Abstract
Model-based drug development (MBDD) is accepted as a vital approach in understanding patients' drug-related benefit and risk by integrating quantitative information integration from diverse sources collected throughout drug development.1 This perspective introduces the activities of the Drug and Disease Model Resources (DDMoRe) consortium, founded in 2011 through the Innovative Medicines Initiative Joint Undertaking (IMI-JU)2 as a European public–private partnership to address a lack of common tools, languages, and standards for modeling and simulation (M&S) to improve model-based knowledge integration.
Perspective
At key stages in the drug-development program, decision makers rely on the best rationale to select targets, drug candidates, trial designs, dose regimens, patient populations, and suitable end point measures. A lack of informed decision making can have detrimental effects to patients, cause attrition of potentially successful development programs, or induce substantial financial losses.3
M&S is seen as the core technology in MBDD to describe and predict the behavior of complex diseases, biological systems, and drug actions; essential to knowledge integration; and through inference providing the quantitative basis for well-informed decision making. But the current path to model-based knowledge integration is hampered by a lack of common tools, languages, and standards for M&S, with limited or time-consuming access to stored information, thereby creating unnecessary hurdles to exploit knowledge.
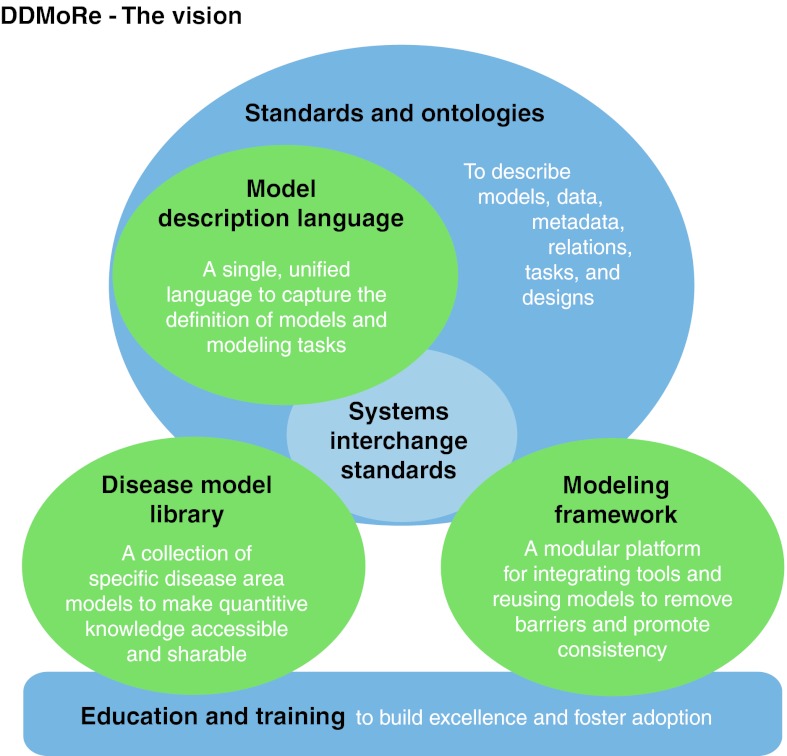
To facilitate knowledge integration for decision making, DDMoRe, a consortium of European Federation of Pharmaceutical Industries and Associations (EFPIA), Academic and Small Medium Enterprise (SME) partners, is developing a public drug and disease model library supported by an open source interoperability framework which will provide access to existing and future modeling tools. This may be ambitious; however, DDMoRe envisages that the standards and tools developed over the course of the project will become the reference for future collaborative work in drug and disease M&S, serving internal and external stakeholders, regulators, academics, and the pharmaceutical industry in addressing the current bottlenecks in the drug-development process. (Figure 1)
Figure 1.
Drug and disease model resources: main deliverables of the project.
Establishing a Drug and Disease Model Library
A drug and disease model library is being created to store and promote the reusability of models. DDMoRe has identified a number of high interest disease areas to exemplify the implementation of its language standards within the open source interoperability framework. A collection of diabetes and oncology models, both existing and requiring development, were selected for their value in drug development. Thirty published pharmacokinetic, pharmacodynamic, pharmacokinetic–pharmacodynamic, physiologically based pharmacokinetic, disease and systems pharmacology/biology models covering differing phases of the drug-development process will initially be stored in the library and new models in key therapeutic areas added over time. In each therapeutic area, relevant models were identified from the literature, but gaps were also highlighted in model coverage which will direct new model development over the course of the DDMoRe project.
