2•tRNAiMet based on recent footprinting experiments (Shin et al., 2011). This overlap suggests that Rio2 blocks binding of eIF2•tRNAiMet.
Figure 2. 80S-like Ribosomes from Gal1:Fap7 Strains Are Not Translation Initiation Intermediates.
(A) 80S ribosomes from Fap7-depleted cells do not contain initiator tRNA. Northern blots for 18S, 25S, and 20S pre-rRNAs as well as for initiator tRNA shows that levels of initiator tRNA do not reflect the levels of 20S rRNA, but are concordant with the levels of mature 18S rRNA.
(B) tRNA levels in 80S peaks averaged from six experiments such as those in panel A.
(C) 80S-like ribosomes contain Rps10, but Rps26, Rps14 and Rps1 are underrepresented. The LC peak area for each peptide identified by MS/MS in 80S-like ribosomes isolated from Fap7 D82AH84A cells is plotted relative to the LC peak area for the same peptide obtained from 80S ribosomes isolated from wild type cells. Data from four independent experiments were averaged and plotted with error bars representing the standard deviation.
Purified 40S assembly intermediates that accumulate in the cytoplasm contain all seven AFs, as well the 18S precursor 20S pre-rRNA, and all 40S ribosomal proteins (Rps), except Rps10 and Rps26, whose binding sites overlap with those for the AFs Ltv1/Enp1 and Pno1, respectively (Strunk et al., 2011). The 80S-like ribosomes that accumulate in the absence of Fap7 lack Ltv1, but retain Pno1 (Figure 1D). Accordingly, the pre-40S subunits in these complexes contain Rps10, as demonstrated by mass-spectrometry (Figure 2C). In contrast, Rps26 is underrepresented relative to mature 40S subunits (Figure 2C), and its level reflects that of the remaining mature 40S ribosomes in these cells (ratio of Rps26 in 80S cells from mutant and wild type cells is 0.25±0.06). These data indicate that ribosomes that accumulate in the absence of functional Fap7 lack Rps26, consistent with their binding to Pno1. Mass-spectrometry also suggests that Rps1 and Rps14 are underrepresented (ratiogal:Fap7/wt of Rps1 and Rps14 is 0.43±0.08 and 0.28±0.10, respectively, Figure 2C). These proteins are at least partially bound in stable cytoplasmic ribosomes (Strunk et al., 2011). Perhaps their association is weak in cells depleted of Fap7 and lost during centrifugation. Consistent with this idea Rps1, Rps14 and Rps26 directly interact on the platform when bound to ribosomes (Rabl, 2011). Furthermore, interactions between the C-terminal tail of Rps14 and rRNA are not made until the very last stages of 40S maturation (Jakovljevic et al., 2004). Thus, the data herein indicate that final incorporation of Rps14 and Rps26 occurs after Fap7 acts, while Rps10 is incorporated prior, possibly in response to dissociation of Ltv1.
Asc1 is the yeast homolog of RACK1, a receptor for protein kinase C that mediates translation regulation, and is stoichiometrically bound to 40S subunits. Interestingly, Asc1/RACK1 is also fully bound to 40S subunits in these 80S-like ribosomes but is not present in steady-state pre-40S subunits purified from yeast (Strunk et al., 2011). Thus, its incorporation into maturing 40S subunits occurs prior to steps dependent on Fap7.
Collectively, these data indicate that the 40S subunits within the 80S-like ribosomes are immature as they contain the 18S-precursor 20S rRNA and lack the essential ribosomal protein Rps26. More importantly, however, the data suggest that the 80S-like ribosomes that accumulate in the absence of Fap7 do not represent translation initiation intermediates, as they do not contain initiator tRNA. This conclusion is further supported by the observation that the pre-40S subunits are found in 80S-like ribosomes and not in polysomes. This finding suggests that the 80S-like ribosomes do not contain mRNA, as it is unlikely that pre-40S subunits should only bind mRNAs not already bound to at least one mature 80S ribosome. Binding of pre-40S subunits to an mRNA would thus shift 80S complexes into the polysomes, as observed for Pno1 in wild type cells (Figure 1C). We do note, however, that it is possible that all 80S-like ribosomes are associated with a special class of mRNAs not found in polysomes (e.g., due to size). We do not think this is likely, as only a few such mRNAs are known, and these are not abundant enough to explain the large accumulation of 80S ribosomes (Arava et al., 2003; Wang et al., 2002). Nevertheless, this possibility cannot be formally excluded. The conclusion that 80S-like ribosomes that accumulate in the absence of functional Fap7 do not contain mRNA and are thus not translation initiation intermediates is also consistent with the previous observation that eIF1A, an initiation factor required for opening of the mRNA channel (Passmore et al., 2007), is not required for 40S maturation (Soudet et al., 2010). Furthermore, Tsr1 and Rio2 on pre-40S assembly intermediates blocks access to the mRNA channel (Strunk et al., 2011). Collectively, the data show that the 80S-like ribosomes that accumulate in the absence of Fap7 are not intermediates in the translation initiation pathway. Finally, in wild type cells, strong polysome association is observed for Pno1 (Figure 1C), indicating that Pno1 does not dissociate until mRNA is bound to pre-40S subunits.
