Abstract
The Mycobacterium smegmatis genome contains six operons designated mce (mammalian cell entry). These operons, which encode membrane and exported proteins, are highly conserved in pathogenic and non-pathogenic mycobacteria. Although the function of the Mce protein family has not yet been established in Mycobacterium smegmatis, the requirement of the mce4 operon for cholesterol utilization and uptake by Mycobacterium tuberculosis has recently been demonstrated. In this study, we report the construction of an M. smegmatis knock-out mutant deficient in the expression of all six mce operons. The consequences of these mutations were studied by analyzing physiological parameters and phenotypic traits. Differences in colony morphology, biofilm formation and aggregation in liquid cultures were observed, indicating that mce operons of M. smegmatis are implicated in the maintenance of the surface properties of the cell. Importantly, the mutant strain showed reduced cholesterol uptake when compared to the parental strain. Further cholesterol uptake studies using single mce mutant strains showed that the mutation of operon mce4 was reponsible for the cholesterol uptake failure detected in the sextuple mce mutant. This finding demonstrates that mce4 operon is involved in cholesterol transport in M. smegmatis.
Keywords: Cell envelope, Mce operons, Mycobacterium smegmatis, Mycobacterium tuberculosis attenuation
1. Introduction
The analysis of the complete sequence of the Mycobacterium tuberculosis H37Rv genome revealed the presence of four paralogous mce genes, all encoded in an operon structure consisting of eight genes (yrbEA, yrbEB, mceA, mceB, mceC, mceD, mceE, mceF ) [1]. There is increasing evidence showing that these operons are important for the virulence of the Mycobacterium tuberculosis complex species. The first indication came from Arruda et al. [2] that described an invasin-like gene, later known as mce1A that conferred on non-pathogenic Escherichia coli the ability to invade and survive within macrophages and human HeLa cells. Flesselles et al. [3] reported that a BCG strain mutated in mce1A exhibited reduced ability to invade the non-phagocytic epithelial cell line HeLa. Moreover, M. tuberculosis knockout mutants deficient in the expression of the mce1, mce2 and mce3 operons showed alteration on their ability to multiply/persist when inoculated in mice [4, 5]. Although this evidence relates the mce operons to the virulence of pathogenic mycobacteria, the completion of the sequencing of the Mycobacterium smegmatis genome indicated that orthologs of the mce genes are present in this specie as well (http:www.tigr.org). Bioinformatic analysis and assembly of information previously published [6, 7] showed that M. smegmatis differs from M. tuberculosis in that it has six mce operons instead of four (mce1, 3, 4, 5, 5 bis and 7). Moreover, a detailed phylogenetic study carried on by Casali & Riley [6] showed the presence of mce loci not only in mycobacteria but also in other actinomycetes. In this study Casali & Riley [6] have hypothesized that the mce operons are transport systems. This hypothesis is supported by the recent finding that the mce4 operon is involved in cholesterol uptake and utilization in M. tuberculosis [8]. In addition, Mohn et al. [9] found that a homologous system performs the same function in Rhodococcus jostii RHA. However, it is unclear which functions the other Mce proteins of M. tuberculosis have and what their functions are in other bacteria such as M. smegmatis. In this work we evaluated the physiological role of the Mce proteins through the construction of M. smegmatis knock-out mutants deficient in the expression of mce operons. We found that the deletion of the mce4 operon impaired cholesterol uptake by M. smegmatis. This finding indicates that, similarly at what happens in M. tuberculosis and R. jostii, the Mce4 proteins are involved in the transport of cholesterol by M. smegmatis. We also found that the lack of all Mce proteins produced subtle alterations in colony morphology, in biofilm formation as well as significant aggregation when cultured in liquid media devoid of tensioactives. Our results suggest that one or more mce operons may be related to cell envelope biosynthesis processes or to the maintenance of its structure.
2. Methods
2.1. Culture media and bacterial growth conditions
M. smegmatis mc2 155 and the isogenic sextuple mce mutant (Δ6 mce) were propagated in different media as follows: Middlebrook 7H9 broth supplemented with glycerol 0.5% (v/v) (G), dextrose 0.2% (w/v) (D), NaCl 0.81% (w/v) (S), Tween 80 0.2% (w/v) (T) and albumin 10% (w/v) (A), (7H9 GADST for short); variations in the composition of the media consisted in the elimination of one or more of the supplements as shown in the denomination of the medium. Muller Hinton (MH) broth and Hartmans-de Bont [10] media supplemented with different components were also used. When indicated, agar (Difco, 1.5% w/v) was added to liquid medium.
