SUMMARY
Optical imaging of the dynamics of living specimens involves tradeoffs between spatial resolution, temporal resolution, and phototoxicity, made more difficult in three-dimensions. Here, however, we report that rapid 3D dynamics can be studied beyond the diffraction limit in thick or densely fluorescent living specimens over many time points by combining ultra-thin planar illumination produced by scanned Bessel beams with superresolution structured illumination microscopy. We demonstrate in vivo karyotyping of chromosomes during mitosis and identify different dynamics for the actin cytoskeleton at the dorsal and ventral surfaces of fibroblasts. Compared to spinning disk confocal microscopy, we demonstrate substantially reduced photodamage when imaging rapid morphological changes in D. discoideum cells, as well as improved contrast and resolution at depth within developing C. elegans embryos. Bessel beam structured plane illumination thus promises new insights into complex biological phenomena that require 4D subcellular spatiotemporal detail in either a single or multicellular context.
INTRODUCTION
Established imaging technologies such as widefield and confocal microscopy play key roles in elucidating structure and function in biological systems (Stephens and Allen, 2003). However, as more detailed information has been needed, particularly in the context of in vivo imaging, their limitations have become increasingly clear. One of these is the two order of magnitude gap between the level at which proteins interact within the cell, and the resolution limit of light microscopy. Another is the difficulty of imaging specimens in their three-dimensional (3D) entirety without sacrificing spatiotemporal information. Yet a third is the damage induced by light in living systems, particularly when higher resolution, larger volumes, and faster or longer imaging are required (Khodjakov and Rieder, 2006; Schroeder, 2011).
In response to these challenges, new technologies have arisen. In particular, several variants of superresolution microscopy have been developed that offer spatial resolution, depending on the method, from ~100 nm down to the near-molecular level (Schermelleh et al., 2010). Additionally, plane illumination microscopy has emerged as a powerful tool for less invasive, long-term 3D imaging in developmental biology (Huisken and Stainier, 2009). Unfortunately, the advantages of these methods are often mutually exclusive: superresolution is limited to specimens with sparsely distributed fluorescent features, either fixed or slowly evolving over only a few time points, while conventional plane illumination sacrifices resolution, particularly along the detection axis, to image at depth in multicellular specimens over broad fields of view. Nevertheless, with a sufficiently thin excitation light sheet, the combination of these methods could prove quite fruitful: the resulting suppression of out-of-focus background could extend widefield cellular superresolution approaches (Gustafsson, 2000; Betzig et al., 2006) to much thicker and/or densely fluorescent specimens, while the more efficient expenditure of the fluorescence photon budget in generating only in-focus signal could lead to improved spatiotemporal resolution and reduced phototoxicity.
Recently, we described the use of a scanned Bessel beam to confine the excitation in plane illumination microscopy more tightly to the detection focal plane than traditional Gaussian beams allow (Planchon et al., 2011). We then introduced two modes of Bessel beam plane illumination to achieve near-isotropic, diffraction-limited 3D imaging of living cells. In the first, two-photon excitation (TPE) is used to eliminate unwanted excitation from side lobes of the Bessel beam, resulting in a thinner light sheet. This mode permits the imaging of living cells over dozens to hundreds of time points at up to 200 slice planes per second, given sufficiently dense fluorescent labeling. In the second mode, the Bessel beam is moved in discrete steps across the focal plane to create a periodic excitation pattern, and an optical sectioning algorithm (OS-SIM, Neil et al., 1997) is applied to a series of images taken at different phases of the applied excitation pattern to computationally eliminate background fluorescence generated by the side lobes. This mode offers better axial resolution than the TPE mode, as well as simple multicolor capability.
Each of these modes has its own limitations. TPE introduces nonlinear photodamage mechanisms, and multicolor imaging is made difficult by the lower TPE photostability of red fluorescent proteins (FPs) compared to green ones, as well as the need for multiple, expensive ultra-fast lasers. On the other hand, the OS-SIM mode is slow and surprisingly far more phototoxic to live cells than the TPE mode, and thus is almost always restricted to imaging fixed cells. In addition, the mathematical operations employed in the OS-SIM algorithm call into question the strict validity of the resulting image reconstructions, a concern reinforced by variations in shape and intensity seen for ostensibly identical structures in such reconstructions.
Here we address all these issues by introducing Bessel beam superresolution structured plane illumination (Bessel plane SR-SIM), which combines the tightly confined planar illumination of a periodically stepped Bessel beam with the principles of widefield 3D superresolution structured illumination microscopy (widefield SR-SIM, Gustafsson et al., 2008) to achieve non-invasive multicolor imaging of rapid 3D biological processes in thick living specimens. Although providing only modest gains in theoretical resolution extension beyond the diffraction limit, we demonstrate, by comparison to Bessel plane OS-SIM and spinning disk confocal microscopy, substantial gains in the practical resolvability, contrast, and signal-to-noise ratio (SNR) on samples ranging from single microtubules to D. melanogaster embryos. Similarly, although the theoretical lateral resolution limits are slightly less than in widefield SR-SIM, we show the superiority of Bessel plane SR-SIM when imaging live samples ~10 µm thick or greater, such as mitotic cells and C. elegans embryos. Furthermore, we demonstrate substantially reduced phototoxicity relative to the Bessel plane TPE mode and spinning disk confocal microscopy when imaging fast membrane dynamics both internally and at the surface of densely fluorescent COS-7 and light-sensitive D. discoideum cells. Lastly, we show that SR-SIM can be extended to image portions of specimens hundreds of microns in size, such as the adult brain of D. melanogaster and the entire C. elegans L1 larval worm, by combining Bessel structured plane illumination with the greater depth penetration afforded by two-photon excitation.