eIF5B Promotes Joining of 60S Subunits During 40S Maturation
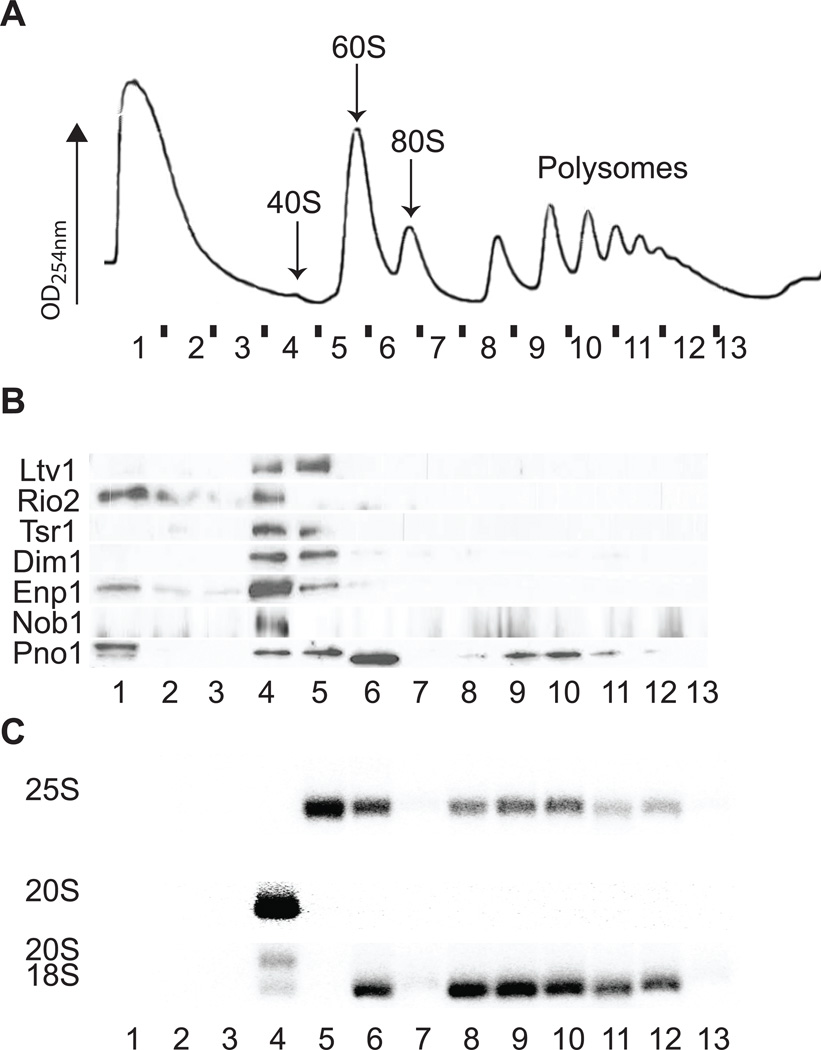
The data indicate that 60S subunits join pre-40S subunits during their maturation to give 80S-like complexes that are not translation initiation intermediates. This model, while surprising, rationalizes the previously unexplained finding of a genome-wide survey showing that eIF5B, the GTPase that promotes 60S subunit joining during translation initiation, is involved in 40S maturation [(Li et al., 2009) and data not shown]. Together, these data suggest that the role of the GTPase eIF5B in 40S maturation might be to promote the formation of 80S-like ribosomes that are the substrate for Fap7. This model predicts that in a strain deleted or depleted for both eIF5B and Fap7, pre-40S ribosomes should no longer accumulate in 80S-like complexes, but rather as free 40S subunits, as formation of 80S-like complexes is slowed in the absence of eIF5B. Indeed, sucrose gradient centrifugation analysis demonstrates that co-depleting eIF5B and Fap7 shifts the AFs bound to pre-40S subunits from an 80S peak (observed in the absence of Fap7 alone) into a 40S peak (Figure 3), where all AFs, as well as 20S rRNA, accumulate. This result strongly supports the model that during 40S maturation, 80S-like ribosomes are formed from 60S subunits and pre-40S subunits in an eIF5B-dependent manner. The sucrose gradient profile of the cells depleted for eIF5B and Fap7 together is also highly similar to that manifest following the deletion of eIF5B alone (Figure S2B&C). Interestingly, in the absence of eIF5B alone, the sedimentation profile for Rio2 is subtly shifted to the 40S ribosome peak, whereas the assembly factor Ltv1 is subtly shifted towards the free fraction. This suggests that Ltv1 dissociates before eIF5B-mediated subunit joining (such that more is free without eIF5B), while Rio2 remains bound until after subunit joining (leading to a stronger peak in 40S subunits without eIF5B). Confirming this finding will require us to identify the factor(s) leading to Rio2 dissociation in the future.
Figure 3. eIF5B Is Required for the Accumulation of 40S Assembly Factors in 80S-like Ribosomes.
10–50% sucrose gradients from cell lysates from ΔeIF5B,Gal1::Fap7 cells grown in glucose for 24h. Longer times for Fap7 depletion were required because due to the absence of eIF5B the strain grows slowly even in galactose.
(A) Absorbance profile at 254 nm.
(B) Western blots for assembly factors.
