Abstract
Vaccines are one of the most cost effective methods to control infectious diseases and at the same time one of the most complex products of the Pharmaceutical industry. Contrary to other drugs, vaccines are used mainly in healthy individuals, many cases children. For this reason, very high standards are set for their production.
Subunit vaccines, especially peptide vaccines, provide a cost effective alternative. Structural studies of peptide-MHC complexes and biochemical characterizations of the peptide-MHC interaction have provided conceptual foundation for the rational design of subunit vaccines and in the other side of the spectrum, development of “immunological silent” therapeutic proteins. In this review, we explain the manner in which the analysis of crystal structures of peptide-class II MHC complexes opened the door to the understanding of the major rules that govern this interaction. We also describe biochemical studies that allowed the development of “virtual matrices” of the peptide-class II MHC interaction and MHC-peptide binding algorithms that incorporate in one or other manner structural and/or biochemical information of the interaction. Finally, and using malaria as a model, we describe the development of a minimal subunit vaccine for the human malaria parasite Plasmodium falciparum.
Introduction
The use of vaccines is one of the most cost effective methods to control infectious diseases as demonstrated by the eradication of smallpox in the 1970’s. In the USA, vaccines for 27 infectious diseases are currently used and many more are in development (http://www.cdc.gov/vaccines/default.htm) [1]. Protective immune responses were generated in the first generation of vaccines by immunization with attenuated or inactivated pathogens. Subsequent vaccines were developed in this manner, but adverse reactions, difficulty in generating attenuated versions of the pathogens, lack of large-scale culture methods, and low protection were found to be roadblocks in the development of vaccines for many important pathogens. The observation that immune responses are elicited to a limited number of all possible targets, and that a subset of these responses confer protection, opened the pathway to the development of subunit vaccines. In this type of vaccines antigens that do not confer protection or induce unwanted responses are excluded in order to enhance the immune response to protective antigens or epitopes. For these reasons a major goal in the development of modern vaccines is the identification of protective antigens and more specifically epitopes, the portions of an antigen actually recognized by the immune system. Knowledge of epitopes can facilitate tracking productive immune responses in vaccine evaluation, and can contribute to selection of antigens for development of subunit vaccines capable of inducing long lasting protective responses without significant adverse effects.
In this review we describe biochemical and structural studies of class II MHC – peptide complexes that are providing an understanding of the mechanisms responsible for selection of T cell epitopes, progress towards computational prediction of such epitopes, and efforts towards development of peptide-based vaccines.
Role of CD4+ T cell responses in vaccine-induced immunity
Multiple studies in animal models have clearly demonstrated the requirement of CD4 T cell help for the generation of protective antibody responses (for example, influenza [2], malaria [3, 4], vaccinia [5, 6]). In humans, the magnitude of the antibody response elicited by vaccination is determined by factors such as age, gender, and the genetic background of the individual [7, 8]. Among the genetic factors, HLA polymorphism plays a critical role (reviewed in [9]). Although correlates of protection with serum and/or mucosal antibodies have been established for many licensed vaccines (reviewed in [1, 10, 11]), correlates of protection with cellular responses have been established only for a few vaccines [10], and for this reason efficacy of vaccination is measured in most cases in terms of antibody titer. The lack of association with cellular responses in part may be due to limitations of the methodology to study T cells, as demonstrated by characterization of T cell responses elicited by a peptide vaccine in which correlation was established only upon expansion of T cells and not by more classical methods [12]. Recent studies have also demonstrated that the role of CD4+ T cells in the immune response is not limited to help for antibody production; CD4+ T cells are also required to generate optimal CD8+ T cell responses [13–16]. Moreover, CD4+ T cells additionally can act as effector cells by the secretion of cytokines and direct killing of infected cells [17–21]. Thus, development of methodologies for facile measurement of CD4+ T cell mediated cellular immune responses in clinical trials, and development of computational methods to predict such responses, should facilitate evaluation of existing vaccines and development of novel ones.
Class II MHC-mediated antigen presentation
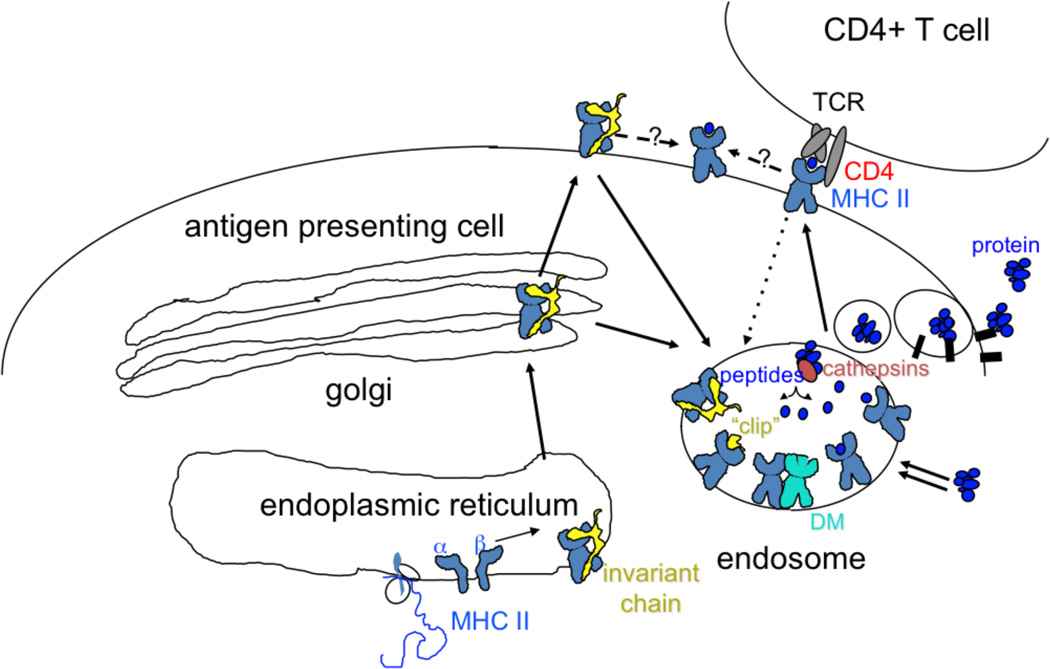
The targets of CD4+ T cells are short peptides bound to class II MHC molecules expressed constitutively on the surface of professional antigen presenting cells (APCs) such as dendritic cells (DCs), macrophages and B cells, or upregulated on the surface of other cells in response to inflammation. Dendritic cells are thought to be crucial to the initial activation of naïve T cells in lymph nodes and specialized lymphatic tissues, while other APC are important in activation of mature effector CD4+ T cells at various sites in the body. Antigen processing pathways, i.e. the manner in which intact proteins are converted to peptides and loaded onto MHC molecules, have been studied in detail [22]. A general scheme is shown in Figure 1. Class II MHC molecules bind peptides generated in endosomal / lysosomal compartments, aided by the peptide exchange factor HLA-DM. Nascent class II MHC molecules enter the endosomal pathway with their peptide binding sites occupied by a segment of the class II MHC-associated invariant chain chaperone. Proteolytic digestion of the chaperone leaves a peptide fragment known as CLIP bound to the MHC molecule; CLIP is a particularly good substrate for HLA-DM and is readily exchanged for endosomal peptides. Class II MHC molecules recycling into the endocytic pathway from the cell surface also can enter this pathway and acquire peptides by an exchange mechanism. Direct cell surface loading of empty class II MHC molecules and cell-surface peptide exchange processes have been described as well, but the efficiency of these pathways relative to the endosomal / lysosomal loading pathways currently is not known. After loading, MHC-peptide complexes traffic to the cell surface where they can be recognized by CD4+ T cells. Class II MHC trafficking and loading pathways and the morphology and contents of the class II MHC containing intracellular compartments, are regulated by cell type and developmental state.
Figure 1.
Class II MHC antigen processing pathway. MHC class II alpha and beta subunits associate with the invariant chain chaperone, which escorts them to an endosomal compartment, perhaps via transient appearance at the cell surface. Proteins destined for processing also arrive at the endosome, by fluid-phase or receptor mediated uptake, or by autophagy (double arrows). Endosomal proteases (cathepsins) cleave the invariant chain, leaving a small remnant “clip” in the MHC peptide binding site. Proteins are also degraded by endosomal cathepsins, perhaps aided by oxidoreductases and unfoldases. HLA-DM catalyzes exchange of clip for endosomal peptides (or partially degraded proteins). Fully processed MHC-peptide complexes traffic to the surface for interaction with TCR on CD4+ T cells. Surface MHC-peptide complexes can recycle to endosomes for reloading (dotted line), or can exchange peptides at the surface (dashed lines).
In the conventional pathway for class II MHC-mediated antigen presentation, peptides destined for antigen presentation derive from antigens taken up from the extracellular medium into the endosomal / lysosomal pathway. Depending on the type of APC, this could involve phagocytosis, fluid-phase endocytosis, or receptor-mediated endocytosis. For example, an inactivated virus vaccine preparation could enter an APC by phagocytosis, most likely mediated by cell surface receptors for virus components, whereas a soluble subunit vaccine would depend on fluid-phase endocytosis unless acquired by specific endocytic receptors. Proteins delivered to the endocytic pathway denature as endocytic vesicles acidify and fuse with lysosomes, and, perhaps assisted by oxido-reductases and unfoldases, are degraded by proteases present in these compartments (cathepsins). Peptide fragments able to compete successfully for binding to class II MHC proteins are protected from terminal degradation, and can traffic to the cell surface for recognition by CD4+ T cells. This conventional pathway thus provides a means for APC to present peptides derived from extracellular antigens.