In short, the theoretical resolution limits of microscopes are difficult to achieve in many biological samples, particularly as one pushes further and further beyond the diffraction limit. In addition, the successful imaging of dynamics in living systems, particularly in 3D, involves tradeoffs of spatial resolution, temporal resolution, and phototoxicity that render irrelevant a single minded focus on spatial resolution alone. Through a unique combination of high speed, low phototoxicity, and volumetric resolution ~2–3× beyond the diffraction limit, Bessel plane SR-SIM permits the investigation of complex biological processes that require fast, high resolution 3D imaging to visualize subcellular detail over many time points, not only in the optically tractable but biologically artificial context of immortalized cultured cells, but also in the more physiologically relevant context of interacting cells within whole multicellular organisms. Among other examples, we use these capabilities to identify and track every chromosome in an aneuploid cell during mitosis, thus opening the door to direct visualization of stereotypical chromosome arrangements and deviations therefrom during cell division, and identify a diverse range of actin dynamics at different planes throughout the volume of motile cells, such as a correlation between the lateral movement of single D. discoideum cells and rapid flows of actin to and from the supporting substrate.
RESULTS
Bessel Plane SR-SIM Images 3D Dynamics Noninvasively at High Spatiotemporal Resolution
Motivated by the surprising phototoxicity, poor SNR, and apparent artifacts in our previous optical sectioning structured illumination mode of Bessel beam plane illumination (Bessel plane OS-SIM, Planchon et al., 2011), we decided instead to adapt the algorithms of 3D widefield superresolution structured illumination microscopy (widefield SR-SIM, Gustafsson et al., 2008) to Bessel microscopy, in part to hopefully address the limitations of OS-SIM, and in part to extend the superresolution capabilities of widefield SR-SIM to thicker and/or more densely fluorescent specimens. The experimental architecture and operation of the resulting method, termed Bessel beam superresolution structured plane illumination microscopy (Bessel plane SR-SIM) are identical to Bessel plane OS-SIM: two microscope objectives sharing a common focal point are mounted at right angles to one another (Figure S1A, S1B) in a medium filled, temperature controlled sample chamber (SC, Figure S1C). The first, or excitation, objective generates the Bessel beam: a narrow yet very long pencil of light surrounded by progressively weaker side lobes ((Figure S2A). At each plane of the specimen, the beam is scanned in discrete steps (Figure S2B) to create a periodic excitation pattern (Figure S2D, left) consisting of a comb of N discrete harmonics in frequency space (Figure S2D, center). N images are recorded per plane as the pattern is translated in N equal fractions of one period and then reconstructed into a single image. This process is then repeated at successive planes to build a 3D image of the specimen.
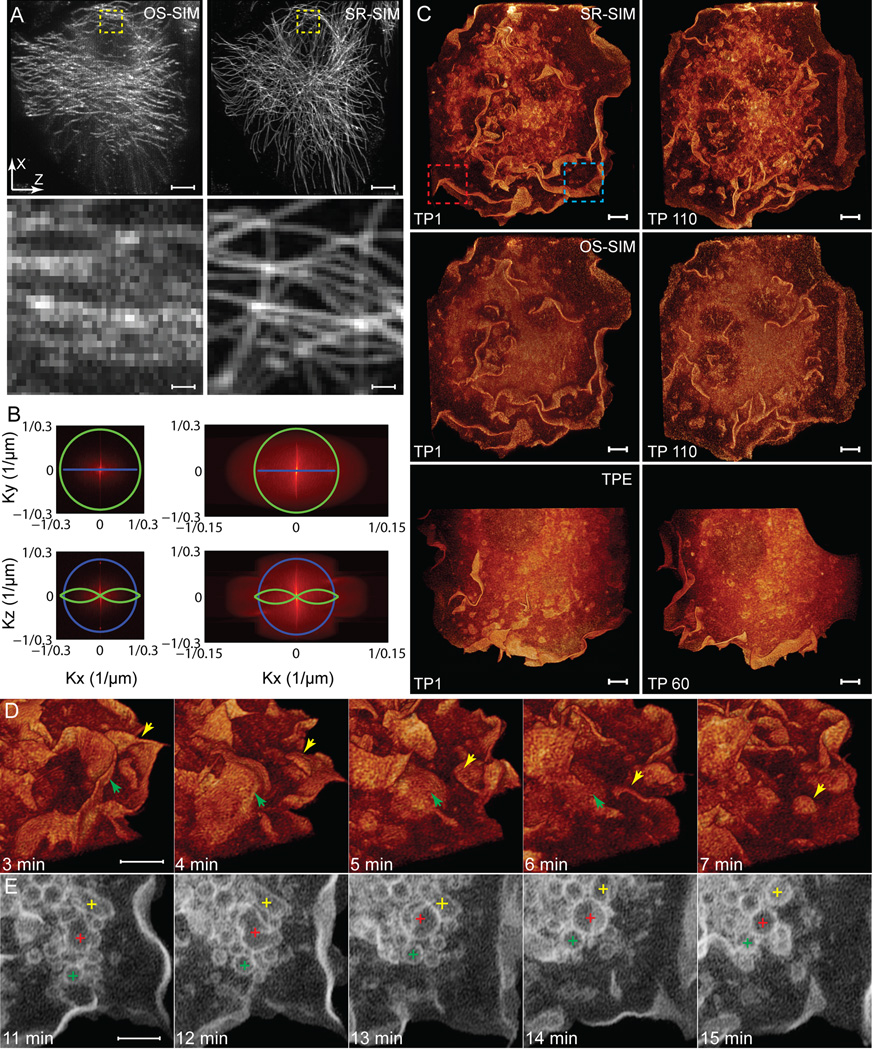
Comarison of the exact same data sets analyzed by either Bessel plane OS-SIM or Bessel plane SR-SIM (Figure 1A–C) confirm the limitations of the former and the superiority of the latter. Indeed, a theoretical analysis of the method of image reconstruction by the two methods reveals why: in OS-SIM, out-of-focus information encoded in the central DC harmonic is eliminated, leading to its optical sectioning capability. However, the information encoded by all the other harmonics is also eliminated, except for one (red arrows to the right of center in Figure S2D, right). Since this harmonic represents only a small fraction of the total spectral energy in the excitation, most of the useful information contained in the original data is discarded. Consequently, much higher excitation power is required to achieve an adequate SNR in the reconstructed image, leading to premature bleaching and photodamage. Furthermore, the phase and amplitude of each spatial frequency in the reconstruction are not correctly recovered, leading, as seen in an image of microtubules in a fixed LLC-PK1 cell (Figure 1A, left), to artifacts such as granularity, asymmetric image contrast, and an overweighting of low spatial frequencies (Figure 1B, left) that makes the resolution appear poorer than the theoretical limit of diffraction (Figure 1B, green and blue curves).