(C) Northern blots for rRNAs and precursors.
See Figure S2 for related data.
Rli1•Dom34 Disrupt 80S-like Ribosomes After Assembly Factors Have Dissociated
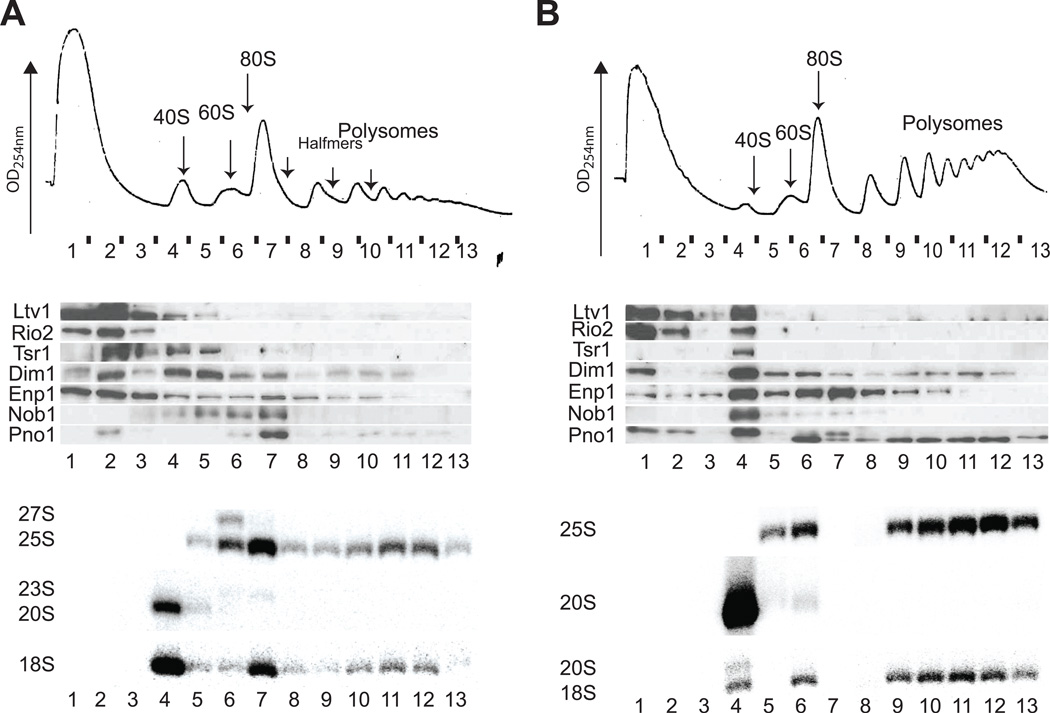
The model where formation of 80S-like ribosomes is necessary for the maturation of 40S subunits also implies that these 80S-like ribosomes have to be broken apart again to allow for translation initiation on mature 40S subunits, as mRNA cannot bind to preformed 80S complexes. After completion of translation, mature 80S ribosomes are dissociated by the ABC-type ATPase Rli1, aided by Dom34 (Khoshnevis et al., 2010; Pisareva et al., 2011; Shoemaker and Green, 2011). Intriguingly, Rli1 has been implicated in 40S assembly, as it co-purifies 20S pre-rRNA (Soudet et al., 2010; Yarunin et al., 2005). Further, depletion of Rli1 leads to reduced levels of 18S and 25S rRNA and to the accumulation of early 23S, 27S and 35S pre-rRNAs (Yarunin et al., 2005 and data not shown), and deletion of Dom34 is synthetically lethal with deletion of many Rps (Bhattacharya et al., 2010). Together, these data suggest that during 40S maturation, Rli1•Dom34 could dissociate 80S-like complexes to prepare maturing 40S ribosomes for translation initiation. To test if Rli1•Dom34 break apart 80S-like ribosomes that contain pre-40S subunits and mature 60S subunits during maturation, sucrose gradient analysis was performed on yeast cells depleted of Rli1 or deleted for Dom34, a non-essential protein. Depletion of Rli1 (Figure 4A) or deletion of Dom34 (Figure 4B) leads to accumulation of Nob1, Pno1, Enp1 and Dim1 in 80S-like ribosomes, demonstrating that Rli1•Dom34 complete the translation-like cycle during 40S maturation by separating 80S-like ribosomes into mature 60S subunits and pre-40S subunits.
Figure 4. Rli1•Dom34 Are Required for Dissociation of 80S-like Ribosomes.
(A) 10–50% sucrose gradients from cell lysates from Gal1::Rli1 cells grown in glucose for 12h. (B) 10–50% sucrose gradients from cell lysates from cells lacking Dom34. Shown are absorbance profile at 254 nm (top), Western blots for assembly factors (middle) and Northern blots for rRNAs and precursors (bottom). Probe b was used to probe for 20S, 23S and 35S rRNA. The 25S probe was used to detect 25S and 27S rRNAs. See Figures S3 and S4 for related data.