Drafting a New Model Description Language
The model coding language (MCL) has been conceived to provide a single, unified language that can capture the definition of models together with their parameters and data attributes, irrespective of the target software for performing the M&S task.
The MCL will bring together features of various languages to code models and is intended to be flexible, extensible and easy to code, understand and use. This will reduce rework and recoding in passing models between software platforms thereby helping to promote consistency and reusability of models and propagate knowledge throughout the drug-development process.
Forming a model description language, the MCL is combined with a complementary task execution language (TEL), which describes how the input from the MCL will be used to generate a desired output. The TEL will allow connecting the users' modeling aims, the immediate task of execution of a modeling-related task, and the overarching analysis workflow within the interoperability framework.
Specifications for both aspects of the model description language (MCL and TEL) have been drafted, reviewed, and will be made public shortly, but a depiction of the key components constituting the model description language is given in Figure 2.
Figure 2.
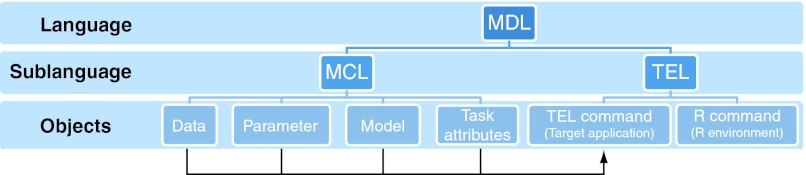
Modeling description language (MDL). Although the MCL will allow the user to define new models, the TEL will provide the functionality to update the objects and execution settings and handles the modeling tasks. MCL, model coding language; TEL, task execution language.
Developing an Open Source Interoperability Framework
For the DDMoRe project to be successful, a platform must be developed that enables the integration of existing software, tools, and desired modeling workflows, as well as improving the integration efficiency for new methodologies.
There will be mechanisms to translate the new modeling language into the specific languages of different softwares such as NONMEM, Monolix, R, and WinBUGS. Once basic commands are supported, an engine will be incorporated that orchestrates the execution of complex modeling workflows, involving simulation of trial designs, parameter estimation, derivation of inferences, and analysis reporting. The framework will be extended to allow interactions with the modeling Ontology KnowledgeBase server that will provide assistance with the annotation of models to facilitate sharing and reuse.
The infrastructure of the framework will also comprise a distributed tool execution framework, connectors within this framework, and rule sets that define constraints on how the tools will be executed (exemplified illustration of process flow given in Supplementary Figure S1 online).
Creating a Modeling Framework Prototype
To demonstrate and facilitate the development of the languages, examples were defined addressing various aspects of the modeling workflow. Although simpler models and modeling tasks will be illustrated using warfarin pharmacokinetic–pharmacodynamic models, more complex models involving integrated physiological systems, disease progression, categorical and time-to-event data, and multilevel random effects models will be illustrated in the oncology, diabetes, and Alzheimer's disease areas. All examples will exercise the main integration points of the framework, namely running an analysis workflow that will allow execution and manipulation of modeling tasks. The prototypes will also be used to address other relevant tasks, such as meta-analyses, simulation, and optimal design problems.
An initial prototype has been developed that provides a client for modeling with MCL objects, and a server for the execution of corresponding workflow tasks.
Creating Standards in Markup Languages, Metadata Frameworks, and Ontologies
To facilitate interoperability of data, models, and related metadata in storage and system-to-system interchange, there is a need for a common set of comprehensive standards. The needs of MBDD were explored in terms of prevailing standards, with respect to markup languages (ML) that allow annotation of documents, controlled vocabularies (ontologies), and model components. A relevant set of MLs was identified (SBML, SED-ML, NuML, CellML, and UncerML) and evaluated to determine whether they could form part of a set for DDMoRe. Beyond providing the modeling community with a complete set of interoperable MLs, with the first schema to be made publically available in March 2013, the eventual goal is to create a coherent ontology-based solution in support of classification and reasoning over data and variable types in databases and model libraries.