Figure 1. Bessel Beam Superresolution Structured Plane illumination Microscopy Improves Signal, Contrast, and Spatial Resolution During the 3D Imaging of Cellular Dynamics.
(A) Top: Maximum intensity projection of the same 3D data set of microtubules in a fixed LLC-PK1 cell, as analyzed by Bessel plane OS-SIM (left, Planchon et al., 2011) and Bessel plane SR-SIM (right). Bottom: Zoom images of the boxed regions in A.
(B) Frequency space representations of the same data, showing that Bessel plane SR-SIM (right) contains information (red) beyond the diffraction limits of the excitation (blue) and detection (green) objectives in two directions (x and z).
(C) Volume renderings of live COS-7 cells expressing mEmerald/c-Src, which promotes membrane ruffles and the formation of internal vacuoles. Top: Bessel plane SR-SIM; Middle: Bessel plane OS-SIM analysis of the same data; Bottom: Two-photon Bessel beam plane illumination, showing cell retraction (right, Planchon et al., 2011) after only 60 image volumes.
(D) Progression of macropinocytosis, as two membrane ruffles (green and yellow arrows) fold over and encapsulate extracellular fluid to form internal vesicles. From the red boxed region in C.
(E) Trafficking of vesicles (colored crosses) at five time points from a series of 110 volumes acquired at 20 sec intervals. From the blue boxed region in C.
Scale bars: 5 µm and 0.5 µm in A; 5 µm in C, D, and E. See also Movie S1.
Bessel plane SR-SIM resolves all these issues (Figure 1A, right): the SNR is improved, since the information encoded by all harmonics is used, and the microtubules are uniform in thickness and intensity, since both the amplitude and phase of all spatial frequencies are reconstructed accurately. Furthermore, the nonzero harmonics encode sample information at spatial frequencies beyond the diffraction limit, and these are also accurately assigned in the SR-SIM reconstruction to achieve superresolution in the scan (x) and axial (z) directions (Figure 1B right), with the amount of resolution improvement determined by the numerical apertures of both the excitation and detection objectives (Figure S1B).
The advantages of the SR-SIM mode are also apparent in 3D images of the dynamics Induced by the signaling kinase cSrc in a living COS-7 cell (Figure 1C, top and Movie S1): far more detail at higher contrast and better SNR is observed for small membrane ruffles and intracellular vesicles, even those larger than the diffraction limit, than when the same data is analyzed by OS-SIM (Figure 1C, middle). In addition, the cell exhibited no noticeable photodamage over 110 3D stacks (Figure 1C, top right), whereas a similar cell imaged in the TPE mode displayed noticeable photo-induced retraction after 60 3D stacks (Figure 1C, bottom right, from Planchon et al., 2011). Further contributing to the lower phototoxicity was the use of a diffractive optical element (DOE, Figure S1C) to produce an array of seven parallel Bessel beams (Figure S1D–F) of proportionally lower peak power. Overall, the combination of SR-SIM with parallel Bessel beams has proved substantially less invasive than either of our earlier TPE or OS-SIM modes for dozens of combinations of cell lines and fluorescent targets, permitting 3D dynamic processes to be followed over more time points and with increased confidence in the physiological nature of the observations.
As an example, we consider macropinocytosis, the process by which membrane ruffles fold over (Figure 1D, yellow and green arrows) and encapsulate extracellular fluid to form intracellular vacuoles (Figure 1E, green, red, and yellow pluses). While complex processes such as this are in principle accessible to conventional methods such as widefield 3D deconvolution and optically sectioned confocal microscopy, Bessel SR-SIM offers at least 2× higher axial resolution, better image contrast at high spatial frequencies, particularly in densely fluorescent cells such as this, and the high imaging speed (21 planes/sec, Movie S1) needed to reveal the stages of this process with sufficient spatiotemporal detail for the quantification of events such as changes in the shape, volume, and velocity of individual vesicles.
Indeed, due to the triple limitations of speed, axial resolution, and photodamage that plague conventional microscopy, much of our knowledge of cellular dynamics comes from 2D imaging of inherently 3D processes. For example, while total internal reflection fluorescence microscopy (TIRFM, Axelrod, 2001) has provided an unambiguous picture of events at the ventral surface of cultured cells, similar clarity at the dorsal surface (which, by virtue of its proximity to the extracellular environment and neighboring cells, may prove more interesting) has been lacking. However, Bessel plane SR-SIM can readily distinguish the dorsal and ventral membranes, and simultaneously image the differing dynamics at each.
Consider, for example, the dynamics of the actin cytoskeleton, whose organization and dynamics relative to the overall 3D cellular architecture is poorly understood. Seen from above (Figure 2A and upper left of Movie S2), a live fox lung cell exhibits waves of centripetally flowing stress fibers, known as actin arcs (Burnette et al., 2011). Cross-sectional views (Figure 2B and upper right of Movie S2) reveal that these contracting arcs are at the dorsal surface, flowing inwards at ~0.3 µm/min, while actin fibers at the ventral surface (magenta arrows) are predominantly static. This suggests that the fibers at the two surfaces play different roles in force generation and the emergence of cell shape and mechanics. As the arcs move farther up the dorsal surface and approach the nucleus, they abruptly spring away from the membrane and straighten, as if under substantial stress (lower right of Movie S2 and green and yellow arrows, Figure 2B). Finally, they dive deeper within the cytosol at a faster rate of ~1.2 µm/min before depolymerizing (upper right of Movie S2 and missing green arrow at 22 min, Figure 2B), highlighting their intrinsic instability away from the cortical zone. Similar behaviors of correlated straightening (Figure 2C) and detachment (Figure 2D) are seen in live U2OS cells, as well as curved arcs for those filaments still localized at the dorsal membrane (Figure 2E). Note that it is only with high speed and high resolution in all directions and over many time points (96 volumes at 30 sec intervals in Movie S2) that these processes can be fully understood.
Figure 2. Differing Cytoskeletal Dynamics at the Dorsal and Ventral Membranes Revealed by Bessel Plane SR-SIM.
(A) Oblique view volume rendering of a live fox lung cell expressing mEmerald/actin, from a series of 96 such volumes recorded at 30 sec intervals.