Dim1 and Enp1 bind to early 90S-like nucleolar pre-40S assembly intermediates. Furthermore, 23S and 35S rRNAs can be found following Rli1 depletion (Figure 4A & data not shown). Thus, it is possible that the sedimentation of these two proteins reflects binding to nucleolar 90S assembly intermediates. However, two lines of evidence suggest otherwise. First, the same sedimentation pattern is observed in cells deleted for Dom34, where no early assembly intermediates accumulate (Figure 4), suggesting that the sedimentation is not explained by the accumulation of early nucleolar pre-40S assembly intermediates. Moreover, the levels of Enp1 in 40S and 80S-sized fractions are equal, while the amount of 23S is >10-fold lower than that of 20S rRNA (Figure 4A&B), indicating that most of the Enp1 is in 80S-like ribosomes and not only in 90S pre-ribosomes. While most of the Dim1 protein is found in 40S subunits after Rli1 depletion, the amount of Dim1 in 80S-like ribosomes is substantial relative to the amount of 23S rRNA present. Furthermore, both Dim1 and Enp1 can be found in the 80S and even the 2-mer ribosome fraction in wild type cells (Figure 1C, Figure S2A), where no 23S or 35S rRNAs are detected. Collectively, these findings are consistent with the notion that Enp1 and Dim1 remain bound to pre-40S ribosomes until after Rli1-directed dissociation of 80S-like ribosomes.
Interestingly, Northern blotting demonstrates that the 80S-like ribosomes that accumulate following Rli1 depletion lack 20S pre-rRNA, but contain mature 18S rRNA (Figure 4A). Similarly, deletion of Dom34 leads to accumulation of most but not all 20S pre-rRNA in 40S-like ribosomes (Figure 4B). This suggests that 20S rRNA is cleaved to produce mature 18S rRNA within 80S-like ribosomes. This is consistent with the observation that 20S rRNA does not accumulate in the absence of Rli1 (Yarunin et al., 2005), and that Rli1 pulls down 20S and 18S rRNAs (Soudet et al., 2010; Yarunin et al., 2005). To further test if Nob1 can cleave 20S pre-rRNA in 80S-like ribosomes and to provide additional evidence for the involvement of Rli1 in separating 80S-like ribosomes prior to translation initiation, we investigated the effect from depletion of Nob1 either alone, or in combination with Rli1 (Figure 5). Nob1 is the endonuclease that is required for cleavage of 20S rRNA to produce mature 18S rRNA (Lamanna and Karbstein, 2009, 2011; Pertschy et al., 2009). Western and Northern analyses demonstrate that upon depletion of Nob1 40S pre-ribosomes containing 20S rRNA and AFs are found in the 40S peak (Figure 5A). Furthermore, and as previously observed (Soudet et al., 2010), a large fraction of 20S rRNA is shifted into the polysomes, indicating that these late 40S pre-ribosomes are competent for translation initiation. In contrast, depletion of Nob1 and Rli1 together leads to accumulation of Enp1 and Pno1 in 80S-like fractions (Figure 5B), consistent with a role for Rli1 in breaking apart 80S-like ribosomes. Moreover, Northern analysis demonstrates that a fraction of the small subunit RNA in the 80S peak is 20S pre-rRNA, as expected if Nob1 can cleave 20S pre-rRNA in 80S-like ribosomes. Most importantly, Northern blotting shows that in the absence of Nob1 and Rli1, 20S rRNA does not shift into the polysomes as observed when Nob1 alone is depleted, establishing that Rli1 is required for release of pre-40S ribosomes into the translating pool (Figure 5B).
Figure 5. Rli1 Is Required for Release of pre-40S Subunits into the Translating Pool.
(A) 10–50% sucrose gradients from cell lysates from Gal1::Nob1 cells grown in glucose for 12h. (B) 10–50% sucrose gradients from cell lysates from Gal1::Nob1,Gal1::Rli1 cells grown in glucose for 12h. Shown are absorbance profile at 254 nm (top), Western blots for assembly factors (middle) and Northern blots for rRNAs and precursors (bottom). Probe b was used to probe for 20S, 23S and 35S rRNA. The 25S probe was used to detect 25S and 27S rRNAs. See Figure S5 for biological replicates.
The overall reduction of both the small and large ribosomal subunits observed in the Rli1-depleted strains [(Yarunin et al., 2005) and Figure S3A] is best explained by degradation of stalled 40S ribosomes (Cole et al., 2009; Soudet et al., 2010), and is consistent with the observation that most assembly factors are substantially depleted in the absence of Rli1 (Figure S3B). This finding also explains why not all 20S rRNA is in 80S ribosomes in the Rli1/Nob1 depleted strain. The pre-40S subunits in 80S-like ribosomes are apparently more susceptible to degradation than pre-40S subunits alone.