However, from the first experimental characterizations of the spectrum of peptides bound to class II MHC proteins purified from APC [23, 24], it has been clear that peptides derived from intracellular sources also are fodder for presentation by class II MHC proteins [25]. While the detailed mechanisms by which such antigens enter the class II MHC mediated antigen presentation pathway are still being elucidated, autophagosomal processes are believe to contribute [26, 27]. Chaperone-medicated autophagy and microautophagy (fluid phase uptake of cytosol into lysosomes) can sample cytosolic proteins, and macroautophagy (sequestering of organelles into double-membrane vacuoles that fuse with lysosomes) can sample nuclear, mitochondrial, and microsomal proteins. These pathways provide a means for peptides derived from intracellular sources to meet class II MHC proteins for presentation. Thus autophagy might be important in presentation of peptides derived from antigens expressed in infected cells but not present in assembled virions, or in cases where viral evasion mechanisms interfere with conventional endosomal processing.
Class II MHC genes
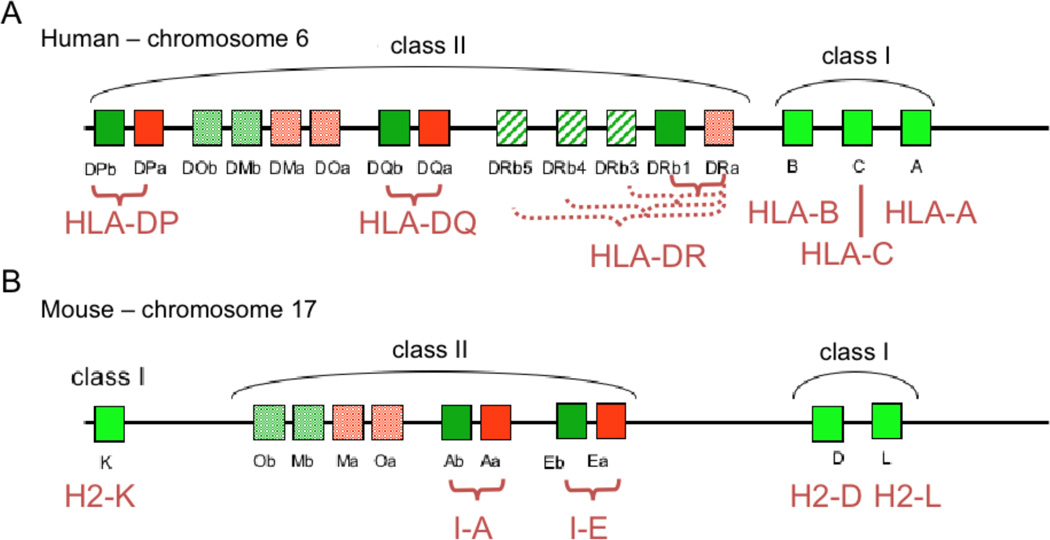
Genes encoding class II MHC proteins are found on the major histocompatibility complex (MHC) region located in chromosome 6 in humans and 17 in mice, together with genes coding for class I MHC molecules and other proteins involved in antigen presentation (Figure 2). In humans the MHC II locus includes genes for alpha and beta subunits of the three class II MHC proteins HLA-DR, HLA-DQ and HLA-DP. In most humans the expression level of class II molecules on APCs follows the order HLA-DR > HLA-DQ > HLA-DP [28, 29]. In mice the corresponding proteins are I-E and I-A, which are expressed at comparable levels. Generally, class II MHC alpha and beta genes are highly polymorphic in the human population, with the exception of HLA-DRA (the alpha subunit of HLA-DR), which is essentially monomorphic. Gene duplication events have resulted in nine DRB genes designated DRB1-DRB9. Most haplotypes characterized to date include a functional HLA-DRB1 gene and one other HLA-DRB gene (DRB3, DRB4 or DRB5), with the remaining genes non-functional. HLA-DRB1 is one of the most polymorphic genes known, with ~500 allelic protein variants described as of this writing, which fall into 15 clusters roughly corresponding to serological reactivity. Polymorphism at the other loci is not as extreme, ranging from 16 variants at DPA to 116 at DPB. Allelic variants are named systematically as gene*number, where the first two digits reflect the cluster number and the second two digits variations within that cluster, as for example HLA-DQB*0302, an allelic variant linked to susceptibility to type II diabetes. (Additional suffixes describe silent, intronic, or non-coding variation (see http://hla.alleles.org/). Older serologically defined names are also in wide use, for example DR1, DR2, DR3, etc. HLA gene names and sequences are assigned by the WHO Nomenclature Committee for Factors of the HLA System and the International Histocompatibility Workshops, and an authoritative sequence database is maintained (IMGT/HLA, at http://www.ebi.ac.uk/imgt/). Allelic variants are co-dominantly expressed so that up to 12 different class II MHC protein variants can be expressed by any particular individual. These different variants have different peptide binding activities (see below), helping to provide a broad spectrum of pathogen-derived epitopes to CD4+ T cells, but leading to considerable complexity in assigning observed T cell responses to particular class II MHC variants.
Figure 2.
The Major Histocompatibility Complex (MHC) region of the human (A) and mouse (B) genome. This region comprises approximately 4 million base pairs. MHC class I heavy chain genes and MHC class II alpha and beta subunit genes are indicated. Note that class I heavy chains associate with the β-2-microglobulin subunit (β-2m); and class II alpha and beta subunits associate transiently with the invariant chain chaperone. Both β-2m and the class II-associated invariant chain are encoded on other chromosomes. The HLA-DRB locus consists of nine duplicated genes, the first five of which can code for functional proteins (although only one or two per haplotype). DMA and DMB (Ma and Mb in mouse) code for the MHC-like peptide exchange factor HLA-DM (H-2M in mouse), and DOA and DOB (Oa and Ob) code for the MHC-like DM inhibitor HLA-DO (H-2O).
Some examples of the significant role played by HLA-DR variation can be found in the response to measles, hepatitis, and mumps vaccines (reviewed in [30, 31]), in autoimmune diseases triggered by Borrelia in Lyme disease [32] and in responses to many experimental vaccines [12, 20, 30, 33, 34]. In the case of the measles vaccine, HLA-DRB1 seems to be a major restriction element for the response to the MV-P and MV-N measles antigens [35], with HLA-DRB1*03 and HLA-DPA1*0201 associated with seronegativity after vaccination [36]. In hepatitis B, vaccine-induced responses are poorly elicited in 5–10% of healthy subjects [37]. The poor responders frequently depict a HLA-DRB1*0701, DQB1*0202 haplotype [38, 39]. In the mumps vaccine low antibody titers are associated with HLA-DQB1*0303 and strong T cell responses with particular alleles of the DRB1 (*0101, *0301, *0801, *1001, *1201, and *1302), DQA1 (*0101, *0105, *0401, and *0501), and DQB1 (*0201, *0402, and *0501) loci [9]. The correlation between HLA class II DR molecules and the immune response are not limited to vaccines. For example HLA-restriction analysis of CD4+ T cells to Mycobaterium tuberculosiswhich are crucial in preventing disease, suggests that HLA-DRB1 alleles are important in presentation of mycobacterial antigens [40, 41].