(B) Internal view at four time points through the yellow cut plane in A. Actin stress fibers at the ventral surface are stationary (magneta arrows), while those at the dorsal membrane flow inwards (green, yellow arrows), before detaching and depolymerizing within the cytosol. See also Movie S2.
(C) Volume rendering of actin in a live U2OS cell, from a series of 30 volumes at 30 sec intervals.
(D,E) Internal views through the yellow cut line in C, showing that detached stress fibers (yellow arrows) are straight, and thus presumably under considerable tension.
(F) Internal view through the blue cut line in C, showing that curved stress fibers (blue arrow) are attached to the dorsal membrane.
Scale bars: 5 µm in B, C; 2 µm in D–F.
Bessel Plane Illumination Extends SR-SIM to Thickly Fluorescent Specimens
Since the performance of Bessel beam plane illumination is enhanced so much by its combination with SR-SIM, it is natural to ask how it compares with the earlier widefield form of 3D SR-SIM (Gustafsson et al., 2008), and whether the Bessel component is even necessary. The answer lies in the imaging of densely fluorescent and/or thick specimens. For example, when imaging densely labeled histones in living LLC-PK1 cells in anaphase, the chromosomes are readily resolved in all three orthogonal projections by Bessel plane SR-SIM (Figure 3A), but are more poorly resolved, particularly in the axial direction, by widefield 3D SR-SIM (Figure 3B) – despite the fact that the theoretical resolution of the latter (120 nm × 120 nm × 360 nm (x,y,z)) is substantially better than that of the former (233 nm × 328 nm × 425 nm in this example, Table S1). Part of the problem is that, since the entire sample is illuminated in the widefield case, the background haze from out-of-focus fluorescence becomes increasingly strong as the sample becomes thicker (i.e., ~15 µm for the cells here) or more densely fluorescent (i.e., the cell in Figure 1C–E), making it increasingly difficult to extract the much weaker high resolution information encoded by the finely structured excitation. On the other hand, in Bessel plane SR-SIM, the excitation is largely confined to the focal plane, independent of the sample thickness, so the resolution gains provided by SR-SIM are retained.
Figure 3. Bessel Plane SR-SIM Outperforms Widefield 3D SR-SIM in Densely Fluorescent or Thick Specimens.
(A) Three orthogonal maximum intensity projections of chromosomes in a live LLC-PK1 cell in anaphase, as seen by Bessel plane SR-SIM.
(B) Similar views in another live LLC-PK1 cell in anaphase, as seenby widefield 3D SR-SIM
(C) Lateral (left) and axial (right) maximum intensity projection views of a living early stage C. elegans embryo, as seen by Bessel plane SR-SIM.
(D) Similar views in another living C. elegans embryo, as seen by widefield 3D SR-SIM.
Scale bars: 2 µm in A, B; 5 µm in C, D.
Similar results are seen in images of live, early-stage, membrane labeled C. elegans embryos: the reduction of out-of-focus excitation with Bessel plane illumination (Figure 3C) leads to higher image contrast and better SNR throughout the embryo, and better effective resolution (theoretically, 194 nm × 238 nm × 419 nm) than widefield 3D SIM (Figure 3D) beyond the first few microns in depth. Indeed, nearly no useful signal remains in widefield SIM in the lower half of the embryo (Figure 3D, right). This may also be due in part to sample-induced aberrations affecting either the excitation pattern or the detection resolution. The steep angle light rays in high numerical aperture objectives are more strongly affected by such aberrations (Ji et al., 2009). Thus, widefield SIM, which depends upon higher numerical aperture excitation than Bessel plane SR-SIM in order to achieve its theoretically higher resolution, is more strongly affected when imaging through microns of heterogeneous biological material. Furthermore, the axial extent of a focused beam grows rapidly with increasing aberration (Ji et al., 2009). While this rapidly degrades the axial resolution in widefield SIM, Bessel plane SR-SIM, which by design exploits the very long axial extent of a Bessel beam, is relatively unaffected.
Of course, for sparsely labeled, index-matched samples or those less than 5–10 microns thick, the practical resolution of widefield 3D SR-SIM approaches its theoretical limit, and outperforms Bessel plane SR-SIM. Indeed, recently widefield SIM at high resolution has been applied to live imaging of thin cells at rates of 2 to 6 planes/sec (Shao et al., 2011, Fiolka et al. 2012). Although the best volumetric resolution we demonstrate here is three-fold poorer, we image at 10 to 27 planes/sec, since only three to five raw images per plane are needed to obtain superresolution in two directions versus 15 images per plane for 3D superresolution in widefield SIM. Thus, Bessel plane SR-SIM is better suited for imaging fast processes in thin cells with reasonably continuity between stacks, minimal motion induced blurring, and accurate multi-color position registration, as well as for thicker specimens, where more planes per stack are required, and where the practical resolution of widefield SIM quickly deteriorates.
It is also useful to compare the performance of Bessel plane SR-SIM to spinning disk confocal microscopy (SDCM) – probably the most common method used today for fast, optically sectioned biological imaging. To do so, we choose the highly dynamic and light-sensitive slime mold amoeba, D. discoideum (Figure 4 and Movie S3). We also introduce a variant of Bessel plane SR-SIM where the light sheet is structured not by moving the beam in discrete steps (Figure S2B), but rather by sweeping it continuously while sinsusoidally modulating its intensity (Figure S2C). This sine mode contains only three harmonics regardless of period (Figure S2E), so only three images are required per plane, leading to even lower phototoxicty at high speed, but at the price of ~20% poorer lateral resolution.