DISCUSSION
40S Maturation Requires a Translation-Like Cycle
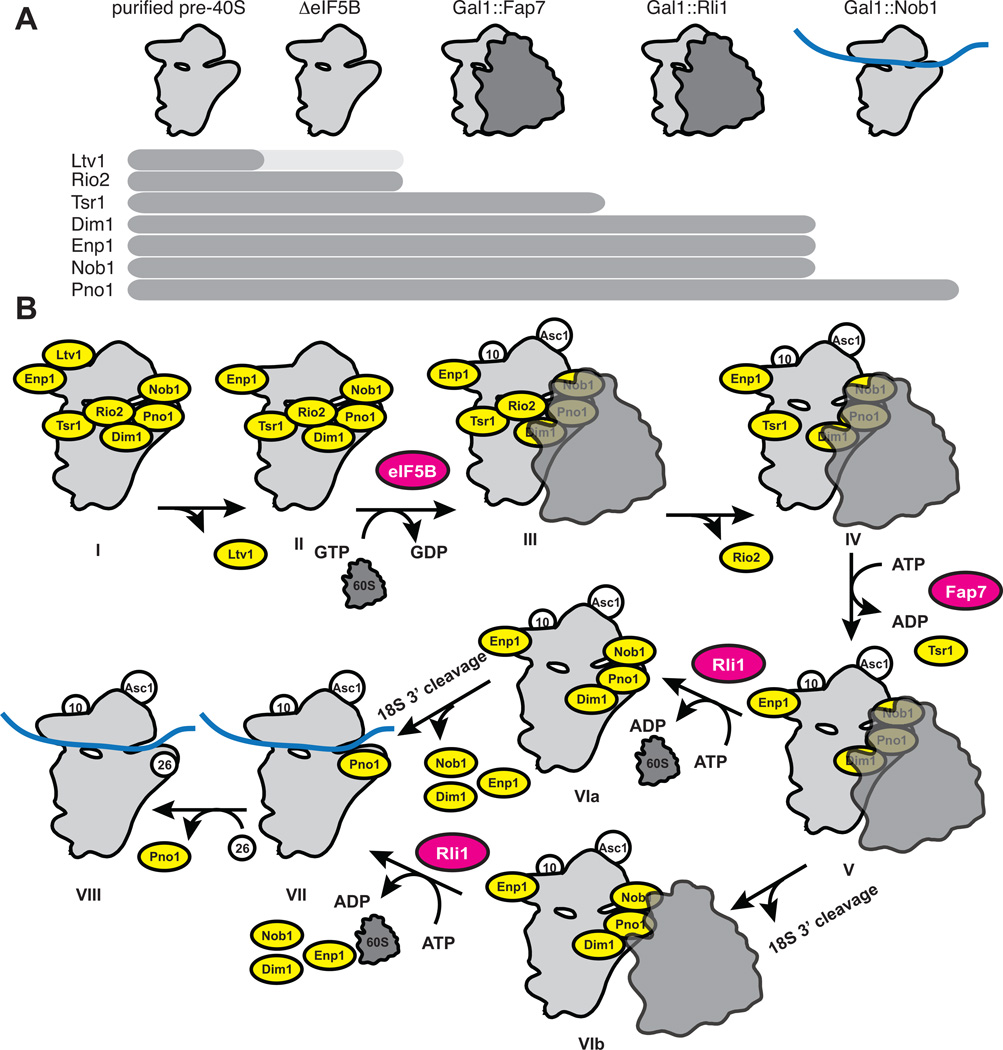
Here we have shown that the final maturation steps of pre-40S ribosomes include a “translation-like” cycle in which 60S subunits join pre-40S subunits in a reaction promoted by the translation initiation GTPase eIF5B, before the resulting 80S-like ribosomes are broken apart by the termination factor Rli1, allowing for mRNA recruitment and translation initiation. This model is based on the data presented in Figures 1–5, which are summarized in Figure 6A. The details of the model shown in Figure 6B will be discussed below.
Figure 6. Model for Late Cytoplasmic Events in 40S Assembly.
(A) Summary of the data presented in Figures 1–5 and Figures S1–S5. On the top are cartoon images to show whether pre-40S subunits are free, or bound to 60S subunits, or mRNA in each accumulated case. The complement of assembly factors used to “stage” the assembly intermediates are below.
(B) Detailed model of cytoplasmic events during small ribosomal subunit maturation. Assembly factors that stably bind to pre-40S ribosomes are shown in yellow, energy-consuming assembly factors that bind transiently to pre-40S ribosomes are shown in magenta, pre-40S ribosomes and 60S ribosomes are shown in light and dark grey, respectively, ribosomal proteins are shown in white and mRNA is shown in blue. Intermediates defined herein are labeled I through VIII. It is likely that additional intermediates are present, yet these remain unknown.
In wild type yeast, intermediate I accumulates in the steady state and can be purified (Strunk et al., 2011). The rate-limiting step in cytoplasmic 40S ribosome maturation must therefore be the conversion to intermediate II. It is possible that an additional intermediate (or several intermediates) precedes intermediate II. However, regardless of how many steps occur between intermediates I and II, the step away from I is rate-limiting. For simplicity, and because thus far there is no evidence for an additional step, we assume that intermediate II directly follows intermediate I. Intermediate II accumulates in the absence of eIF5B, a GTPase that promotes the formation of 80S ribosomes. In the absence of eIF5B, the sedimentation of Rio2 is slightly shifted toward the 40S fraction while more Ltv1 is in the free fraction. This suggests that Ltv1, but likely not Rio2, dissociates before eIF5B-mediated joining of 60S subunits. However, further experiments will be required to confirm this notion. Independent confirmation for a step in 40S maturation that precedes the joining of 60S subunits comes from an analysis of the rate constants observed for individual steps. Cytoplasmic 40S maturation occurs with a rate-constant of about 0.3 min−1 (Granneman et al., 2005; Kos and Tollervey, 2010), which represents a working estimate for the rate-limiting step between I and II. In contrast, eIF5B-mediated 60S subunit joining in vitro is at least 10-fold faster (Acker et al., 2009), suggesting 60S subunit joining is not the rate-limiting step in 40S maturation. After eIF5B promotes 60S subunit joining to form intermediate III, Rio2 must somehow dissociate from the resulting 80S-like ribosomes to give intermediate IV, which is the substrate for the ATPase Fap7, and has been observed here directly. While intermediate I lacks Rps10 and the ribosome associated protein Asc1 (Strunk et al., 2011), both of these are fully bound in intermediate IV. The binding site for Rps10 overlaps the binding site for the Enp1/Ltv1 complex (Strunk et al., 2011), explaining its absence in intermediate I. In contrast, intermediate IV lacks Ltv1 but contains Rps10, consistent with an exchange between these two proteins; it is not known how this exchange is mediated.