To achieve external acceptance and compatibility of the developed ontology and meta standards, collaborations have been established with the Clinical Data Interchange Standards Consortium, the Ricordo4 project to scope the integration of semantic relationships within the framework, the Pistoia Alliance to achieve endorsement, and the IMI consortia Open PHACTS (http://www.openphacts.org) and EHR4CR (http://www.ehr4cr.eu) to ensure compatibility of standards.
Integrating Existing Modeling Software into the Open Source Interoperability Framework
DDMoRe will integrate some existing softwares such as NONMEM, Monolix, and WinBUGS, and various R and Matlab-based tools into the interoperability framework. It will also provide means to integrate other existing softwares by adopting standards for the design, analysis, and interpretation phases of the M&S workflow and embedding the application programming interface and modeling language developed.
Developing New Modeling Approaches and Software
The participation of numerous partners provides DDMoRe with the unique opportunity to integrate existing modeling tools and address gaps in the current array of modeling software tools and related methodological aspects to facilitate MBDD.
Clinical trial simulation is at the center of quantitative decision making to derive an optimal drug development path for future trials or optimal portfolio management in the pharmaceutical industry. DDMoRe is developing a clinical trial simulation engine to address most realistic trial scenarios, for which a prototype has already been released, and has explored demand among industry partners for adaptive optimal design methodologies to increase trial informativeness. New methodologies for parameter estimation in complex models, new strategies for covariate and random effects modeling, and new diagnostics for time-to-event data and random effects components have already been developed and are among the 16 DDMoRe scientific contributions to the 2012 PAGE conference.5
Training and Education
An initiative such as DDMoRe can only show its potential if it is accompanied early by a comprehensive training program to influence the drug-development environment in a persistent way. Training will cover methodological and technological aspects of the project, and model-based training exercises will cover a number of drug-development scenarios across a variety of therapeutic areas. To this end, a survey on the technical and conceptual requirements in drug/disease M&S was conducted to identify opportunities for training and education purposes and will be published shortly.
Outlook
This is an abbreviated summary of the activities of DDMoRe for the first of the 5 years of this partnership. It is by no means a complete detailed account of all deliverables and tangible outcomes, which will be provided through subsequent publications.
It was a considerable effort to bring more than two dozen organizations into a common enterprise, with many more colleagues being actively involved in the various developments, but it is also important to notice that this initiative can only succeed on its set goals if the community not directly involved is kept informed, allowed to comment, and interact. In this regard, we conducted an Advisory Board meeting with external stakeholders to gain feedback on plans and progress. Because external acceptance and contribution is vital for the long-term success of this initiative, we will continue to engage with stakeholders and the community by making outcomes public at http://www.ddmore.eu and through communications at conferences and workshops.
Conflict of interest
L.H. is an employee of Pfizer Ltd, Sandwich, UK. I.M. is an employee of Novartis, Basel, Switzerland. J.C. is an employee of Mango Solutions, Chippenham, Wiltshire, UK, and received funding through Drug and Disease Model Resources/Innovative Medicines Initiative (DDMoRe/IMI). M.O.K received funding through DDMoRe/IMI
Supplementary Material
References
- Lalonde R.L., et al. Model-based drug development. Clin. Pharmacol. Ther. 2007;82:21–32. doi: 10.1038/sj.clpt.6100235. [DOI] [PubMed] [Google Scholar]
- Innovative Medicines Initiative Strategic Research Agenda, 2008/2011. <http://www.imi.europa.eu/content/research-agenda>.
- Paul S.M., et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat. Rev. Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- de Bono B., Hoehndorf R., Wimalaratne S., Gkoutos G., Grenon P.The RICORDO approach to semantic interoperability for biomedical data and models: strategy, standards and solutions BMC Res. Notes 4313.2011. <http://www.biomedcentral.com/1756-0500/4/313>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAGE 21 (2012) Abstracts 2337, 2372, 2376, 2377, 2404, 2428, 2440, 2449, 2455, 2527, 2529, 2538, 2564, 2578, 2586 and 2595. <http://www.page-meeting.org/?abstract=xxxx>.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.