For lipid extractions, the strains were cultured on Sauton minimal medium (0.5 g/L K2HPO40.5 g/L MgSO4. 7H2O, 2 g/L citric acid, 4 g/L L-asparagine, 0.05 g/L ferric ammonium citrate, 0.025% (vol/vol) tiloxapol, pH 6.9) containing 0.1% (wt/vol) glycerol as unique carbon source. When the bacteria were grown with unmarked cholesterol a Sauton minimal medium containing 0.4 g/L l-asparagine (Sauton low asparagine for short) was used. Cholesterol was dissolved in 100% ethanol and used at a 0.01% (wt/vol) final concentration.
Biofilm formation was assessed in M63 minimal medium supplemented with casaminoacids (0.5%), dextrose (2%), MgSO4 (1 mM) and CaCl2 (0.7 mM).
Triton WR1139, a non-metabolizable tensioactive agent, was added at 0.125 µg/ml to prevent clumping when Tween 80 was omitted from liquid media.
2.2. Construction of mce mutants in M. smegmatis and generation of the Δmce4 complemented strain
For the deletion of the six mce operons, upstream and downstream regions of each operon were amplified by PCR using the primers described in Table 1 (Supplementary material 1). The 12 resultant regions were cloned in the pGEM-T vector (Promega) and then subcloned in the p2Nil vector [11]. The final delivery vectors were obtained by ligating the PacI cassette from pGOAL17s into each p2Nil delivery plasmid.
The vectors generated were pretreated with UV light (100 mJ × cm−2) and used to electroporate M. smegmatis mc2 155 or the different intermediate strains constructed. The non-marked mutants were obtained using a two-step strategy described previously [11] and they were identified by colony PCR, using the primers described in Table 2 (Supplementary material 1).
Intents to clone the whole mce4 operon from M. smegmatis were unsuccessful. For this reason the mce4 deletion in Δmce4 was complemented by the addition of pmce4 plasmid vector (GenBank accession no. DQ823233), which contains the orthologous mce4 operon from M. tuberculosis. The complemented strain was used in cholesterol uptake experiments.
2.3. Macroscopic and microscopic studies
Colony morphology was analyzed by plating 2 µl spots or aliquots (≈ 100 colony forming units (CFU)) of the parental and the Δ6 mce mutant strains on solid Mueller Hinton, Hartmans-de Bont or Middlebrook 7H9 media with the addition of glycerol, NaCl, albumin and dextrose in different combinations. Congo Red, an azo-dye with a well known affinity for lipids and lipoproteins, was added to improve visualization of colony morphology alterations as described by Cangelosi et al. [12]. Plates were inspected after three days at 37 °C either by naked eye or under a binocular scope at low magnification. Congo Red binding assays were done as described by Cangelosi et al. [13]. Each determination was done at least three times by duplicate.
The study of the sliding motility of the wild-type and mutant strain was carried out as described by Martínez et al. [14].
Biofilm formation was analyzed in multititer plates containing 3 ml of supplemented M63, inoculated with 2 µl of a fresh stationary culture of each strain followed by incubation at 37 °C for 18 days. After this time, colony forming units for each well were determined and biofilm formation was assessed by the addition of Crystal Violet (1% w/v in water) for 30 min, washing of the wells and extraction of the bound dye with 3 ml of ethanol for 1 h as described prevously [14]. The extracted Crystal Violet was quantitated spectrophotometrically at 570 nm; CFU present in each well were determined by plating serial dilutions of each culture in Mueller Hinton agar media. Each determination was done at least three times in triplicate. Final CFUs were similar between wild-type and mutant strain therefore Abs570 values were compared directly.