Tsr1 is present in 80S-like ribosomes that accumulate in the absence of functional Fap7 (intermediate IV), but not in 80S-like ribosome that accumulate in the absence of Rli1 (intermediate VIb). It is thus tempting to speculate that Fap7 ATPase activity directs displacement of Tsr1 from pre-40S ribosomes. However, Fap7 and Tsr1 do not directly interact (data not shown), which is consistent with the location of the Tsr1 binding site on the left of helix 44 [viewed from the 60S subunit, (Strunk et al., 2011)] and the position of Rps14, the only known binding partner for Fap7 (Granneman et al., 2005), on the platform. Furthermore, archaeal genomes encode a homolog for Fap7, but not for Tsr1 (data not shown), making it very unlikely that their function is directly linked.
Intermediate VIb accumulates in the absence of the ABC-type ATPase Rli1, or its partner Dom34. Rli1•Dom34 break apart 80S ribosomes into 40S and 60S subunits after translation termination (Khoshnevis et al., 2010; Pisareva et al., 2011; Shoemaker and Green, 2011), and we conclude that it also carries out an identical function in disrupting 80S-like ribosomes during maturation to release the resulting pre-40S ribosomes into the translating pool. Consistent with this notion, in the absence of Nob1, the nuclease for 18S rRNA production, 20S rRNA can enter the translating pool and is found in polysomes, while in the absence of Rli1 and Nob1, 20S rRNA is barred from entering the polysomes and stalled in 80S-like ribosomes. Further, the data suggest that the Nob1 and Rli1-dependent steps can occur in either order. In one pathway, the 40S assembly intermediate that accumulates in the absence of Rli1 or Dom34 (intermediate VIb) contains 18S rRNA, suggesting that Nob1-dependent 18S rRNA production can occur within 80S-like ribosomes. In the other pathway, the 40S assembly intermediate that accumulates in the absence of Nob1 (intermediate VIa) contains 20S rRNA, but sediments as a 40S particle. This late 40S assembly intermediate can initiate translation, thereby explaining why Nob1 and 20S pre-rRNA can enter polysomes as part of translation initiation complexes [(Soudet et al., 2010) and Figure 5A]. Finally, Rli1 immunoprecipitates both 20S and 18S rRNAs (Soudet et al., 2010; Yarunin et al., 2005), further suggesting that 20S cleavage to produce 18S rRNA can occur before or after Rli1-dependent dissociation of 80S-like ribosomes.
Finally, following Rli1-dependent dissociation of 80S-like ribosomes and Nob1-dependent cleavage of 20S rRNA, Nob1, Enp1 and Dim1 (if present) dissociate via an unknown mechanism. Pno1 is the last factor to dissociate and is found deep in the polysome fractions in wild type yeast. Intriguingly, the Pno1 binding site overlaps the Rps26 binding site (Strunk et al., 2011), suggesting that the Pno1-bound pre-40S ribosomes lack Rps26, as shown for intermediates I and IV. Therefore, the last step in 40S maturation appears to be the incorporation of Rps26 (intermediates VII to VIII).
In summary, these data reveal intimate connections between the 40S maturation and translation initiation pathways whereby a translation-like cycle occurs during 40S maturation. This new framework will be instrumental in understanding how 40S ribosomes are matured in the cytoplasm and how the dissociation of assembly factors is achieved. Interestingly, the majority of assembly factors described herein are conserved from archaea to humans, including eIF5B, Rli1 and Fap7, suggesting that this translation-like mechanism in small subunit maturation is similarly conserved.
80S-like Ribosomes Formed During 40S Maturation Are not Translation Initiation Intermediates
While our data demonstrate that 80S-like ribosomes are formed during pre-40S maturation, they indicate that these 80S ribosomes are not translation initiation intermediates. Specifically, Northern blotting shows that they are not bound to initiator tRNA, and mass spectrometry indicates that eIF2 is similarly lacking. Furthermore, pre-40S ribosomes accumulate as 80S-like ribosomes and not in the polysome fraction, indicating that they also are not bound to mRNA. This is consistent with findings that binding of translation initiation factors and mRNA to pre-40S subunits is blocked by AFs (Strunk et al., 2011), and with the observations that eIF1A and eIF3, key factors during translation initiation, are not required for 40S maturation (Soudet et al., 2010). In contrast, the data demonstrate that eIF5B is required for 60S joining during 40S maturation as it is during translation initiation, and they also establish dual roles for Rli1, which functions to dissociate 80S and 80S-like ribosomes into 40S and 60S subunits during canonical translation termination and 40S maturation, respectively. Interestingly, the requirement for a small subset of translation initiation factors for this translation-like step in 40S maturation is reminiscent of viral or IRES-mediated translation initiation, which often requires only a subset of canonical translation factors (Filbin and Kieft, 2009).