Class II MHC protein structure
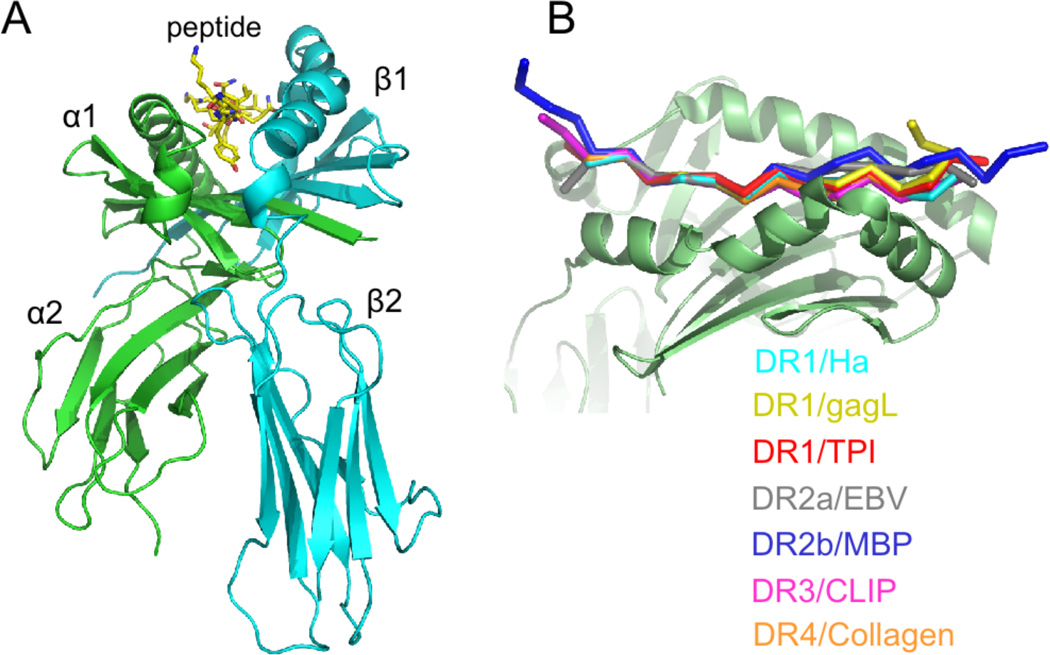
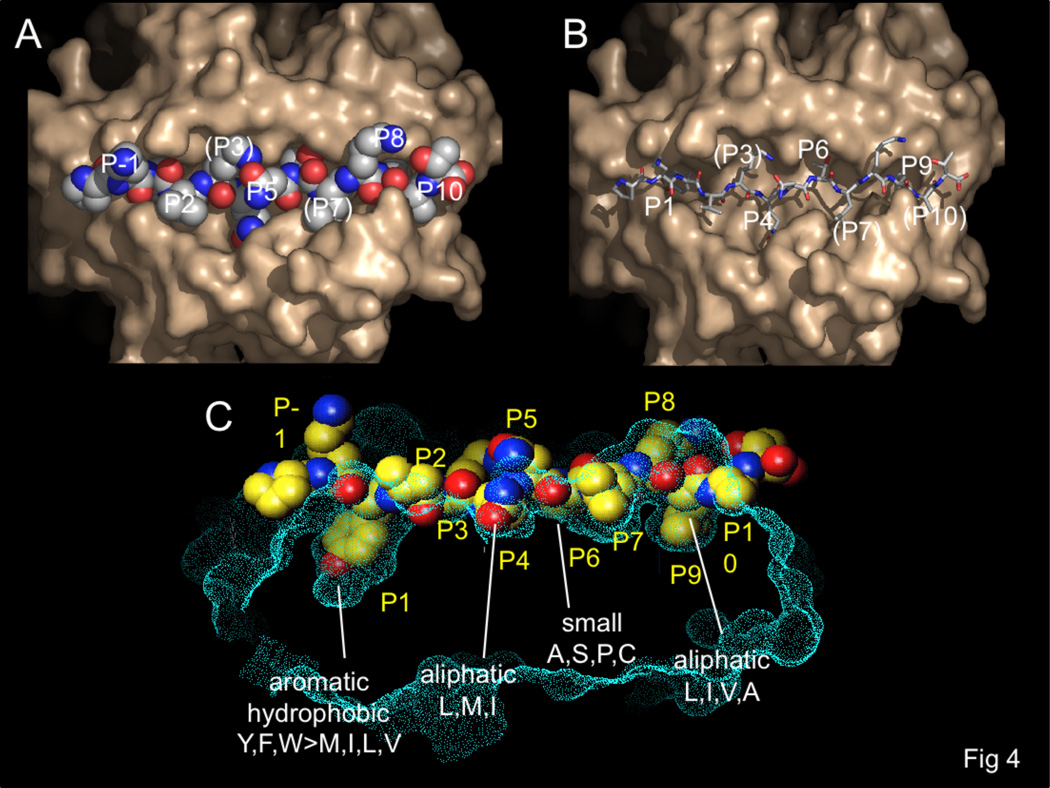
At the protein level, class II MHC proteins are heterodimeric membrane glycoproteins composed of one alpha (34KD) and one beta (30KD) subunit. Each subunit contributes one half of the binding site, one immunoglobulin domain, and transmembrane and short cytosolic portions. The extracellular portion, consisting of the peptide binding and immunoglobulin domain, has been characterized by X-ray crystallography and other physical methods (see Figure 3A). At this writing approximately thirty human class II MHC structures have appeared, for peptide complexes of HLA-DR1 (DRB1*0101), DR2b (DRB1*1501), DR3 (DRB1*0301), DR4 (DRB1*0401), DR52a (DRB3*0101, 0102), and DR2a (DRB5*0101), and for HLA-DQ2 (DQA1*0501, DQB1*0501) and DQ6 (DQA1*0601, DQB1*0602). No crystal structure has been determined for an HLA-DP molecule, but the degree of sequence homology and patterns of conserved residues indicate that it is very likely to share the canonical class II MHC structure. All the class II MHC structures reveal essentially the same protein folding and mode of peptide binding. The peptide binding site is formed by two roughly parallel alpha helical regions atop a beta sheet. Peptides bind in the groove between the helices, with their terminal residues extending from the site. In all cases characterized to date, the peptide adopts a similar extended polyproline II-like conformation that exposes the peptide backbone for interaction with conserved MHC hydrogen-bonding residues lining the groove, with the peptide termini projecting from the ends of the site. This conformation places several of the peptide side chains into pockets within the binding site, and directs others outward for interaction with the T cell receptor antigen binding site (Figure 3B). The peptide side chains in the 1, 4, 6, and 9 positions (conventionally numbered from the aromatic or large hydrophobic side chain near the peptide N-terminus that binds into a prominent pocket in DR1) bind into MHC pockets, peptide side chains at the -1, 5, and 8 positions project away from the site, and peptide side chains at the 2, 3, 7, and 10 positions are accommodated by shallow pockets or shelves (Figure 4).
Figure 3.
Structure of the class II MHC – peptide complex. (A) Extracellular domain containing alpha (green) and beta (cyan) subunits and bound peptide. Short peptide sequences connecting the extracellular domain to the membrane spanning regions extend from the alpha and beta termini at the bottom of the figure. (B) Overlay of DR1-peptide complexes, with peptide N-terminus to the left and the MHC beta subunit in front. All peptides adopt essentially the same conformation, although variation at the peptide termini can be seen.
Figure 4.
Pockets in the HLA-DR binding site.. HLA-DR1 surface shown in tan (A,B) or cyan (C). Bound influenza peptide shown in CPK ball (A,C) or stick (B) representation. (B) Peptide side chains accessible to TCR interaction are visible in this top view, at positions P-1, P2, P5, and P8, with partially buried side chains at P3, P7, and P10. (B) Peptide side chains bound into MHC pockets at the P1, P4, P6, and P9 pockets are evident when the peptide is represented by sticks. Peptide side chains at P3, P7, and P10 are partially buried. (C) Side cut-away view of the same complex, with side chain preferences of the P1, P4, P6, and P9 pockets indicated.
The first crystal structures of class II MHC-peptide complexes helped to resolve contradictory descriptions of the sequence determinants for binding peptides, which in retrospect were caused by the difficulty in aligning anchor residues of known peptide binders. Since for class II MHC proteins the peptide termini extend out of the binding site, and typical naturally-processed or experimentally determined minimally immunogenic peptides average 15 residues in length, much longer than the 9–10 residue stretch making specific MHC-peptide contacts, any particular peptide potentially can bind in multiple registers. However, with the realization that the structure of the class II MHC binding site and the conformation of bound peptides restrict peptide-MHC contacts to particular positions (Figure 4), it became possible to more easily align peptide sequences to reveal key residues. MHC-bound peptides adopt a characteristic polyproline type II like conformation, with approximately three-residue repeating pattern, because of the location and chemical identity of key MHC hydrogen bounding residues [42]. These interactions all involve conserved aspects of the peptide (i.e. the main chain amide bonds) and conserved class II MHC residues, and would therefore be expected to direct the same peptide conformation and pattern of key MHC-peptide contacts in all complexes regardless of allelic variants or peptide sequences. This prediction has been confirmed in all crystal structures determined to date, and is the basis for motif-based prediction of peptide binding preferences of allelic variants that have not been directly characterized experimentally, and for predicting T cell epitopes from self and foreign peptide sequences.
To date, no bulges or buckles interrupting the canonical polyproline-II pattern characteristic of class II MHC-bound peptides have been observed. This contrasts the situation with class I MHC proteins, where peptide bulges and conformational variation are common, a result of the major MHC-peptide contacts being clustered at the peptide termini rather than distributed throughout the length of the peptide [43]. The most variant conformer for an MHC-bound peptide to date is for the complex of HLA-DR1 with a 17 residue peptide derived from HIV-gag, in which the residues in the P10-P14 position adopt a hairpin conformation as the peptide exits the binding site, aligning the P14 residue near the position usually occupied by the P8 side chain [44]. Nonetheless, the conformation within the binding site and the pattern of key MHC-peptide contacts are conventional for this complex. In a few cases of auto-immune and allo-immune class II MHC complexes, the bound peptides exit the site prematurely with weakened interactions in the P7-P10 region [45–47] and for an murine autoimmune complex (I-Au bound to an N-acetylated peptide from myelin basic protein) the bound peptide also leaves the P1 and P2 sites empty [47]. Whether the altered immunoreactivity observed in these peptides is related to their unconventional binding arrangements remains a matter of conjecture, but in the portions actually bound in the site, the interactions are similar to those observed in conventional complexes. However, in one case, Unanue and colleagues have provided extensive characterization of a peptide that apparently can adopt different conformations in a class II MHC binding site. The peptide, derived from hen egg lysozyme, is binds to I-A(k) in two different conformers, one recognized by conventional T cells and another recognized by “type B” T cells [48]. While there is no direct structural characterization of the type B-reactive species, it appears to be a less-stable conformer that is sensitive to editing by HLA-DM, and is observed only under conditions where the influence of HLA-DM is minimized. The possibility of such alternative, HLA-DM-sensitive conformations has implications for the development of peptide-based vaccines, since a vaccine peptide might load onto class II MHC molecules on the cell surface, or in HLA-DM-negative intracellular compartments, and elicit type B-like T cell responses that would not cross-react with pathogen-derived peptides conventionally loaded onto the MHC molecules in the presence of HLA-DM.
MHC peptide binding preferences
The chemical and steric character of the residues that line the side-chain binding pockets determine which peptide sequences can be accommodated in the binding site. Because much of the polymorphism in class II MHC sequence is localized in the pocket regions, different class II MHC allelic variants have different peptide binding preferences. Such peptide binding preferences, or “motifs” have been described, with varying degrees of accuracy, for many human class II allelic variants, and are a great help in understanding and predicting CD4+ responses to pathogenic and vaccine challenge.
Peptide binding motifs for class II MHC proteins have been characterized by various methods (Table I). For some class II MHC variants, mass spectrometry and microchemical separations have been used to characterize the spectrum of peptides bound to mature MHC proteins present on the surface of antigen presenting cells, usually as part of studies characterizing antigen processing pathways. Sequence motifs present in the resultant lists of naturally processed peptides reflect MHC binding preferences (along with sequence preferences in the processing machinery), and have also been used to develop allele-specific binding motifs [49]. Many of these motifs have been collected in the Syfpeithi database (http://hwww.syfpeithi.de) In general, these studies of naturally processed peptides have been performed mostly in B lymphobastoid lines, although occasional studies of other sorts of antigen-presenting cells have appeared [50].