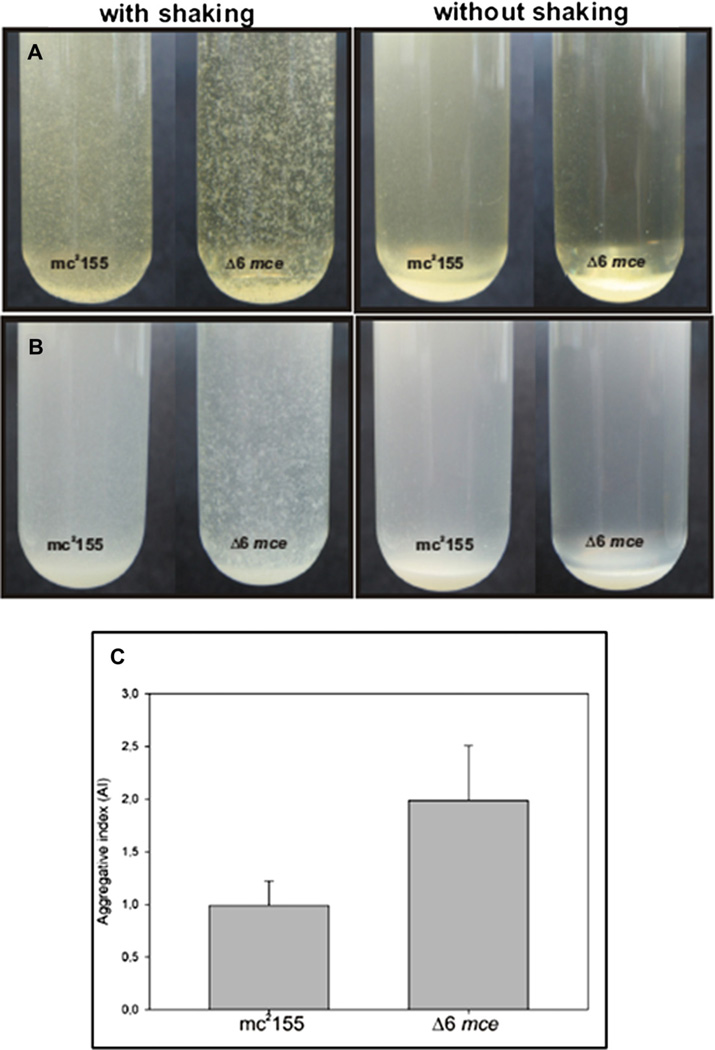
Biofilms were also analyzed by confocal microscopy. Parental and Δ6 mce M. smegmatis strains expressing GFP were grown in 7H9 GADST until stationary phase. These cultures were used to inoculate M63 supplemented minimal medium contained in Lab-Tek Chamber Slides (Thermo Scientific) at an initial OD of 0.005. Bacteria were cultured at 37 °C in humid chambers and biofilms examined daily using a Leica confocal system with a 63× water immersion objective and the 488 nm argon laser line. Images were obtained and processed using the Leica Confocal Software. The analysis of cell aggregation was carried out as described by Deshayes et al. [15]. Briefly, duplicate cultures of the parental and the mutant strains were grown in MH broth or Middlebrook 7H9 GADS broth in the absence or presence of tensioactives (the latter culture used as an estimate of the OD of the former). When an OD600nm = 0.8 was reached in the cultures grown in the presence of tensioactives, aliquots of the cultures devoid of tensioactives were withdrawn and left standing for 10 min at room temperature. After that, the supernatants were removed and the cell sediments were vortexed with 4 mm glass beads and resuspended in the original volume of medium. An aggregation index (AI) was calculated as a ratio of optical densities (OD600) of the aggregated cells resuspended by mechanical agitation and dispersed cells in the supernatants. Assays were performed at least three different times by triplicate.
2.4. Cholesterol uptake
M. smegmatis parental and mutant strains were grown in Middlebrook 7H9 GADST medium and used to inoculate a Sauton low asparagine medium containing unmarked cholesterol. The strains were growth at 37 °C in agitation until stationary state phase. Then, the cells were washed twice with PBS buffer and diluted to an OD600 of 0.5–0.6 in a Sauton low asparagine medium containing 0.02 µCi/ml [4-14C] cholesterol. The bacteria were incubated at 37 °C and 1 ml culture samples were taken at various time points. The OD600 was measured to each sample and then they were centrifuged to 12,000 rpm for 5 min at 4 °C, washed twice with TE buffer and resuspended in 100 µl. A 10 µl aliquot of cells was mixed with 100 µl liquid scintillation solution and the cell-associated [14C] cholesterol was measured using a 1450 Microbeta TriLux Liquid Scintillation and Luminescence Counter (Perkin Elmer). The assay was performed three times.
The uptake experiment was modified when the complemented strain was evaluated. The cells were grown in Sauton medium containing glycerol, instead of cholesterol, and cultivated at room temperature without agitation until stationary state phase. Then, the uptake experiment was performed as described above but the washes were done with Sauton low asparagine medium, instead of TE buffer.