The Translation-like Cycle as a Licensing Step in 40S Maturation
The extraordinary fidelity of the ribosomal decoding process requires long-range communication within the ribosome, and thereby relies on correctly assembled ribosomes. In fact, it is known that even minor deficiencies, such as the lack of even a single RNA modification, have measureable effects on translational fidelity (Baudin-Baillieu et al., 2009; Liang et al., 2007). It thus appears sensible that assembling ribosomes be inspected before they are released into the translating pool.
When pre-40S ribosomes arrive in the cytoplasm, the presence of assembly factors on pre-40S subunits blocks premature translation initiation (Strunk et al., 2011). Later 40S assembly intermediates are competent for translation initiation [(Soudet et al., 2010) and Figure 5A). The data herein demonstrate a complex series of events between these two stages, in which 60S subunits join pre-40S ribosomes in an eIF5B-dependent manner before the resulting 80S-like subunits are dissociated by the ABC-type ATPase Rli1. This translation-like cycle precedes the canonical translation initiation pathway, which involves mRNA binding and the renewed joining of 60S subunits. We hypothesize that this translation-like cycle is a final licensing step in ribosome assembly, before the block towards translation initiation is released and 40S subunits enter the translating pool. In this cycle, critical activities of the maturing 40S subunits are tested: the ability to bind to 60S subunits, the ability to bind to and activate the GTPase eIF5B and the ATPase Rli1, and the ability to bind Dom34, a tRNA mimick, which binds the A-site (Becker et al., 2012). For all three factors, eIF5B, Dom34 and Rli1, the ribosome binding site includes the 40S subunit (Becker et al., 2012; Shin et al., 2011). Interestingly, a similar licensing step has been documented during maturation of tRNAs (Lund and Dahlberg, 1998).
Tsr1 and Rli1 both bind to the GTPase site (Becker et al., 2012). Thus, Tsr1 is expected to block access of Rli1 to this site. Therefore, the dissociation of Tsr1 from the subunit interface marks pre-40S containing 80S complexes to be ready for Rli1-mediated disassembly and subsequent translation initiation. When Rli1 cannot act on pre-40S subunits (because it is depleted as herein, or because subunits are not functional), the resulting 80S-like subunits are rapidly degraded, as expected from a quality control mechanism. Thus, release of the GTPase like Tsr1 activates a molecular timer: If Rli1•Dom34 cannot act in time, the resulting pre-40S subunits will be degraded. Tsr1-mediated block of the GTPase site in the absence of Fap7 also explains why 20S rRNA is so extraordinarily stable in this strain: the degradation enzymes Hbr1/Dom34 also bind to this site to initiate degradation of stalled ribosomes (Becker et al., 2011; Cole et al., 2009), and their access is thus similarly blocked by the presence of Tsr1 in the 80S ribosomes stalled in the absence of Fap7.
We speculate that Rli1 and Fap7 could also be the regulated under certain stresses: Perhaps inactivation of Fap7 under short term stress could preserve 40S assembly intermediates to later resume their assembly, while regulation of Rli1 would efficiently block 40S maturation, and lead to the degradation of stalled assembly intermediates. Further experiments will be required to test this idea.
This translation-like cycle also allows for the dissociation of AFs without leading to premature translation initiation. Tsr1 is the largest AF bound to the subunit interface of the pre-40S subunits (Strunk et al., 2011). Its presence blocks binding of eIF1A, mRNA and impedes the spontaneous association of 60S subunits (Strunk et al., 2011). Rio2 also binds to the subunit interface (Strunk et al., 2011) and is predicted to block the binding of initiator tRNA (Shin et al., 2011) and mRNA. These two assembly factors are therefore key to prevent premature translation initiation. The data here suggest that both proteins dissociate from 80S-like ribosomes. Because mRNAs or initiator tRNA cannot bind to or dissociate from 80S ribosomes, the formation of 80S-like ribosomes during 40S maturation allows dissociation of these assembly factors without leading to premature translation initiation. Thus, 60S subunits are revealed to function as chaperones of pre-40S subunits that prevent translation initiation, akin to assembly factors.
Finally, the requirement for 60S subunits in the maturation of 40S subunits links the production of the two ribosomal subunits. Indeed, it is tempting to speculate that 60S maturation similarly requires joining to 40S subunits, a notion supported by the observation that depletion of Rli1 affects the accumulation of 60S subunits, as previously reported (Kispal et al., 2005) and indicated here by the halfmers observed in the Rli1 depletion strain.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
Saccharomyces cerevisiae strains used herein are listed in Table S1 and were obtained from Open Biosystems or created by standard recombination techniques. Strain identity was verified by PCR and Western blotting.