Table I.
Some approaches to predicting HLA-DR peptide binding
| Approach | Name | Notes | References |
|---|---|---|---|
| Endogenous peptide sequences | SYFPEITHI | Preferred residues at particular positions | [1] |
| IC50 assay of single amino acid variants of test peptides | TEPITOPE ProPred | Includes “virtual” matrices for some alleles by pocket combination | [2, 3] |
| IC50 assay of large sets of varied peptide sequences | ARB, SMM - align | Position-specific scoring matrices | [4, 5] |
| Analysis of known T cell epitopes | RANKPEP | Position-specific scoring matrices | [6] |
| Analysis of known T cell epitopes | MULTIPRED | Neural network and Hidden Markov model algorithms, identification of promiscuous epitopes | [7] |
| Direct binding of peptide displayed on a phage libraries | “anchor combination” | Position-specific scoring matrices | [8] |
| Binding inhibition by a positional scanning libraries | “undecapeptide library screen” | 10-residue motif developed | [9] |
| Known T cell epitopes, MHC structures and sequences | NetMHCIIPan | Neural network approach to associate binding preferences and MHC sequences | [10] |
Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999; 50:213-9.
Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999; 17:555-61.
Singh H, Raghava GP. ProPred: prediction of HLA?DR binding sites. Bioinformatics. 2001; 17:1236-7.
Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, et al. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005; 57:304-14.
Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007; 8:238.
Reche PA, Glutting JP, Reinherz EL. Prediction of MHC class I binding peptides using profile motifs. Hum Immunol. 2002; 63:701-9.
Zhang GL, Khan AM, Srinivasan KN, August JT, Brusic V. MULTIPRED: a computational system for prediction of promiscuous HLA binding peptides. Nucleic Acids Res. 2005; 33:W172-9.
Hammer J, Belunis C, Bolin D, Papadopoulos J, Walsky R, Higelin J, et al. High-affinity binding of short peptides to major histocompatibility complex class II molecules by anchor combinations. Proc Natl Acad Sci U S A. 1994; 91:4456-60.
Fleckenstein B, Kalbacher H, Muller CP, Stoll D, Halder T, Jung G, et al. New ligands binding to the human leukocyte antigen class II molecule DRB1*0101 based on the activity pattern of an undecapeptide library. Eur J Biochem. 1996; 240:71-7.
Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, et al. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 2008; 4:e1000107.
In another approach, the effects of single amino acid substitutions on MHC binding affinity of a test peptide are evaluated using an in vitro binding assay. Typically, this assay is performed as a set of competition reactions between the test and mutated peptides, with the effect reported as a relative IC50 for every possible substitution at each position in the peptide. The final result is a matrix giving the effect of each possible amino acid at every position in a nine (or ten) residue binding frame, corresponding to the portion of a bound peptide making contact with class II MHC residues. One potential complication of this approach is that the mutated peptides might shift register in the binding site, scrambling the positional specificity of the effects of the substitution. To avoid this, Sinigaglia, Hammer, and colleagues introduced their mutations into a polyalanine-based test peptide designed to bind to DRB1*0401 only in a single register, by virtue of having a tyrosine at first position in the peptide, which was known from earlier studies to be preferred at the P1 but disfavored at the P2 and P3 positions [51]. This technique was applied to several other alleles as sources of purified proteins became available and sufficient information was collected to design test peptides (DR1, DR4). However, because of the great variation of MHC sequences, and the difficulty in producing recombinant proteins for the binding assays, binding motifs have been determined by this method only for a small fraction of class II MHC allelic variants.
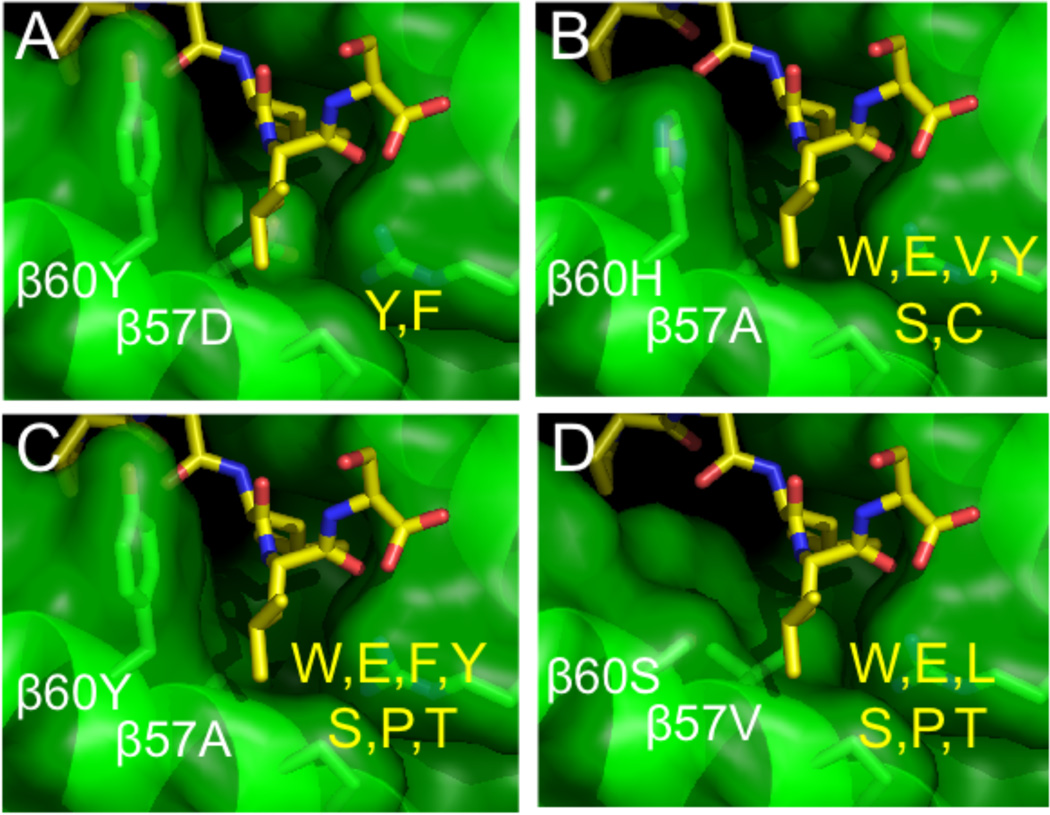
The method was extended by Sturniolo et al. to allelic variants that were not available for direct experimental testing, but which shared pocket residues with well-characterized variants [52]. Under the assumption that the peptide binding pockets have independent effects, pocket-specific binding preferences were combined to form “virtual matrices” for new alleles. For example, DRB1*1321 has all the same P1 pocket residues as DRB1*0101, P4 residues as DRB1*1101, P6 residues as DRB1*0301, and P9 residues as DRB1*0801. As each of these variants has been characterized in detail, their pocket preferences can be combined to predict those of new DRB1*1321 variant. In this manner “virtual” binding motifs for ~50 common DR allotypes were developed, and are available in the “TEPITOPE” program [52] and in a similar form from the ProPred server (http://www.imtech.res.in/raghava/propred/) [53]. It should be noted that this approach implicitly considered that contributions of the various pockets be independent, but cooperativity has been observed in particular cases [54], and would be expected to complicate motif-based analysis of peptide binding preferences. We have used a more explicitly structure-based approach to extend known binding preferences to additional pockets and allelic variants [55]. In a study of binding specificity in the shallow P10 pocket or “shelf”, for which MHC-peptide contacts were observed but for which specificity had not been analyzed in detail, MHC variants carrying substitutions in the P10 pocket spanning the range of natural sequence variation were constructed and tested against series of peptides carrying P10 substitutions (Figure 5). The resultant sequence preferences were added to a TEPITOPE-like algorithm, resulting in a small but significant improvement in binding predictions [56].
Figure 5.
Variability and side-chain binding preferences of the P10 pocket. (A) Crystal structure, and (B–D), computational models, of the P10 “minor” pocket or shelf. Polymorphism at beta subunit residues 57 and 60 leads to different side chain binding preferences at this position (shown in yellow). From Zavala-Ruiz et al [55].
In yet another approach to characterize allele-specific peptide binding motifs, binding data obtained for large sets of natural peptide sequences are studied instead of a series of designed point mutations. Patterns among the binding peptides are used to determine position-specific peptide binding preferences. In some studies, quantitative binding data from hundreds to thousands of in vitro competition assays have been used. With the development of publicly-available databases of MHC-peptide binding data, a variety of computational approaches have been used to derive binding motifs from these data [57, 58]. Similar approaches have been applied to development of motifs from more qualitative datasets, such as lists of known epitopes [59, 60] or hits in positional scanning [61] and phage display [62] libraries. This approach also has been expanded to predict binding preferences for allelic variants not directly studied in the NetMHCIIPan algorithm, which used a neural network approach to associate sequences with binding specificities [63], rather than an explicit pocket mapping as in the TEPITOPE approach.
Identification of class II MHC epitopes
Often in vaccine research one is interested in defining the targets of CD4+ T cell responses elicited by vaccination or natural infection. Classically, CD4+ T cell responses are identified by challenging PBMC (peripheral blood mononuclear cells) or PBMC-derived cells lines with a series of overlapping synthetic peptides that cover the entire sequence of a protein or the whole set of proteins expressed by an organism. The overlaps are designed so that every potential T cell epitope is present on at least one peptide. Peptides able to induce CD4+ T cell proliferation or cytokine production (or occasionally other T cell responses) are considered candidate epitopes. Additional studies are required to establish the MHC specificity, since PBMCs from most individuals express multiple class II MHC proteins. Epitope validation studies can include inhibition studies using antibodies specific for HLA-DR, HLA-DP or HLA-DQ, isolation of epitope specific T cell lines or clones, studies of cross-reactivity with epitopes processed from native proteins or pathogens, and class II MHC tetramer binding studies. Many epitopes from influenza, HIV, and other small-genome pathogens have been identified in this way. A database of known epitopes has recently been developed (IEDB, http://www.immuneepitope.org) [64].