3. Results
3.1. Construction of M. smegmatis mutants lacking mce operons
The mce operons from M. smegmatis mc2 155 were mutated using the gene knock-out system described by Parish & Stoker [11]. In order to delete each mce operon completely, upstream and downstream regions from each operon were cloned in the delivery vector, as described in the methodology section. This strategy allowed the construction of unmarked mutants. The first deletion made was that of mce4 operon, then double (mce4+3), triple (mce4+3+5bis), quadruple (mce4+3+5bis+5), quintuple (mce4+3+5bis+5+1) and sextuple (mce4+3+5bis+5+1+7) mutants were constructed. The deletion of the mce operons was confirmed by PCR (Fig. 1) using primers that amplify the wild type and mutant allele from each operon. These results suggest that the mce operons are not essential in M. smegmatis, at least under the standard growth conditions used in this study. The first examination of the deletion mutants by naked eye failed to reveal any noticeable phenotype affecting size or morphology of the colonies on Middlebrook 7H9 agar medium (data not shown).
Fig. 1.
PCR reactions to screen for mutant strains. Different sets of primers were designed to amplify the wild type and mutant alleles from each operon, as shown in panel A, and they were used to screen for mutants. Vertical arrows represent the beginning and the end of each mce operon and delimit the fragments that were deleted from the genome. Panel B shows PCR reactions done with one clone (called 25) of the sextuple mutant to confirm the deletion of each operon. Colony PCR was performed to this clone with the set of primers “A” to amplify the mutant allele and with the set of primers “B” and “C” to amplify the wild type allele from each mce operon. +A: positive control of the mutant allele using the delivery plasmid to mutate the corresponding operon as template. +B and +C: positive controls of the wild type allele using genomic DNA from M. smegmatis mc2 155 as template.
3.2. M. smegmatis sextuple mce deletion mutant is altered in colony morphology
In order to further characterize the sextuple Δmce mutant strain, we analyzed its ability to grow in liquid and solid media of different composition at 37 °C. That was tested on the chemically defined Middlebrook 7H9 broth medium to which glycerol, dextrose, albumin and NaCl were added in different combinations. Mueller Hinton medium was used as a complex, rich medium with and without dextrose or glycerol, while Hartmans-de Bont medium with glycerol or dextrose as carbon source was used as minimal medium. In all cases Tween or Triton were present to avoid clumping. No differences were observed between growth of the wild-type strain and the sextuple mce deletion mutant, at any stage of the growth curve, although a slightly slower growth rate on 7H9 –GADST was reproducibly seen for the mutant strain (data not shown). Thus, the loss of all the mce operons does not have any significant consequence on growth in the most commonly used liquid media, ruling out their involvement in the import of nutrients or export of metabolic end products or any major structural components required for viability or optimal growth under laboratory conditions.
We next set out to analyze colony morphology and size on different solid media with various carbon sources. For that purpose, Middlebrook 7H9 agar and Mueller Hinton agar were used as rich media with the addition of carbon sources in different combinations as described above. Hartmans-de Bont minimal medium was again included to test other substrates. In all these cases Congo Red was used with a dual purpose, in first place as an aid to improve the visualization of possible alterations in cell morphology and secondly as a mean to assess changes in the lipid and lipoproteins contents of the cell envelope [12]. Surprisingly, the sextuple mce mutant consistently produced “hairy” colonies on all of the media tested (Fig. 2). These projections, similar to short spikes and hairs, could be easily detected under a binocular scope at low magnification. This phenotype was specially noticeable when plated on Mueller Hinton and –although less remarkable- on Middlebrook media devoid of dextrose. When Hartmans-de Bont media was used, while parental colonies were smooth and lacked the usual colonial morphology, mutants produced rough colonies bearing some projections, albeit fewer than in Mueller Hinton medium (Fig. 2). In all cases, independent of the medium used, the sextuple mce deletion mutant had a drier appearance than the wild-type strain. Altogether, the observed changes in morphology, suggest cell envelope alterations of unknown nature associated to the loss of the mce operons.
Fig. 2.