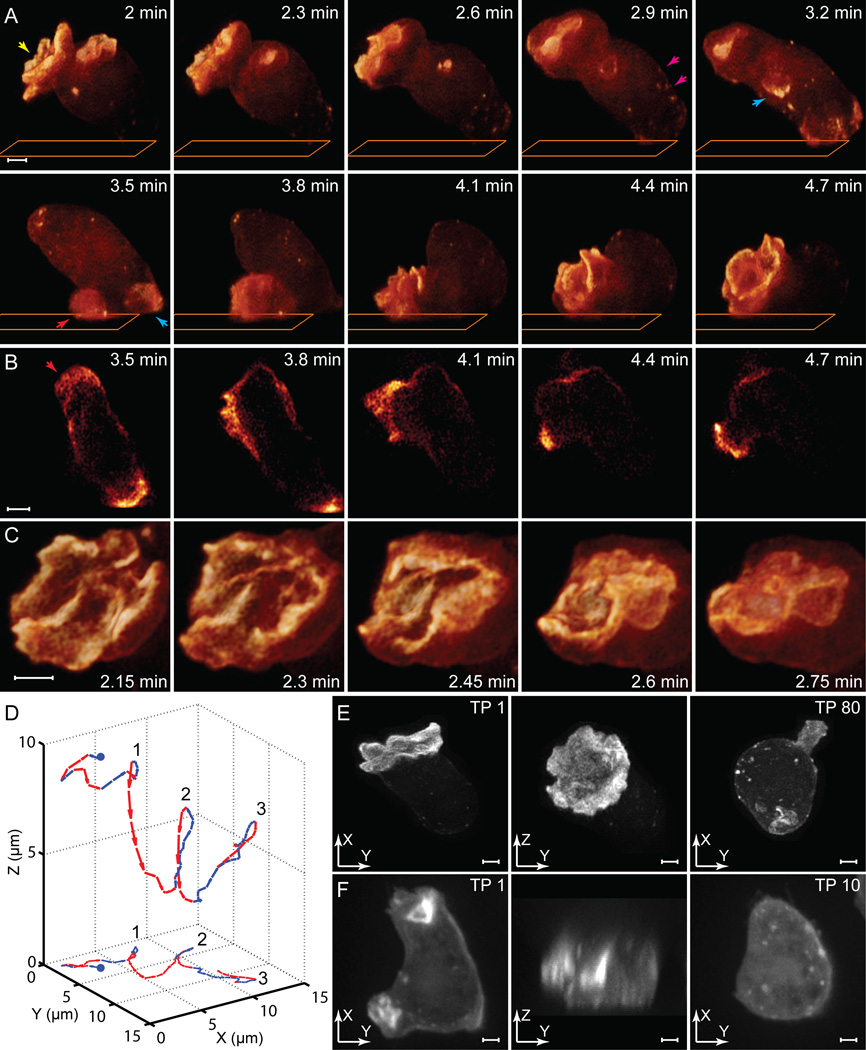
Figure 4. 3D Actin Flow, Membrane Remodeling, and Motility in D. discoideum.
(A) Volume renderings at ten time points extracted from a series of 80 volumes recorded by Bessel plane SR-SIM, showing actin flowing from the distal to the proximal end of the cell, and crown shaped protrusions both collapsing (yellow arrow) and growing (red arrow).
(B) 2D slices taken from the data near the cover slip plane (orange rectangle in A).
(C) Expanded view at 9 sec intervals of the collapsing crown-shaped protrusion in A.
(D) 3D map (top) of the center of fluorescence intensity over all 80 time points, showing repeated cycles of motion toward (red) and away from (blue) the cover slip, with lateral motility (bottom 2D projection) correlated to the downward rush of actin.
(E) Lateral (left) and axial (middle) maximum intensity projections at the first time point, and lateral view at the 80th time point (right). Rapid protrusive motion is still observed, indicative of cell health.
(F) Similar views for another D. discoideum ameboid, taken with spinning disk confocal microscopy, showing slightly poorer lateral (left) and substantially poorer axial (center) resolution. After the 10th time point (right), the cell is sessile and round in shape, indicative of cell damage.
Scale bars: 2 µm. See also Movie S3.
3D images in this mode at 9 second intervals over 12 minutes (80 total stacks, Movie S3) of a single ameboid stably expressing GFP / actin reveal a back and forth flow of actin from the ends of the cell distal and proximal to the cover slip (orange rectangle, Figure 4A). To reduce photodamage, such cells are often studied in 2D by TIRFM or confocal microscopy. However, as demonstrated by a series of 2D slices extracted near the cover slip from the 3D Bessel stacks (Figure 4B), such a picture is incomplete and potentially misleading – what appear to be actin enriched, 2D lamellipodia driving cellular motion (e.g., red arrow, Figure 4B) are actually the ventral surfaces of large, 3D pseudopods that develop as mature crowns at the distal and proximal ends (yellow and red arrows, Figure 4A), or as smaller blebs along the shank of the cell (blue arrows, Figure 4A). Viewed at higher magnification and with greater temporal detail (Figure 4C), the actin-enriched edges of the mature crowns are seen to fold over as they collapse, incorporating extracellular fluid as macropinosomes within the cell. Nevertheless, they are indeed correlated to cell motion: a 3D plot of the centroid of the actin signal (Figure 4D) over time shows that the cell stalls in its motion across the cover slip when the actin is concentrated furthest from the cover slip (points 1, 2, and 3, Figure 4D), and then resumes motion in a different direction as the actin rushes downward (red segments, Figure 4D).
Understanding this process fully requires fast, high resolution imaging in all directions over an extended period of time (Figure 4E). While SDCM can image D. discoideum at similar speeds in 3D, its axial resolution (Figure 4F, center) is inadequate to observe morphological details or to similarly quantify actin flow within the cell. Furthermore, D. discoideum cells typically become round and sessile, indicating significant photodamage, after ~10 stacks of SDCM imaging (Figure 4F, right) -- too short a period to capture the essential features of the cell’s behavior.
We finally note that, for these cells, the image contrast and practical lateral resolution (Figure 4F, left) of SDCM are inferior to Bessel plane SR-SIM, even though the theoretical lateral resolution is superior (203 nm × 203 nm × 645 nm versus 223 nm × 238 nm × 348 nm). This again points to the difficulty of achieving optimal resolution in 3D samples. It also illustrates how poor resolution in one dimension (e.g., axial) can negate the advantages of high resolution in another (e.g. lateral): complex 3D objects such as the membrane ruffles in Figure 4F that might be resolvable with isotropic resolution instead merge into an unresolvable haze when averaged over an axially elongated voxel. More generally, 4D imaging (x,y,z,t) of cellular dynamics is only as strong as its weakest dimension. Thus, Bessel plane SR-SIM, which offers a good balance of resolution in all four dimensions, is often better suited to such problems than other microscopes, superresolution and otherwise, which sacrifice resolution in certain dimensions (e.g., z or t) to achieve nominally superior resolution in others (e.g., x, y).