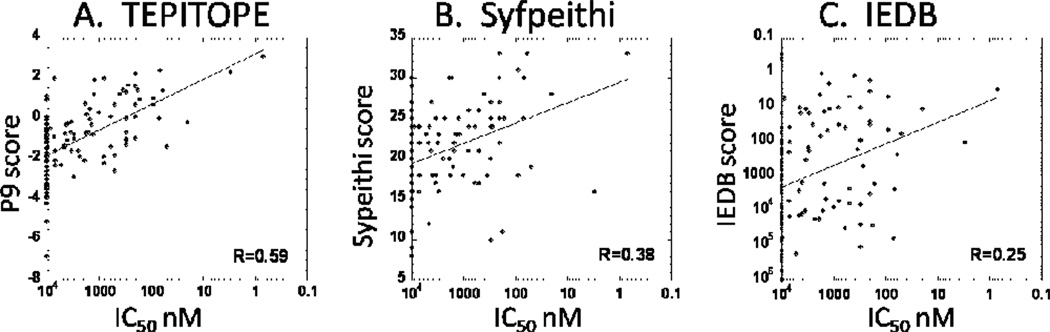
For pathogens with large genomes this systematic approach for epitope mapping is not practical due to the large number of peptides required. For example, a large DNA virus like vaccinia can have as many as 50,000 potential 9-mer epitopes, which would require ~5000 overlapping 20-mers. Even if the peptides were assayed in pools, practical and ethical considerations limit the amount of PBMC available for epitope determination. Bacterial and protozoan pathogens, with much larger genomes, are even more impractical. In many cases investigators have used MHC-peptide binding prediction approaches described above to limit the number of peptides to be screened in cellular assays with samples from immune donors. This approach obviously will be limited by the accuracy of the predictions. Somewhat surprisingly, there have not been many systematic examinations of this issue for class II MHC binding predictors, despite the widespread use of these motifs in epitope prediction. In part this is due to the difficulty in finding independent datasets for statistically valid testing, particularly for algorithms designed using all available published data, and by the difficulty in identifying the relevant 9-mer binding frames within the longer peptides tested experimentally. In Figure 6 we show HLA-DR1 (DRB1*0101) binding predictions and experimental HLA-DR1 binding data for a series of peptides derived from the gE surface protein of varicella zoster (chicken pox) virus gE (our unpublished data), from human glutamic acid decarboxylase [65], a suspected diabetes autoantigen, and from a major honeybee venom allergen [66]. The prediction algorithms evaluated are representative of the motifs developed from different sources of information: panel A shows a motif derived from binding studies of single amino acid variants of a test peptide (TEPITOPE), panel B from alignment of naturally processed peptides eluted from purified MHC molecules (Syfpeithi), and panel C from binding studies of a large series of synthetic peptides (IEDB). In all cases, there is significant non-random correlation between the predicted and observed binding behavior, but the overall correspondence is fairly low. Tight binders are found more frequently among the high-scoring peptides for all algorithms, but are also found among the low scoring sets. Very recently two systematic evaluations of class II MHC binding predictions have appeared, including a comprehensive test of several available algorithms against all peptide binding data collected to date in the Immune Epitope database [67], and one testing more algorithms against a smaller set of high-quality binding data [68]. The predictive power observed in those studies is similar to that shown in Figure 6.
Figure 6.
Comparison of DR1 binding predictions with actual binding data. IC50 vales shown were determined for overlapping peptide series from Varicella (chickenpox) virus glycoprotein E antigen (A. Arvin, D. DeOliveira, L. Stern, unpublished), glucose decarboxylase autoantigen [65] and honeybee venom allergen [66]. Values were normalized with the influenza HA307–319 peptide common to all studies. Syfpeithi scores are on an arbitrary linear scale, IEDB scores reflect estimated IC50 values, and TEPITOPE scores are on a linear scale reflecting the binding relative to a test polyalanine-based peptide. None of the algorithms provide highly reliable predictions of binding affinity as measured by IC50 values, although consensus approaches based on these algorithms have proven useful in efficient prediction of T cell epitopes.
Accurate prediction of class II MHC binding preference is limited by several intrinsic and extrinsic factors. Approaches based on competition assay data have been criticized because the assay cannot be conducted under equilibrium conditions due to the extremely long lifetimes (>200 hrs) of tight-binding test peptides [69], and so can give different results for different investigators depending on experimental conditions. In addition, the molecular property measured, the equilibrium dissociation constant, might be less relevant to immunogenicity than other properties of the MHC-peptide interaction, such as the dissociation lifetime [70]. However, kinetic data which might more suitable are available only for few MHC-peptide complexes. As noted above, many peptides can bind to class II MHC proteins in multiple registers, and uncertainly in the actual register bound complicates straightforward use of any experimental binding data. For the naturally processed peptide data, factors other than MHC-peptide interaction influence the selection of peptides, including peptide abundance in the antigen presenting cell, susceptibility to endosomal proteolysis, and different effects of the peptide exchange factor HLA-DM on different peptides [71]. As these factors differentially affect the various predictive approaches, they might, to some extent, be mitigated by a consensus approach that combines data from different sources. Such consensus algorithms recently have been implemented [56, 67] and appear to provide significantly improved predictive performance as compared to single algorithm approaches.
Despite these uncertainties, motif-based predictions can contribute significantly to epitope identification. In a recent study of CD4+ vaccinia immunity induced by vaccination, we used a consensus approach based on Syfpeithi and TEPITOPE-like algorithms to identify 36 epitopes strongly predicted to be bound by HLA-DR1, of which 25 were recognized by immune donors [56]. This very high predictive accuracy was unexpected, but appears to result from the very stringent selection of epitopes scoring in the top range by multiple algorithms, and was also observed in several independent test cases [56]. Two of the high scoring peptides additionally subsequently were observed to be present among the set of naturally processed vaccinia-derived peptides eluted from infected cells [72], Iending additional credence to this approach.
Malaria, a widespread disease of the tropics that is transmitted by mosquitoes infected with parasites of the genus Plasmodium and for which as many as 500 million cases result in 1 million deaths per year [73] is a particularly challenging case for epitope identification, as more than 5000 proteins are encoded by the genome, and there are hundreds of thousands of possible CD4+ T cell epitopes. Moreover, identification of CD4+ epitopes from malaria is urgently required to track various vaccine approaches, and to evaluate candidates for subunit vaccines. In a study of malaria epitopes recognized by immune donors, Doolan et al first used proteomic approaches to identify 27 highly-expressed candidate antigens, and then used HLA-DR binding predictions to identify 723 predicted HLA-DR binders [74]. Of these, 39 peptides binding tightly to HLA-DR variants derived from four newly-identified antigenic targets were identified [74]. This application of proteomics and bioinformatics to epitope identification seems particularly powerful, and is likely to prove useful in other applications, particularly as consensus motif prediction approaches evolve and become accessible to a wider subset of the immunological community.
Peptide vaccines
We have shown how structural studies provided a conceptual framework for understanding of the peptide-MHC interaction and the development of MHC binding motifs. Chemical synthesis of peptides provides a potential method to develop low cost and well defined subunit vaccines based on these motifs. Studies in animal models in the 1980’s demonstrated the viability of synthetic peptides as the ultimate subunit vaccine [75]. Malaria was, and probably is, for several reasons the perfect model to evaluate this nascent technology. First, naturally-induced immunity to malaria provides some degree of protection to the disease, but not to the infection. Second, although robust immunity can be obtained with inactivated parasites, there are not yet methods to obtain large numbers of this organism for formulation in an affordable vaccine that could be easily distributed and used in malaria-infested regions. Peptide-based vaccines inducing responses directed at the same epitopes as elicited by inactivated parasites might be expected to provide similar immunity. Third, and most important, short linear peptide sequences that could be synthesized in large quantities at low cost were recognized by neutralizing antibodies, suggesting that protective antibody as well as T-cell responses might be induced by peptide vaccination.
The first clinical trials of peptide vaccines were carried out at the end of the 1980’s, not surprisingly with peptides based on malaria parasites protein [76–78]. These trials were designed to test in humans the ability of the synthetic peptides to induce protective antibodies. We will refer here to the results of a vaccine designed to target the sporozoite, the stage of the parasite inoculated by infected mosquitoes. The target of protective anti-sporozoite antibodies are short repetitive units located in the central part of the circumsporozoite (CS) protein. In the CS protein of the most important human malaria parasite, P. falciparumthe repeats are represented by the sequence (NANP) [79, 80] and the minimal B cell epitope consists of 3 repetitive units. Although antibodies recognizing the (NANP) repeats are observed in most individuals, the antibody levels T cell responses to the repetitive units are usually low. This limitation was solved by coupling the synthetic (NANP)3 peptide to tetanus toxoid protein as a carrier to generate the (NANP)3-TT conjugate. In the first clinical trial of the (NANP)3-TT conjugate, a large fraction of the immunized volunteers generated antibodies that recognized the (NANP)3 peptide and the CS protein on the parasite surface [21, 78]. Even more importantly, some volunteers were protected against challenge with parasites. These findings were considered a significant development and the basis for a much larger trial that included more than 200 individuals [81]. However in this second trial, the (NANP)3-TT conjugated elicited low anti-parasite antibody titers that could not be boosted and the tetanus toxoid carrier enhanced and suppressed responses in volunteers to the (NANP)3 peptide. These findings suggested that subunit vaccines should preferentially include pathogen-derived T cell epitopes that could be boosted by natural infection.