Impact of the deletion of the six mce operons in colony morphology. Aliquots of fresh cultures of either M. smegmatis mc2 155 (left) and M. smegmatis Δ6 mce (right) were plated on MH 0.2% dextrose (panel A); MH 0.5% glycerol (panel B); MH (panel C); Middlebrook 7H9 10% ADS (panel D); Middlebrook 7H9 0.5% glycerol, 0.2% dextrose (panel E); Hartmans-de Bont 0,5% glycerol, 0.2% dextrose (panel F). Congo Red (100 µg/ml) was added to all the tested media. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. M. smegmatis sextuple Δmce mutant reveals alterations in biofilm formation and clumping
Based on the results described above, we set out to investigate possible changes in the cell envelope structure and composition of the mutant. An approach consisted in comparing the binding of Congo Red [12] to wild-type and mutant colonies. Quantitation of the extracted Congo Red showed no significant difference between parental and mutant strains (data not shown). Other indirect approaches for the analysis of the cell envelope included the examination of the ability for sliding motility [14] and biofilm formation [16] of the mutant and parental strains of M. smegmatis. These phenotypic traits were chosen because of their relationship to the cell envelope composition [14, 17]. No differences in the motility assay were detected between both strains (data not shown). However, we were able to observe variations in the biofilms produced by the sextuple deletion mutant compared to that of the wild-type strain (Fig. 3). Although the amount of biofilm -quantitated by Crystal Violet binding- was similar in the mutant and wild- type strains (Supplementary material 2), two important differences were observed. In the first place, alteration in the biofilm surface was observed in the mutant as compared to the parental strain (Fig. 3A), with the sextuple mce mutant showed a smoother surface than the parental strain. A close examination of the biofilm structure by confocal microscopy showed that the mutant biofilm contained bigger bacterial clumps than those of the wild-type biolfim (Fig. 3B). The second difference was related to the amount of time that the biofilm could maintain its organization on the medium/air interface before collapsing, probably due to aging. While the biofilms produced by the wild-type strain detached from the walls of the well and were covered by the culture medium by day 8, the biofilm generated by the sextuple deletion mutant could stay on the interface, attached to the walls of the well for more than 26 days without collapsing. Another evidence suggestive of cell envelope alterations generated by the loss of the mce operons was observed when the mutant was grown in 7H9 –GADS devoid of Tween 80; in these conditions, the degree of clumping was twice higher than the one observed for the wild-type strain (Fig. 4, panel B and C). The same phenotype was seen when Mueller Hinton was used, although in this case, the aggregation was markedly increased leading to clumps that we were not able to break up mechanically by vortexing with glass beads (Fig. 4A). Thus, these results strongly reinforce our idea that at least one of the mce operons is functionally related to cell envelope structure.
Fig. 3.
Biofilm formation of M. smegmatis mc2 155 and its Δ6 mce mutant derivative. Panel A: Cultures from both strains were grown in supplemented minimal salts M63 medium in 12-well multititre plates for up to eighteen days at 37 °C. For the sake of simplicity only the time points up to seven days are shown. Panel B: Sections derived from confocal microscopy of the biofilms developed by parental and Δ6 mce strains expressing GFP on chamber slides.
Fig. 4.
Loss of mce operons increases clumping. Aggregation of M. smegmatis mc2 155 and its Δ6 mce mutant derivative was determined as described by Deshayes et al. [15] Panel A: MH; Panel B: Middlebrook 7H9-ADS-glycerol. Panel C: determination of the aggregation index (AI, see text for details) for cultures grown in Middlebrook 7H9-ADS-glycerol; cultures of the Δ6 mce mutant grown in MH were extremely difficult to resuspend and thus their aggregation index could not be quantified.
3.4. Deletion of the mce4 operon in M. smegmatis impaired the uptake of cholesterol
A recent report by Pandey & Sassetti [8] showed that a M. tuberculosis H37Rv Δmce4 knock-out strain was defective in cholesterol transport. Thus, it seemed reasonable to test the proficiency of our sextuple mce deletion mutant to transport this steroid. The role of the Mce proteins in cholesterol uptake was examined by measuring [4-14C] cholesterol uptake in mutant and wild-type strains. The rate at which the sextuple mce mutant accumulated [4-14C] cholesterol was lower than that of the parental strain, indicating that at least one of the mce operons participates in the cholesterol uptake. Given that mce4 operon participates in the transport of cholesterol in M. tuberculosis and in R. jostii RHA we compared the uptake of [4-14C] cholesterol by a single mce4 mutant of M. smegmatis to those of the sextuple mce mutant and the parental strains. A single mce1 mutant of M. smegmatis was included as control. As it is shown in Fig. 5A, mutant mce4 accumulated [4-14C] cholesterol at a similar rate than that of the sextuple mce mutant, while the accumulation of [4-14C] cholesterol in mutant mce1 was equal to that of the parental strain (Supplementary material 3). In addition, we tested the cholesterol uptake in the Δmce4 strain complemented with an intact copy of the mce4 operon from M. tuberculosis. The growth of the complemented strain either in the presence of cholesterol as sole carbon source or at 37 °C, or in both conditions was significantly impaired (data not shown). For these reasons, the cultures for the complementation experiments were grown in a cholesterol-free media, at 25 °C. Fig. 5B shows that the reintroduction of mce4 restored the uptake of cholesterol in the mutant strain at similar level to that of the parental strain. However, the cholesterol uptake kinetic in the complemented strain showed to be delayed as compared to that of the wild type, suggesting differences in the Mce4 transport system between M. smegmatis and M. tuberculosis. Together these results demonstrate that, as it happens in M. tuberculosis and R. jostii RHA, the Mce4 proteins of M. smegmatis participate in the transport of cholesterol. Besides, these proteins seem to be the only ones among Mce proteins involved in cholesterol uptake.