Bessel Plane SR-SIM of Mitosis: Application to In Vivo Karyotyping
Mitosis represents a critical juncture in the cell cycle, involving the coordinated, dynamic interplay of numerous proteins. Imaging mitosis with sufficient spatiotemporal detail to identify and track individual chromosomes and their molecular partners throughout might reveal new insights into chromosome arrangements and how these arrangements change between healthy and diseased conditions, such as those leading to trisomies. However, this has proven difficult for conventional epi-fluorescence methods such as widefield and confocal microscopy, due to their inadequate axial resolution, the sensitivity of the mitotic cell to light (Khodjakov and Rieder, 2006), its thickness from prophase to telophase, and the high density of labeled protein when histones in condensed chromosomes or tubulin in the mitotic spindle are imaged. It is, however, a problem well suited to the high resolution, low photodamage, and reduced out-of-focus background of Bessel plane SR-SIM.
We consider two examples involving dual color imaging of chromosomes (green) and kinetochores (red) in living U2OS cells expressing mEmerald/H2B and tdTomato/CENP-B. In the first, we studied the separation of chromatids from late metaphase through late telophase at 30 sec intervals over 36 min (Figure S3 and Movie S4, part 1). At the end of metaphase, all chromatid pairs remain joined (Figure S3A, first column), and all kinetochores are tightly aligned within 1–2 µm of the metaphase plate (MP, Figure S3B). In early anaphase (second column), small gaps appear between many sister chromatids as they and their kinetochore pairs begin to separate. Over the next 2.5 min, most of the chromosomes separate completely. At this point (fifth column), the two sets of kinetochores are, with a few exceptions, approximately equidistant from the MP. As mitosis continues (Movie S3, part 1), many daughter chromosomes unravel even before the final chromatids separate.
The high spatiotemporal resolution in 4D of Bessel plane SR-SIM enables the segmentation of any desired chromatid pair (e.g. yellow arrow, Figure S3A, and Figure S3C), to better study how and when that particular pair separates. Furthermore, with our parallel Bessel beam configuration, dual color imaging is fast enough (10 planes/sec) that each kinetochore is correctly co-localized with its partner chromatid and thus accurately marks the location of the centromere, even during rapid movement in anaphase. Combined with the accurate segmentation and shape information provided, we can therefore uniquely identify and track each chromosome – in essence, allowing complete in vivo karyotyping of the cell as it evolves through mitosis.
We demonstrate this in a second example showing the alignment of chromosomes to the MP, starting from prometaphase. Initial orthogonal views (left and right columns of Movie S4, part 2B) show the chromosomes and kinetochores distributed over a spherical volume. They then align in a ring as viewed along the axis perpendicular to the MP (stack 16, left column), with most centromeres facing inward. At this point, they still remain widely distributed above and below the MP (right column). However, soon thereafter certain chromosomes move inward to fill the void, while simultaneously all chromosomes migrate to the MP, signaling the start of metaphase (stack 50). While all 77 chromosomes of this massively polyploid cell can be identified at any time point, such as shown for the initial stack (Figure 5B and Movie S4, part 2A), three specific chromosomes (magenta, yellow, brown) were manually segmented and tracked across all 50 stacks, as shown at six time points in various orientations (Figure 5A). Such in vivo karyotyping, coupled with automated segmentation and tracking across multiple cells, may determine the degree of stereotypy in the spatiotemporal relationship of specific chromosomes during mitosis (Magidson et al., 2011), and how deviations in such stereotypy lead to chromosome abnormalities (Kitajima et al., 2011).
Figure 5. In Vivo Karyotyping of a Mitotic U2OS Cell.
(A) Dual color imaging of chromosomes (green) and kinetochores (red) at six different time points from prometaphase to metaphase, taken from a series of 50 volumes acquired at 60 sec intervals. Three specific chromosomes (colored magenta, brown, and yellow) were identified by shape and tracked at all times.
(B) In vivo karyotyping by manual segmentation of all chromosomes at the initial time point. In each case, kinetochore pairs are shown correctly co-localized with their corresponding chromatids, and can be used to help identify specific chromosomes. The cell exhibits extreme polyploidy, with 76 diploids and one possible triploid (red arrow).
Scale bars: 5 µm in A; 2 µm in B. See also Movie S4.
Bessel Plane SR-SIM of Multicellular Specimens
Much of our knowledge of sub-cellular dynamics comes from the study of immortalized cells cultured on glass cover slips, because this is one of the few scenarios amenable to diffraction-limited imaging. However, it is also an artificial situation far removed from the more evolutionarily relevant one of cells interacting within a living multicellular organism. Conventional plane illumination microscopy has made important inroads in this regard, but sub-cellular details are often masked by its limited axial resolution. Spinning disk confocal microscopy is also frequently applied to multicellular systems, but is often hampered by photobleaching, photodamage, limited speed, and rapid loss of signal and resolution at increasing depth. Finally, as shown above (Figure 3D), out-of-focus background and sample induced aberrations usually preclude the use of widefield 3D SIM (and other superresolution methods) in thick specimens.
In contrast, the high speed, high axial resolution, and low photodamage of Bessel plane SR-SIM make it well suited to the study of 3D sub-cellular dynamics in multicellular systems, at least for whole small embryos or superficial cells in larger cellular assemblages. Such capabilities may prove essential in understanding the detailed cellular and molecular mechanisms by which tissues and organs are shaped: due to the limitations of instrumentation, current morphogenetic studies are often restricted to much lower magnification and resolution (Metzger, et al. 2008; Ewald, et al, 2008).
We demonstrate these capabilities by imaging the 3D dynamics of plasma membranes throughout the C. elegans embryo in orthogonal views from the 8 to the 23 cell stage in Movie S5, part 1 (top), as well as during the early stages of gastrulation at the 26 cell stage in another embryo (bottom). A particular view at the 12-cell stage (Figure 6A) reveals sheet-like membrane extensions resembling lamellipodia at several cellular interfaces, including a broad extension, less than 0.5 µm thick, of the ABprp cell over the curved surface of the neighboring E cell (Figure 6B).
Figure 6. 3D Development in C.elegans Embryos.