Antibodies
Antibodies against recombinant 40S assembly factors were raised in rabbits by Josman LLP. Anti-TAP antibodies (Open Biosystems) were used in the case of Rio2, where appropriate. All primary antibodies were used at a 1:1000 dilution, HRP-conjugated anti-rabbit secondary antibody (Rockland Immunochemicals) was used at a 1:10000 dilution.
Sucrose Density Gradient Analysis
0.1 mg/ml cycloheximide was added to cells in mid-log phase prior to harvesting by centrifugation. Cells were mechanically lysed in 1.5 ml/g dry weight gradient buffer (20 mM Hepes, pH 7.4, 5 mM MgCl2, 100 mM NaCl, 0.1 mg/ml cycloheximide, 3 mM DTT, 1 mM PMSF, 1 mM benzamidine, Complete EDTA-free Protease Inhibitor Cocktail (Roche), 1 ng/µl pepstatin, 1 ng/µl leupeptin, 1 ng/µl aprotinin, and 1 U/µl RNAsin (Promega). 50 µl of cleared lysate were applied to 11 ml 10–50% sucrose gradients and centrifuged for 2 hours at 40,000 RPM in an SW41 rotor. Gradients were fractionated with a fraction collector (Brandel Inc.) and absorbance at OD 254 was monitored continuously. For Northern analysis, RNA was extracted from 100 µL of fractions after 1:1 dilution with water, ethanol precipitated and resuspended in 10 µL of water. rRNAs were analyzed on an agarose/formaldehyde gel run overnight and tRNAs were analyzed using an 8% TBE/8M urea polyacrylamide gel, run for 1h at 25W prior to transfer onto a Hybond-N membrane.
In vivo Formaldehyde Crosslinking and Purification of Fap7-TAP-Associated Complexes
Either 2 l cells expressing TAP-tagged Fap7 from a galactose promoter were grown to mid-log phase in YP media supplemented with 0.2% galactose and 2% raffinose, or 10 l of cells expressing Fap7TAP from its native promoter were grown to mid-log phase in YP media supplemented with 2% dextrose. In vivo cross-linking was performed according to (Valasek et al., 2007). Crosslinked cells were lysed and affinity-purification of TAP-tagged Fap7 was performed essentially as described (Strunk et al., 2011).
Mass Spectrometry
For protein identification, samples were subjected to proteolytic digestion with trypsin and proteins were analyzed using LC-MS/MS by the Proteomics Core at Scripps Florida. Product ion data were searched against the SwissProt database using the Mascot search engine. Mascot output files were parsed into Scaffold for filtering. Quantitative analysis was carried out by analysis of the area under the LC peaks.
Supplementary Material
HIGHLIGHTS.
60S ribosomes join maturing pre-40S subunits in a translation-like cycle
This cycle is conserved, eIF5B-dependent and provides a fidelity checkpoint
The resulting 80S-like ribosomes are not translation initiation intermediates
Rli1•Dom34 disassemble 80S-like ribosomes for translation initiation
ACKNOWLEDGEMENTS
We thank A. Hinnebusch and T. Dever for providing antibodies and plasmids, and J. Hershey, C. Fraser, K. Nettles and J. Cleveland for critical review of the manuscript. We also thank M. Chalmer and C. Steckler for carrying out mass spectrometry experiments. B.S.S. was partially supported by an NSF predoctoral fellowship. M.N.N. was supported by a SURF fellowship from the Kellogg School of Science and Technology. This work is funded by NIH grant R01-GM086451.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acker MG, Shin BS, Nanda JS, Saini AK, Dever TE, Lorsch JR. Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol. 2009;385:491–506. doi: 10.1016/j.jmb.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 2009;37:7665–7677. doi: 10.1093/nar/gkp816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol Cell Biol. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Nandineni MR, Baserga SJ. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol Cell Biol. 2005;25:10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM, Leger-Silvestre I, Gas N, Woolford JL., Jr The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol Cell. 2004;14:331–342. doi: 10.1016/s1097-2765(04)00215-1. [DOI] [PubMed] [Google Scholar]
- Karbstein K. Inside the 40S ribosome assembly machinery. Curr Opin Chem Biol. 2011;15:657–663. doi: 10.1016/j.cbpa.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janaky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. Embo J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna AC, Karbstein K. Nob1 binds the single-stranded cleavage site D at the 3'-end of 18S rRNA with its PIN domain. Proc Natl Acad Sci U S A. 2009;106:14259–14264. doi: 10.1073/pnas.0905403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna AC, Karbstein K. A Conformational Change Regulates Pre-18S Cleavage. J Mol Biol. 2011;405:3–17. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lee I, Moradi E, Hung NJ, Johnson AW, Marcotte EM. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000213. e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnadig M, Schafer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284:35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. Embo J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide S, Haag A, Ban N. Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Walker SE, Dong J, Lorsch JR, Dever TE. Initiation factor eIF2gamma promotes eIF2-GTP-Met-tRNA(i)(Met) ternary complex binding to the 40S ribosome. Nat Struct Mol Biol. 2011;18:1227–1234. doi: 10.1038/nsmb.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. Embo J. 2010;29:80–92. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Szamecz B, Hinnebusch AG, Nielsen KH. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007;429:163–183. doi: 10.1016/S0076-6879(07)29008-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci U S A. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. Embo J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.