Fig. 5.
The mce4 operon is required for cholesterol uptake in M. smegmatis. A) The indicated strains of M. smegmatis were grown on Sauton medium containing cholesterol at 37 °C, washed and incubated with [4-14C]-cholesterol for the indicated times. The cell-associated radioactivity remaining after extensive washing was quantified and expressed as . B) The cholesterol uptake was measeared in a conplementated M. smegmatis Δmce4 mutant. The strains were grown on Sauton medium containing glycerol at 25 °C, and the cholesterol uptake was evaluated as described above. One of three independent experiments with similar result is shown.
3.5. The cell lipid profile is not altered in the sextuple mce null mutant
Based on functional experiments, Santangelo et al. [18] have recently found that the regulator of the mce3 locus, Mce3R, negatively regulates the expression of two additional transcriptional units, predicted to be involved in lipid metabolism or redox reactions. These findings, together with other published reports [19] and our observations linking M. smegmatis mce4 to cholesterol uptake, led us to hypothesize that the mce operons could be implicated in the transport of lipids. Thus, we set out to analyze the lipid profile of the sextuple mce null mutant, grown in Middlebrook 7H9-GAD or Mueller Hinton. The lipids from whole cell extracts, cytoplasm plus membrane and cell wall fractions of the sextuple mce deletion mutant and of the wild-type strain were analyzed by 1D-TLC with various solvent systems to resolve different lipids from M. smegmatis: (phosphatidylinositol (PI), phosphatidyletha-nolamine (PE), cardiolipin (CL), phosphatidylinositol mannosides (PIM), trehalose monomycolates (TMM), trehalose dimycolates (TDM) and glycopeptidolipids (GPLs) (see Supplementary material 4). Against our expectations, no differences in the lipid profiles were observed under the conditions tested between the two strains (Fig. S4). Analysis of extractable lipids and cell wall-bound mycolic acid contents also failed to show any significant differences between mutant and wild-type strains (data not shown). Therefore, although a phenotype compatible with cell envelope alterations was noticeable, we were not able to identify its molecular basis.
4. Discussion
The role of mce operons in mycobacteria has been an intriguing enigma for several years. So far, mce operons 1, 2, 3, 4 and the combination 3 + 4 have been knocked-out in M. tuberculosis [3, 20]. In all cases except for mce1, the deletion strains showed a decrease in virulence. Interestingly, all of the attenuated strains behaved in vivo in a slightly different way, with respect to the progression of disease, pathology and immune responses, suggestive of their non-redundant functions. With the goal of elucidating the roles of the mce operons in mycobacterial physiology and their relationship to the pathogenesis of TB, we undertook the generation of a M. smegmatis mutant deficient in the expression of all mce operons.