(A) Live embryo at the 12 cell stage, from a series of 31 volumes recorded at 1.1 min intervals from the 8 to 23 cell stage, as seen using a GFP membrane marker.
(B) Expanded view from within the box in A, showing, at four time points, the evolution of a membrane protrusion from the ABprp cell over the surface of the neighboring E cell.
(C) Another live embryo, at the 8 cell stage, expressing both GFP/myosin (green) and a mCherry membrane marker (orange).
(D) Expanded view from within the box in C, showing the contractile cytokinetic ring between two dividing AB-derived cells.
(E) Cross sectional view in the plane of the contractile ring from D.
(F) Lateral maximum intensity projection (left) and single axial slice (right, from along the green line at left) of a live embryo seen by Bessel plane SR-SIM.
(G) Similar views for an embryo at a similar stage, as seen by spinning disk confocal microscopy, showing rapid loss of signal and resolution at depth.
(H) Comparative photobleaching rates for Bessel plane SR-SIM and spinning disk confocal microscopy in early embryos. Bleaching is reduced ~2× in the Bessel case, even though twice as many image planes are acquired per time point to achieve higher axial resolution, and even though the time interval between 3D stacks is three fold faster.
Scale bars: 5 µm (A, C, F, G), 2 µm (B,D,E). See also Movie S5.
Similar membrane extensions and their correlation to F-actin have been observed previously by SDCM (Pohl and Bao, 2010), but with Bessel SR-SIM they can be studied in uncompressed embryos, eliminating the possibility of mechanotransduced perturbations, with 3–5× greater axial resolution (Table S1). Even though a proportionally larger number of image planes are required per stack are required to achieve this higher resolution, the volumetric imaging time of the two methods is similar, while photobleaching in Bessel plane SR-SIM is substantially reduced. Furthermore, comparative views of early embryos reveal that Bessel plane SR-SIM retains good resolution and signal at all depths (Figure 6F), whereas the performance of SDCM drops precipitously after a depth of approximately five microns (Figure 6G). As a result, SDCM 3D data presented in the common format of a 2D maximum intensity projection in the lateral plane are heavily weighted to the brightest features closest to the surface (e.g., Figure 6G, left), whereas Bessel plane SR-SIM provides a view of the complete 3D architecture.
As another example, we viewed at several stages of development the 3D relationship between membrane morphology and myosin throughout the volume of another live embryo using dual color Bessel SR-SIM (Figure 6C and Movie S5, part 2). In particular, at the point of division of the AB-derived cells in the 8-cell embryo (Figure 6C), even buried features such as the subsurface contractile cytokinetic rings between daughter cells can be viewed at high resolution from any orientation, such as perpendicular to (Figure 6D) or within the plane of (Figure 6E) each ring.
Imaging even larger specimens is more challenging, due to aberrations and scattering that affect both the detection (Figure S4A) and excitation (Figure S4B) pathways. Nevertheless, nuclei within 10–20 microns of the surface can still be viewed with the full extended resolution of Bessel plane SR-SIM, as shown in an image of anaphase chromosomes of multiple nuclei of a living syncytial embryo of D. melanogaster (Figure 7A, taken from a time series from prophase to telophase). As with single cultured cells (Figure 5), in vivo karyotyping is possible (Figure 7B): the sex chromosomes identify the embryo as female (Figure 7C), and the relative locations of autosomes 2, 3, and 4 can be determined (Figures 7D, 7E, and 7F, respectively). This is possible only with high resolution in all directions (Figure 7G): the poor axial resolution of multibeam confocal microscopy (Figure 7H) precludes such segmentation and identification.
Figure 7. Two-Photon Bessel Beam Plane SR-SIM of Larger Multicellular Specimens.
(A) Chromosomes in mitotic nuclei at the surface of a D. melanogaster embryo.
(B) Zoom view of the boxed chromosome in A, with chromatids individually colored.
(C) Sex chromosomes isolated from B, identifying the embryo as female.
(D, E, F) Autosomes 2, 3, and 4 isolated from B.
(G) Lateral and axial maximum intensity projections by Bessel plane SR-SIM.
(H) Similar views of chromosomes at a similar stage, from another embryo as viewed by multibeam confocal microscopy.
(I) Stochastically labeled subset of neurons within the lobula plate of the optic lobe of a brain extracted from an adult D. melanogaster, including the HSN (blue arrow) and VS2 (green arrow) giant neurons.
(J, K) Dendrites within the box in I, viewed from different angles.
(L) Cross-sectional view of dendrites at the cut plane in J, showing the ability to resolve the intracellular space.
(M) All nuclei within a fixed L1 stage C. elegans larva.
(N) View into the mouth of the nematode.
(O) Zoom in view of the blue box in M.
(P) Cross section through nuclei at the cut plane in O.
Scale bars: 5 µm in A; 1 µm in B–H, L; 10 µm in I, M; 2 µm in J, K, N, O, P. See also Movie S6.
In order to image even deeper within large specimens, one solution is to adapt two-photon excitation to Bessel plane SR-SIM: the infrared light used in TPE penetrates tissue with reduced scattering and aberration and the out-of-focus emission from the side lobes of the Bessel beam is suppressed (Figure S5A vs. Figure S2A). As before, the beam is moved in discrete steps (Figure S5B) to create the necessary periodic pattern (Figure S5C). However, the nonlinearity of TPE creates extra sidebands (Figure S5D) at spatial frequencies beyond the diffraction limit (solid blue circle, FigureS5D) defined by the numerical aperture and wavelength of the excitation. Although weak (Figure S5E), with sufficient signal-to-noise, they enable lateral resolution extension comparable to that in linear Bessel SR-SIM, despite the longer wavelength used. Similarly, the suppression of the concentric rings confines the excitation more closely to a single plane (Figure S5C), resulting in strong sidebands at intermediate spatial frequencies within the diffraction limit (Figure S5E, right) and permitting substantial axial resolution improvement when applied to SR-SIM.