A search for T cell epitopes in the CS protein that could be used instead of TT resulted in the identification of strong T helper epitopes in the C-terminal region of the protein. One of these T cell epitopes was recognized by CD4+ T cells in the context of multiple MHC class II molecules from humans and mice [82] -- a novelty, since at this time only a few peptides were observed to bind to more than one MHC allele [83–85]. Shortly after, other epitopes with similar properties were identified [86–89]. These epitopes were designated as promiscuous or universal T cell epitopes [90]. Proof that such universal epitopes could be used to provide broadly based immunity in individuals of varied MHC haplotype was provided some years later by immunization of humans with a second generation of synthetic malaria vaccine [12]. This minimal subunit vaccine included just a universal T helper epitope (different from the one mentioned above and designated as a T*) and repeats of the P. falciparum CS protein. This vaccine induced high antibodies titers in 7/10 randomly selected volunteers.
The discovery of universal T helper epitopes mitigated a major potential limitation of peptide vaccines in application to open human population, that the extremely high variability of human MHC genes might restrict T cell recognition of particular peptide sequences to individuals carrying only certain MHC allelic variants. Such epitopes can bind to a wide variety of MHC variants with affinity sufficient to allow immune recognition; to be termed universal epitope the promiscuity should extend to a sufficient number of MHC variants that a substantial fraction of a population of interest will carry those variants. Initially, studies with truncated peptides suggested that universal helper epitopes contained a series of close overlapping frames for class II MHC antigen presentation [91]. However, based on current understanding of MHC structure and MHC-peptide interactions, the broad MHC-reactivity of universal peptide epitopes can be seen to be due also to degeneracy in binding motifs, particularly for HLA-DR restricted epitopes. For most if not all HLA-DR alleles, the major determinant of binding is the P1 pocket. Of the 9 MHC residues that line the P1 pocket, six (a7I, a24F,a31I,a32F,a34F, a52A) are contributed by the non-polymorphic alpha chain [92], and one from the beta chain (b89F) is non-polymorphic among the 559 HLA-DRB alleles described to date. Of the remaining two positions, b86 is di-morphic, with all known alleles having either Gly or Val, while b85 usually is Val but occasionally Ala. The 85 and 86 substitutions are known to direct the specificity of the P1 pocket: Gly85/Val86 (GV) alleles prefer large aromatic and aliphatic side chains, whereas AV and VV alleles prefer small aromatic and aliphatic side chains. At the other P4, P6, P7, and P9 pockets, much of the binding specificity is negative, i.e. certain side chains interfere with binding, and consensus residues able to bind to many alleles can be found. Thus, universal HLA-DR epitopes tend to have Tyr, Phe or Leu at the P1 position, and small or uncharged residues at the P4, P6, P7, and P9 positions.
A totally synthetic pan DR helper T cell epitope (PADRE) designed along these lines carries a non-native cyclohexylalanine observed to bind well to all P1 pocket variants at the P1 position and small consensus side chains at the P3, P4, P6, and P7 positions, as well as bulky and charged side chains at T cell contact positions P2, P5, and P8. PADRE was shown to bind to 15 of 16 MHC alleles tested [93], and to induce in vitro T cell responses in PBMCs from 90% of the normal donors tested [94]. The strong helper activity of this epitope has been shown to be of great use in the generation of optimal CTLs responses in subunit vaccines for cancer [95, 96], in overcoming tolerance for the generation of CTLs in a HBV vaccine model [97] and antibody responses for TNF-a [98], and recently in Alzheimer’s disease [99]. In this last case, the use of PADRE as T helper epitope in an experimental vaccine that includes also a B cell epitope from the B-amyloid protein have shown to induce in mice protective antibodies without the autoimmune T cell responses generated when T cell epitopes from the β-amyloid protein are included [99]. Similar considerations can be used to help identification of naturally occurring universal or near-universal epitopes in native pathogen and cancer antigens, and to help guide design of improved T cell epitopes for inclusion in peptide and subunit vaccines [100–102].
Conclusions
Structural and biochemical studies of peptides bound to HLA-DR molecules provided a conceptual frame for the understanding of the factors that determine the binding of peptides to class II molecules. This information has been used to develop virtual matrixes incorporated in algorithms that predict peptide binding to MHC molecules. Although many researchers consider these programs to be of limited use, the predictive power recently has improved considerably with the development of consensus approaches, and published studies demonstrate that these programs can be of great use in cases where comprehensive screening of all potential epitopes is not possible. These studies also have provided a rational course to select potent and broadly recognized T cell epitopes for inclusion in subunit vaccines, to enhance suboptimal epitopes, and to develop immunotherapeutic approaches for cancer and other diseases. Continued improvement in class II MHC epitope prediction strategies is likely to lead to continued advances in these areas.
Acknowledgements
The authors thank Maria-Dorothea Nastke, Walter Kim, and Corrie Painter for critical reading of the manuscript. We also thank Daniel DeOliveira for peptide binding studies and Zarixia Zavala-Ruiz for preparation of some figures.
This work was supported by NIH-U19-057319.
References
- 1.Amanna IJ, Messaoudi I, Slifka MK. Protective immunity following vaccination: how is it defined? Hum Vaccin. 2008;4:316–319. doi: 10.4161/hv.4.4.5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambkin R, Novelli P, Oxford J, Gelder C. Human genetics and responses to influenza vaccination: clinical implications. Am J Pharmacogenomics. 2004;4:293–298. doi: 10.2165/00129785-200404050-00002. [DOI] [PubMed] [Google Scholar]
- 3.Molano A, Park SH, Chiu YH, Nosseir S, Bendelac A, Tsuji M. Cutting edge: the IgG response to the circumsporozoite protein is MHC class II-dependent and CD1-dindependent: exploring the role of GPIs in NK T cell activation and antimalarial responses. J Immunol. 2000;164:5005–5009. doi: 10.4049/jimmunol.164.10.5005. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira GA, Kumar KA, Calvo-Calle JM, Othoro C, Altszuler D, Nussenzweig V, et al. Class II-restricted protective immunity induced by malaria sporozoites. Infect Immun. 2008;76:1200–1206. doi: 10.1128/IAI.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu R, Johnson AJ, Liggitt D, Bevan MJ. Cellular and humoral immunity against vaccinia virus infection of mice. J Immunol. 2004;172:6265–6271. doi: 10.4049/jimmunol.172.10.6265. [DOI] [PubMed] [Google Scholar]
- 6.Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, Davies DH, Johnston RJ, et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008;28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poland GA, Ovsyannikova IG, Jacobson RM. Personalized vaccines: the emerging field of vaccinomics. Expert Opin Biol Ther. 2008;8:1659–1667. doi: 10.1517/14712598.8.11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poland GA, Ovsyannikova IG, Jacobson RM. Vaccine immunogenetics: bedside to bench to population. Vaccine. 2008;26:6183–6188. doi: 10.1016/j.vaccine.2008.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ovsyannikova IG, Jacobson RM, Dhiman N, Vierkant RA, Pankratz VS, Poland GA. Human leukocyte antigen and cytokine receptor gene polymorphisms associated with heterogeneous immune responses to mumps viral vaccine. Pediatrics. 2008;121:e1091–e1099. doi: 10.1542/peds.2007-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 11.Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 12.Nardin EH, Calvo-Calle JM, Oliveira GA, Nussenzweig RS, Schneider M, Tiercy JM, et al. A totally synthetic polyoxime malaria vaccine containing Plasmodium falciparum B cell and universal T cell epitopes elicits immune responses in volunteers of diverse HLA types. J Immunol. 2001;166:481–489. doi: 10.4049/jimmunol.166.1.481. [DOI] [PubMed] [Google Scholar]
- 13.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho LH, Sano G, Hafalla JC, Morrot A, Curotto de Lafaille MA, Zavala F. IL-4-secreting CD4+ T cells are crucial to the development of CD8+ T-cell responses against malaria liver stages. Nat Med. 2002;8:166–170. doi: 10.1038/nm0202-166. [DOI] [PubMed] [Google Scholar]
- 15.Kemball CC, Pack CD, Guay HM, Li ZN, Steinhauer DA, Szomolanyi-Tsuda E, et al. The antiviral CD8+ T cell response is differentially dependent on CD4+ T cell help over the course of persistent infection. J Immunol. 2007;179:1113–1121. doi: 10.4049/jimmunol.179.2.1113. [DOI] [PubMed] [Google Scholar]
- 16.Marzo AL, Vezys V, Klonowski KD, Lee SJ, Muralimohan G, Moore M, et al. Fully functional memory CD8 T cells in the absence of CD4 T cells. J Immunol. 2004;173:969–975. doi: 10.4049/jimmunol.173.2.969. [DOI] [PubMed] [Google Scholar]