Our results showed that the M. smegmatis sextuple mce deletion mutant was still able to grow at rate comparable to that of the parental strain in all of the different culture media tested. Thus, the transport (import or export) of essential structural components or nutrients by the mce operons could be ruled out. In order to determine whether the loss of the entire mce gene repertoire could have a more delicate impact on the cell envelope organization, we compared wild-type and mutant colonial morphology on different culture media, their ability to bind Congo Red, their sliding motility, and capacity to form biofilms, all tests directly or indirectly associated to cell envelope alterations. Surprisingly, mutant colonies showed a “hairy” appearance. This phenotype was more clearly observed in the richest medium used, Mueller Hinton. Since colony morphology reflects bacterial cell interactions, this result pointed out to a possible modification of cell envelope surface components. Consistently with this presumption, the distribution to the bacteria in the biofilm formed by the mutant showed to be more aggregated than that of the parental strain. In addition, the mutant biofilm could remain at the surface of the medium without collapsing longer than the parental biofilm; thus, together with the colonial morphology alterations, these results were suggestive of cell envelope changes. More evidence of alterations in cell envelope was provided by aggregation studies, in which the mutant strain formed more clumps than the parental strain when both were grown in liquid culture medium without added tensioactives. Thus, taken altogether these results suggest that the mce operons of M. smegmatis are implicated in the maintenance of the surface properties of the cell. In spite of this we still don’t know which operons are responsible for the observed phenotypes (biofilm alterations, cell aggregation and altered colony morphology) and more work is needed in order to elucidate this.
In a previous work, Santangelo et al. [18] used microarray analysis to compare the transcriptional profile of an M. tuberculosis mutant strain defective in the production of Mce3R, the repressor of the mce3 operon, to parental M. tuberculosis H37Rv. In that work they found that Mce3R controls the expression of a number of genes involved in lipid metabolism and β-oxidation in M. tuberculosis. In addition, the lipid profiles of both strains were analyzed by TLC and variations have been observed between the mutant and the wild-type strain. These results, which suggest that the mce operons from M. tuberculosis encode lipid transporters, are supported by those of Pandey & Sassetti [8] and Mohn et al. [9], who demonstrated that the mce4 operon is involved in the transport of cholesterol in M. tuberculosis and R. jostii RHA, respectively. Here we found that the role of mce4 operon in cholesterol transport is also conserved in M. smegmatis and that the uptake of this steroid seems to be an exclusive function of mce4 among the six mce operons. However, as in the case of M. tuberculosis [8], there is same residual cholesterol uptake in the mce4 mutant of M. smegmatis, which could be due to another less efficient import system or may simply reflect passive diffusion across the membrane. Complementation with an intact copy of mce4 from M. tuberculosis restored the uptake of cholesterol in M. smegmatis Δmce4, but a clear delay in the cholesterol uptake kinetic was detected in the complemented strain when compared to that of the parental strain. In addition, the expression of Mce4 proteins from M. tuberculosis significantly impaired the growth of M. smegmatis when bacteria were growth either in the presence of cholesterol or at 37 °C. Therefore, in spite of the fact that the function of Mce4 transporters are conserved in M. tuberculosis and M. smegmatis, there would be differences in the cholesterol affinity, transport efficiencies or regulation of the expression between both Mce4 systems.
The failure in cholesterol uptake, together with a recent report by Dunphy et al. [21] in which they found that mce1 fadD5 mutant of M. tuberculosis was diminished in growth in minimal medium supplied only with mycolic acid as a carbon source, lead us to analyze if the sextuple mutant was diminished in growth in minimal medium supplied only with different steroids (oleic acid, palmitic acid, stearic acid, testosterone and cholesterol, all prepared in ethanol) as a sole carbon source (data not shown). However, no obvious differences were observed between the mutant and parental strains, al least in the conditions and with steroids tested here. Of course, we cannot exclude that mce operons play a role in the transport of other types of host lipid or fatty acid substrates that have not been tested here. In summary, our results suggest that the mce operons may have a role in transport of molecules that either provide M. smegmatis with carbon sources or form part of the cell envelope architecture. Thus, along with the fact that the mce4 operon of M. smegmatis is involved in cholesterol transport, it is a challenging hypothesis to postulate that mce operons have two functions, with some of them performing import, modification and export of steroids and others transporting cell wall compounds in mycobacteria.
All together, the results obtained in this study and the availability of the herein reported M. smegmatis mce null mutants will help to decipher the biological significance of the Mce protein family in mycobacteria.
Supplementary Material
Acknowledgments
This work was supported by grants NIH/NIAID 1R01AI083084-01 to FB, INTA AEBIO 243512 to FB, ANPCyT PICT 38198 to HRM, NIH/NIAID grant AI64798 to MJ. FB, AAC and MPS are career members of CONICET, HRM is a career member of CIUNR. EJS is a fellow of CONICET. We thank Valeria Rocha for technical assistance and Luis Fernandez for the bibliography provided.
Footnotes
Appendix. Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.micinf.2012.01.007.