- 17.van de Berg PJ, van Leeuwen EM, ten Berge IJ, van Lier R. Cytotoxic human CD4(+) T cells. Curr Opin Immunol. 2008;20:339–343. doi: 10.1016/j.coi.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Johnson AJ, Chu CF, Milligan GN. Effector CD4+ T-cell involvement in clearance of infectious herpes simplex virus type 1 from sensory ganglia and spinal cords. J Virol. 2008;82:9678–9688. doi: 10.1128/JVI.01159-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elyaman W, Kivisakk P, Reddy J, Chitnis T, Raddassi K, Imitola J, et al. Distinct functions of autoreactive memory and effector CD4+ T cells in experimental autoimmune encephalomyelitis. Am J Pathol. 2008;173:411–422. doi: 10.2353/ajpath.2008.080142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuji M, Romero P, Nussenzweig RS, Zavala F. CD4+ cytolytic T cell clone confers protection against murine malaria. J Exp Med. 1990;172:1353–1357. doi: 10.1084/jem.172.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington LE, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol. 2006;18:349–356. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. doi: 10.1146/annurev.immunol.22.012703.104538. [DOI] [PubMed] [Google Scholar]
- 23.Chicz RM, Urban RG, Lane WS, Gorga JC, Stern LJ, Vignali DA, et al. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992;358:764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 24.Reich EP, von Grafenstein H, Barlow A, Swenson KE, Williams K, Janeway CA., Jr Self peptides isolated from MHC glycoproteins of non-obese diabetic mice. J Immunol. 1994;152:2279–2288. [PubMed] [Google Scholar]
- 25.Malnati MS, Marti M, LaVaute T, Jaraquemada D, Biddison W, DeMars R, et al. Processing pathways for presentation of cytosolic antigen to MHC class II-restricted T cells. Nature. 1992;357:702–704. doi: 10.1038/357702a0. [DOI] [PubMed] [Google Scholar]
- 26.Vanderwolf CH. Effects of water temperature and core temperature on rat's performance in a swim-to-platform test. Behav Brain Res. 1991;44:105–106. doi: 10.1016/s0166-4328(05)80244-x. [DOI] [PubMed] [Google Scholar]
- 27.Strawbridge AB, Blum JS. Autophagy in MHC class II antigen processing. Curr Opin Immunol. 2007;19:87–92. doi: 10.1016/j.coi.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Glimcher LH, Kara CJ. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 29.Guardiola J, Maffei A. Control of MHC class II gene expression in autoimmune, infectious, and neoplastic diseases. Crit Rev Immunol. 1993;13:247–268. [PubMed] [Google Scholar]
- 30.Ovsyannikova IG, Dhiman N, Jacobson RM, Poland GA. Human leukocyte antigen polymorphisms: variable humoral immune responses to viral vaccines. Expert Rev Vaccines. 2006;5:33–43. doi: 10.1586/14760584.5.1.33. [DOI] [PubMed] [Google Scholar]
- 31.Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA. Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. J Infect Dis. 2006;193:655–663. doi: 10.1086/500144. [DOI] [PubMed] [Google Scholar]
- 32.Guerau-de-Arellano M, Huber BT. Development of autoimmunity in Lyme arthritis. Curr Opin Rheumatol. 2002;14:388–393. doi: 10.1097/00002281-200207000-00009. [DOI] [PubMed] [Google Scholar]
- 33.Tuttle TM, Anderson BW, Thompson WE, Lee JE, Sahin A, Smith TL, et al. Proliferative and cytokine responses to class II HER-2/neu-associated peptides in breast cancer patients. Clin Cancer Res. 1998;4:2015–2024. [PubMed] [Google Scholar]
- 34.Rohn TA, Reitz A, Paschen A, Nguyen XD, Schadendorf D, Vogt AB, et al. A novel strategy for the discovery of MHC class II-restricted tumor antigens: identification of a melanotransferrin helper T-cell epitope. Cancer Res. 2005;65:10068–10078. doi: 10.1158/0008-5472.CAN-05-1973. [DOI] [PubMed] [Google Scholar]
- 35.Ovsyannikova IG, Vierkant RA, Poland GA. Importance of HLA-DQ and HLA-DP polymorphisms in cytokine responses to naturally processed HLA-DR-derived measles virus peptides. Vaccine. 2006;24:5381–5389. doi: 10.1016/j.vaccine.2006.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poland GA, Ovsyannikova IG, Jacobson RM, Vierkant RA, Jacobsen SJ, Pankratz VS, et al. Identification of an association between HLA class II alleles and low antibody levels after measles immunization. Vaccine. 2001;20:430–438. doi: 10.1016/s0264-410x(01)00346-2. [DOI] [PubMed] [Google Scholar]
- 37.Zuckerman JN. Protective efficacy, immunotherapeutic potential, and safety of hepatitis B vaccines. J Med Virol. 2006;78:169–177. doi: 10.1002/jmv.20524. [DOI] [PubMed] [Google Scholar]
- 38.Desombere I, Willems A, Leroux-Roels G. Response to hepatitis B vaccine: multiple HLA genes are involved. Tissue Antigens. 1998;51:593–604. doi: 10.1111/j.1399-0039.1998.tb03001.x. [DOI] [PubMed] [Google Scholar]
- 39.Kruger A, Adams P, Hammer J, Bocher WO, Schneider PM, Rittner C, et al. Hepatitis B surface antigen presentation and HLA-DRB1*-lessons from twins and peptide binding studies. Clin Exp Immunol. 2005;140:325–332. doi: 10.1111/j.1365-2249.2005.02765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mustafa AS. HLA-restricted immune response to mycobacterial antigens: relevance to vaccine design. Hum Immunol. 2000;61:166–171. doi: 10.1016/s0198-8859(99)00137-8. [DOI] [PubMed] [Google Scholar]
- 41.Shams H, Klucar P, Weis SE, Lalvani A, Moonan PK, Safi H, et al. Characterization of a Mycobacterium tuberculosis peptide that is recognized by human CD4+ and CD8+ T cells in the context of multiple HLA alleles. J Immunol. 2004;173:1966–1977. doi: 10.4049/jimmunol.173.3.1966. [DOI] [PubMed] [Google Scholar]
- 42.Jardetzky TS, Brown JH, Gorga JC, Stern LJ, Urban RG, Strominger JL, et al. Crystallographic analysis of endogenous peptides associated with HLA-DR1 suggests a common, polyproline II-like conformation for bound peptides. Proc Natl Acad Sci U S A. 1996;93:734–738. doi: 10.1073/pnas.93.2.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 44.Zavala-Ruiz Z, Strug I, Walker BD, Norris PJ, Stern LJ. A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition. Proc Natl Acad Sci U S A. 2004;101:13279–13284. doi: 10.1073/pnas.0403371101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parry CS, Gorski J, Stern LJ. Crystallographic structure of the human leukocyte antigen DRA, DRB3*0101: models of a directional alloimmune response and autoimmunity. J Mol Biol. 2007;371:435–446. doi: 10.1016/j.jmb.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Li H, Martin R, Mariuzza RA. Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins. J Mol Biol. 2000;304:177–188. doi: 10.1006/jmbi.2000.4198. [DOI] [PubMed] [Google Scholar]
- 47.He XL, Radu C, Sidney J, Sette A, Ward ES, Garcia KC. Structural snapshot of aberrant antigen presentation linked to autoimmunity: the immunodominant epitope of MBP complexed with I-Au. Immunity. 2002;17:83–94. doi: 10.1016/s1074-7613(02)00340-0. [DOI] [PubMed] [Google Scholar]
- 48.Lovitch SB, Unanue ER. Conformational isomers of a peptide-class II major histocompatibility complex. Immunol Rev. 2005;207:293–313. doi: 10.1111/j.0105-2896.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 49.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 50.Rohn TA, Boes M, Wolters D, Spindeldreher S, Muller B, Langen H, et al. Upregulation of the CLIP self peptide on mature dendritic cells antagonizes T helper type 1 polarization. Nat Immunol. 2004;5:909–918. doi: 10.1038/ni1108. [DOI] [PubMed] [Google Scholar]
- 51.Hammer J, Bono E, Gallazzi F, Belunis C, Nagy Z, Sinigaglia F. Precise prediction of major histocompatibility complex class II-peptide interaction based on peptide side chain scanning. J Exp Med. 1994;180:2353–2358. doi: 10.1084/jem.180.6.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, et al. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 53.Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 54.Anderson MW, Gorski J. Cooperativity during the formation of peptide/MHC class II complexes. Biochemistry. 2005;44:5617–5624. doi: 10.1021/bi048675s. [DOI] [PubMed] [Google Scholar]
- 55.Zavala-Ruiz Z, Strug I, Anderson MW, Gorski J, Stern LJ. A polymorphic pocket at the P10 position contributes to peptide binding specificity in class II MHC proteins. Chem Biol. 2004;11:1395–1402. doi: 10.1016/j.chembiol.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 56.Calvo-Calle JM, Strug I, Nastke MD, Baker SP, Stern LJ. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3:1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, et al. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- 58.Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reche PA, Glutting JP, Reinherz EL. Prediction of MHC class I binding peptides using profile motifs. Hum Immunol. 2002;63:701–709. doi: 10.1016/s0198-8859(02)00432-9. [DOI] [PubMed] [Google Scholar]
- 60.Zhang GL, Khan AM, Srinivasan KN, August JT, Brusic V. MULTIPRED: a computational system for prediction of promiscuous HLA binding peptides. Nucleic Acids Res. 2005;33:W172–W179. doi: 10.1093/nar/gki452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fleckenstein B, Kalbacher H, Muller CP, Stoll D, Halder T, Jung G, et al. New ligands binding to the human leukocyte antigen class II molecule DRB1*0101 based on the activity pattern of an undecapeptide library. Eur J Biochem. 1996;240:71–77. doi: 10.1111/j.1432-1033.1996.0071h.x. [DOI] [PubMed] [Google Scholar]