References
- 1.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris CD, Gordon SV, Eiglmeier K, Gas S. other authors, Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261:1454–1457. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 3.Flesselles B, Anand NN, Remani J, Loosmore SM, Klein MH. Disruption of the mycobacterial cell entry gene of Mycobacterium bovis BCG results in a mutant that exhibits a reduced invasiveness for epithelial cells. FEMS Microbiol. Lett. 1999;177:237–242. doi: 10.1111/j.1574-6968.1999.tb13738.x. [DOI] [PubMed] [Google Scholar]
- 4.Gioffre A, Infante E, Aguilar D, Santangelo MP, Klepp L, Amadio A, Meikle V, Etchechoury I, Romano MI. other authors, Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 2005;7:325–334. doi: 10.1016/j.micinf.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Shimono N, Morici L, Casali N, Cantrell S, Sidders B, Ehrt S, Riley LW. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl. Acad. Sci. 2003;100:15918–15923. doi: 10.1073/pnas.2433882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casali N, Riley LW. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics. 2007;8:60. doi: 10.1186/1471-2164-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haile Y, Bjune G. H.G.Wiker, Expression of the mceA, esat-6 and hspX genes in Mycobacterium tuberculosis and their responses to aerobic conditions and to restricted oxygen supply. Microbiology. 2002;148:3881–3886. doi: 10.1099/00221287-148-12-3881. [DOI] [PubMed] [Google Scholar]
- 8.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohn WW, Van der GR, Stewart GR, Okamoto S, Liu J, Dijkhuizen J, Eltis LD. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 2008;283:35368–35374. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmans S, De Bont J. The Genus Mycobacterium - Nonmedical, the Prokaryotes. Springer; New York: 2009. pp. 1215–1237. [Google Scholar]
- 11.Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 12.Cangelosi GA, Palermo CO, Laurent JP, Hamlin AM, Brabant WH. Colony morphotypes on Congo red agar segregate along species and drug susceptibility lines in the Mycobacterium avium-intracellulare complex. Microbiology. 1999;145:1317–1324. doi: 10.1099/13500872-145-6-1317. [DOI] [PubMed] [Google Scholar]
- 13.Cangelosi GA, Do JS, Freeman R, Bennett JG, Semret M, Behr MA. The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium Antimicrob. Agents Chemother. 2006;50:461–468. doi: 10.1128/AAC.50.2.461-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez A, Torello S, Kolter R. Sliding motility in mycobacteria. J. Bacteriol. 1999;181:7331–7338. doi: 10.1128/jb.181.23.7331-7338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshayes C, Laval F, Montrozier H, Daffe M, Etienne G, Reyrat JM. A glycosyltransferase involved in biosynthesis of triglycosylated glycopeptidolipids in Mycobacterium smegmatis: impact on surface properties. J. Bacteriol. 2005;187:7283–7291. doi: 10.1128/JB.187.21.7283-7291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Recht J, Martinez A, Torello S, Kolter R. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 2000;182:4348–4351. doi: 10.1128/jb.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kocincova D, Singh AK, Beretti JL, Ren H, Euphrasie D, Liu J, Daffe M, Etienne G, Reyrat JM. Spontaneous transposition of IS1096 or ISMsm3 leads to glycopeptidolipid overproduction and affects surface properties in Mycobacterium smegmatis . Tuberculosis. 2008;88:390–398. doi: 10.1016/j.tube.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Santangelo MP, Blanco FC, Bianco MV, Klepp LI, Zabal O, Cataldi AA, Bigi F. Study of the role of Mce3R on the transcription of mce genes of Mycobacterium tuberculosis . BMC Microbiol. 2008;8:38. doi: 10.1186/1471-2180-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Chandolia A, Chaudhry U, Brahmachari V, Bose M. Comparison of mammalian cell entry operons of mycobacteria: in silico analysis and expression profiling. FEMS Immunol. Med. Microbiol. 2005;43:185–195. doi: 10.1016/j.femsim.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 20.Senaratne RH, Sidders B, Sequeira P, Saunders G, Dunphy K, Marjanovic O, Reader JR, Lima P, Chan S. other authors,Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J. Med. Microbiol. 2008;57:164–170. doi: 10.1099/jmm.0.47454-0. [DOI] [PubMed] [Google Scholar]
- 21.Dunphy KY, Senaratne RH, Masuzawa M, Kendall LV, Riley LW. Attenuation of Mycobacterium tuberculosis functionally disrupted in a fatty acyl-coenzyme A synthetase gene fadD5 . J. Infect. Dis. 2010;201:1232–1239. doi: 10.1086/651452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.