- 62.Hammer J, Belunis C, Bolin D, Papadopoulos J, Walsky R, Higelin J, et al. High-affinity binding of short peptides to major histocompatibility complex class II molecules by anchor combinations. Proc Natl Acad Sci U S A. 1994;91:4456–4460. doi: 10.1073/pnas.91.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, et al. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 2008;4:e1000107. doi: 10.1371/journal.pcbi.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geluk A, van Meijgaarden KE, Schloot NC, Drijfhout JW, Ottenhoff TH, Roep BO. HLA-DR binding analysis of peptides from islet antigens in IDDM. Diabetes. 1998;47:1594–1601. doi: 10.2337/diabetes.47.10.1594. [DOI] [PubMed] [Google Scholar]
- 66.Texier C, Pouvelle-Moratille S, Busson M, Charron D, Menez A, Maillere B. Complementarity and redundancy of the binding specificity of HLA-DRB1, -DRB3, -DRB4 and -DRB5 molecules. Eur J Immunol. 2001;31:1837–1846. doi: 10.1002/1521-4141(200106)31:6<1837::aid-immu1837>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 67.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin HH, Zhang GL, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics. 2008;9( Suppl 12):S22. doi: 10.1186/1471-2105-9-S12-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McFarland BJ, Beeson C. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med Res Rev. 2002;22:168–203. doi: 10.1002/med.10006. [DOI] [PubMed] [Google Scholar]
- 70.Lazarski CA, Chaves FA, Jenks SA, Wu S, Richards KA, Weaver JM, et al. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 71.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 72.Strug I, Calvo-Calle JM, Green KM, Cruz J, Ennis FA, Evans JE, et al. Vaccinia peptides eluted from HLA-DR1 isolated from virus-infected cells are recognized by CD4+ T cells from a vaccinated donor. J Proteome Res. 2008;7:2703–2711. doi: 10.1021/pr700780x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Malaria WG, Programme. World malaria report 2008: World Health Organization. 2008
- 74.Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Audibert F, Jolivet M, Chedid L, Arnon R, Sela M. Successful immunization with a totally synthetic diphtheria vaccine. Proc Natl Acad Sci U S A. 1982;79:5042–5046. doi: 10.1073/pnas.79.16.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herrington DA, Clyde DF, Losonsky G, Cortesia M, Murphy JR, Davis J, et al. Safety and immunogenicity in man of a synthetic peptide malaria vaccine against Plasmodium falciparum sporozoites. Nature. 1987;328:257–259. doi: 10.1038/328257a0. [DOI] [PubMed] [Google Scholar]
- 77.Patarroyo ME, Amador R, Clavijo P, Moreno A, Guzman F, Romero P, et al. A synthetic vaccine protects humans against challenge with asexual blood stages of Plasmodium falciparum malaria. Nature. 1988;332:158–161. doi: 10.1038/332158a0. [DOI] [PubMed] [Google Scholar]
- 78.Etlinger HM, Felix AM, Gillessen D, Heimer EP, Just M, Pink JR, et al. Assessment in humans of a synthetic peptide-based vaccine against the sporozoite stage of the human malaria parasite, Plasmodium falciparum. J Immunol. 1988;140:626–633. [PubMed] [Google Scholar]
- 79.Dame JB, Williams JL, McCutchan TF, Weber JL, Wirtz RA, Hockmeyer WT, et al. Structure of the gene encoding the immunodominant surface antigen on the sporozoite of the human malaria parasite Plasmodium falciparum. Science. 1984;225:593–599. doi: 10.1126/science.6204383. [DOI] [PubMed] [Google Scholar]
- 80.Weber JL, Hockmeyer WT. Structure of the circumsporozoite protein gene in 18 strains of Plasmodium falciparum. Mol Biochem Parasitol. 1985;15:305–316. doi: 10.1016/0166-6851(85)90092-1. [DOI] [PubMed] [Google Scholar]
- 81.Herrington DA, Clyde DF, Davis JR, Baqar S, Murphy JR, Cortese JF, et al. Human studies with synthetic peptide sporozoite vaccine (NANP)3-TT and immunization with irradiated sporozoites. Bull World Health Organ. 1990;68(Suppl):33–37. [PMC free article] [PubMed] [Google Scholar]
- 82.Sinigaglia F, Guttinger M, Kilgus J, Doran DM, Matile H, Etlinger H, et al. A malaria T-cell epitope recognized in association with most mouse and human MHC class II molecules. Nature. 1988;336:778–780. doi: 10.1038/336778a0. [DOI] [PubMed] [Google Scholar]
- 83.Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. Nature. 1985;317:359–361. doi: 10.1038/317359a0. [DOI] [PubMed] [Google Scholar]
- 84.Guillet JG, Lai MZ, Briner TJ, Smith JA, Gefter ML. Interaction of peptide antigens and class II major histocompatibility complex antigens. Nature. 1986;324:260–262. doi: 10.1038/324260a0. [DOI] [PubMed] [Google Scholar]
- 85.Buus S, Colon S, Smith C, Freed JH, Miles C, Grey HM. Interaction between a"processed" ovalbumin peptide and Ia molecules. Proc Natl Acad Sci U S A. 1986;83:3968–3971. doi: 10.1073/pnas.83.11.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol. 1989;19:2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- 87.Busch R, Strang G, Howland K, Rothbard JB. Degenerate binding of immunogenic peptides to HLA-DR proteins on B cell surfaces. Int Immunol. 1990;2:443–451. doi: 10.1093/intimm/2.5.443. [DOI] [PubMed] [Google Scholar]
- 88.O'Sullivan D, Sidney J, Appella E, Walker L, Phillips L, Colon SM, et al. Characterization of the specificity of peptide binding to four DR haplotypes. J Immunol. 1990;145:1799–1808. [PubMed] [Google Scholar]
- 89.Moreno A, Clavijo P, Edelman R, Davis J, Sztein M, Sinigaglia F, et al. CD4+ T cell clones obtained from Plasmodium falciparum sporozoite-immunized volunteers recognize polymorphic sequences of the circumsporozoite protein. J Immunol. 1993;151:489–499. [PubMed] [Google Scholar]
- 90.Panina-Bordignon P, Demotz S, Corradin G, Lanzavecchia A. Study on the immunogenicity of human class-II-restricted T-cell epitopes: processing constraints, degenerate binding, and promiscuous recognition. Cold Spring Harb Symp Quant Biol. 1989;54 Pt 1:445–451. doi: 10.1101/sqb.1989.054.01.053. [DOI] [PubMed] [Google Scholar]
- 91.Kilgus J, Jardetzky T, Gorga JC, Trzeciak A, Gillessen D, Sinigaglia F. Analysis of the permissive association of a malaria T cell epitope with DR molecules. J Immunol. 1991;146:307–315. [PubMed] [Google Scholar]
- 92.Murthy VL, Stern LJ. The class II MHC protein HLA-DR1 in complex with an endogenous peptide: implications for the structural basis of the specificity of peptide binding. Structure. 1997;5:1385–1396. doi: 10.1016/s0969-2126(97)00288-8. [DOI] [PubMed] [Google Scholar]
- 93.Alexander J, Fikes J, Hoffman S, Franke E, Sacci J, Appella E, et al. The optimization of helper T lymphocyte (HTL) function in vaccine development. Immunol Res. 1998;18:79–92. doi: 10.1007/BF02788751. [DOI] [PubMed] [Google Scholar]
- 94.Alexander J, Sidney J, Southwood S, Ruppert J, Oseroff C, Maewal A, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 95.Daftarian P, Mansour M, Benoit AC, Pohajdak B, Hoskin DW, Brown RG, et al. Eradication of established HPV 16-expressing tumors by a single administration of a vaccine composed of a liposome-encapsulated CTL-T helper fusion peptide in a water-in-oil emulsion. Vaccine. 2006;24:5235–5244. doi: 10.1016/j.vaccine.2006.03.079. [DOI] [PubMed] [Google Scholar]
- 96.Bettahi I, Dasgupta G, Renaudet O, Chentoufi AA, Zhang X, Carpenter D, et al. Antitumor activity of a self-adjuvanting glyco-lipopeptide vaccine bearing B cell, CD4+ and CD8+ T cell epitopes. Cancer Immunol Immunother. 2009;58:187–200. doi: 10.1007/s00262-008-0537-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Livingston BD, Alexander J, Crimi C, Oseroff C, Celis E, Daly K, et al. Altered helper T lymphocyte function associated with chronic hepatitis B virus infection and its role in response to therapeutic vaccination in humans. J Immunol. 1999;162:3088–3095. [PubMed] [Google Scholar]
- 98.Nielsen FS, Sauer J, Backlund J, Voldborg B, Gregorius K, Mouritsen S, et al. Insertion of foreign T cell epitopes in human tumor necrosis factor alpha with minimal effect on protein structure and biological activity. J Biol Chem. 2004;279:33593–33600. doi: 10.1074/jbc.M403072200. [DOI] [PubMed] [Google Scholar]
- 99.Agadjanyan MG, Ghochikyan A, Petrushina I, Vasilevko V, Movsesyan N, Mkrtichyan M, et al. Prototype Alzheimer's disease vaccine using the immunodominant B cell epitope from beta-amyloid and promiscuous T cell epitope pan HLA DR-binding peptide. J Immunol. 2005;174:1580–1586. doi: 10.4049/jimmunol.174.3.1580. [DOI] [PubMed] [Google Scholar]
- 100.Okazaki T, Pendleton CD, Sarobe P, Thomas EK, Iyengar S, Harro C, et al. Epitope enhancement of a CD4 HIV epitope toward the development of the next generation HIV vaccine. J Immunol. 2006;176:3753–3759. doi: 10.4049/jimmunol.176.6.3753. [DOI] [PubMed] [Google Scholar]
- 101.Berzofsky JA, Ahlers JD, Belyakov IM. Strategies for designing and optimizing new generation vaccines. Nat Rev Immunol. 2001;1:209–219. doi: 10.1038/35105075. [DOI] [PubMed] [Google Scholar]
- 102.Cifuentes G, Bermudez A, Rodriguez R, Patarroyo MA, Patarroyo ME. Shifting the polarity of some critical residues in malarial peptides' binding to host cells is a key factor in breaking conserved antigens' code of silence. Med Chem. 2008;4:278–292. doi: 10.2174/157340608784325160. [DOI] [PubMed] [Google Scholar]