We demonstrate the ability of TPE Bessel plane SR-SIM to achieve high 3D resolution over large multicellular volumes in two systems: the brain extracted from a D. melanogaster fly (Figure 7I–L and Movie S6, part 2), and a fixed C. elegans L1-stage larva (Figure 7M–P and Movie S6, part 1). For both systems, such large image volumes were attained by combining four adjacent sub-volumes, each 85 × 85 × 30 to 40 µm in extent. In the fly, several neurons stochastically expressing membrane-bound GFP immunolabeled with AF488 are seen within the lobular plate of the optic lobe. Most dominant, as identified by their morphology (Scott et al. 2002), are the HSN and VS2 giant neurons of the visual system (Figure 7I). Views of dendrites from different angles (Figures 7J and 7K) and in cross-section (Figure 7L) demonstrate isotropic resolution sufficient to resolve the specificity of the label to their outer membranes. This should enable the unambiguous identification of intracellular versus membrane-bound proteins in dendrites and similarly narrow structures branching through tissues, which previously had not been possible. In the second example, DAPI-stained nuclei show uniform, nearly isotropic resolution across the entire larval nematode (Figure 7M), as seen in views down the axis of the gut (Figure 7N), transversely near the nerve ring (Figure 7O), or in cross-sections of nucleoli within individual nuclei (Figure 7P).
A key challenge in biology today is to understand how specific spatiotemporal patterns of gene expression drive development from zygote to adult. In C. elegans embryos, this has been studied at single cell resolution using confocal microscopy for several genes up through the 350 cell stage (Murray et al. 2008), but is hampered at this point by limited resolution and SNR. It has also been mapped in chronograms of expression along the body axis from the early larval stage to adulthood for ~900 promoters, using a high-throughput flow cytometry based assay (Dupuy et al. 2007), but not at single cell resolution. Linear or TPE Bessel plane SR-SIM offer the possibility of noninvasively mapping gene expression onto C. elegans anatomy at subcellular resolution and many stages of development, even for weakly expressed proteins otherwise difficult to detect and localize in the autofluorescent background.
DISCUSSION
Bessel plane SR-SIM offers a unique combination of high spatial resolution, high speed, low phototoxicity and improved depth penetration that make it well suited to the study of 3D dynamics in a variety of single and multicellular systems. Compared to confocal microscopy, it has ~2× better axial resolution. Its thin light sheet produces substantially less photodamage and photobleaching in light sensitive or thick specimens, even compared to spinning disk systems, and without the problem of pinhole crosstalk that can arise in such systems when imaging densely fluorescent or thick specimens. Compared to our earlier optical-sectioning form of Bessel beam plane illumination (Planchon et al. 2011), it provides ~1.5 – 1.9× improved resolution in the beam scan direction, an artifact-free 3D representation of the sample (Figure 1A), much improved SNR at comparable excitation power (Figure S5D–S5G), and greatly reduced phototoxicity. Compared to our earlier TPE mode, it offers ~1.5 – 1.9× improved resolution in both the scan and axial directions. Unlike the TPE mode, it is readily adapted to multicolor imaging (Figures 5, 6, and S6), and is less phototoxic (Figure 1C, bottom row). Lastly, compared to widefield 3D SR-SIM, it can be applied to densely fluorescent thin cells (Figure 1), thick cells (Figure 5), embryos (Figure 6), and tissues (Figure 7) where out-of-focus background would otherwise be prohibitive (Figure 3). While its lateral resolution is not as high as in widefield 3D SR-SIM, it is at least 3 – 5× faster, which is necessary to capture fast 3D dynamic processes (Figure 4). It may also prove less damaging when imaging thick samples, due to the fewer number of images per plane plus the tight, planar confinement of Bessel beam excitation.
Notably, Bessel plane SR-SIM is able to maintain signal and resolution at greater depth than either widefield 3D SR-SIM (Figures 3C, 3D) or spinning disk confocal microscopy (Figures 6F, 6G) in optically heterogeneous samples such as early stage C. elegans embryos. This is likely due to its lower excitation numerical aperture and the orthogonality of its excitation and detection pathways, which make the method less sensitive to aberrations affecting the axial focus of either. Even greater depth penetration is possible by combining it with two photon excitation (Figure 7). However, for multicolor imaging or to achieve minimal phototoxcity, linear excitation is likely needed. In this case, optimal resolution might still be retained in large, fixed specimens and tissues by using clearing agents (Dodt et al., 2007; Hama et al., 2011) that homogenize the sample refractive index and thus remove aberrations. However, for linear imaging deep within living multicellular specimens, adaptive optics (Booth, 2007; Ji et al., 2010; Ji et al., 2012) will likely be needed to compensate for sample induced aberrations.
In addition to being compatible with optical clearing, another key advantage of studying fixed specimens is that the entire photon budget defined by photobleaching can be expended in generating a single high resolution, low noise 3D image, as opposed to having to spread the signal across many 3D data stacks at different time points. This is particularly helpful when visualizing smaller, dimmer objects in a sea of much brighter sources or large cytosolic background. Figure S6 and Movie S7, for example, show the 3D architecture of chromosomes and the mitotic spindle in a series of six fixed cells from prophase through cytokinesis.
Recently, the concept of using patterned excitation for resolution improvement has been applied to multifocal confocal microscopy of multicellular samples, with modest claimed improvement (~1.4×, York et al., 2012). However, the method is too slow (1 plane/sec) to follow many dynamic processes in 3D. Furthermore, it relies on conventional epi-illumination, and thus will suffer from the same limitations of photobleaching, phototoxicty, and high sensitivity to aberrations in thick specimens as shown above for spinning disk confocal microscopy (Figures 4F and 6G). Likewise, multicellular imaging has been demonstrated using a combination of superresolution photoactivated localization microscopy with traditional Gaussian sheet illumination (Cella Zanacchi et al., 2011). However, the approach is very slow (0.5 – 5.0 min /plane), requires extended exposure to high intensity light (5–12 kW/cm2), and has a practical resolution far worse than its theoretical limit due to aberrations, out-of-focus background from the thick light sheet, and low localization density. On the other hand, while the theoretical resolution limits of Bessel plane SR-SIM are not as high most superresolution methods, we demonstrate that these limits are achievable in a wide variety of specimens, and at greater depth, without compromising the speed and noninvasiveness necessary for in vivo, physiological imaging of dynamics.
Indeed, life is defined by change: metabolism, growth, adaptation, stimulus response, and reproduction. Thus, just as a 2D image provides an incomplete and possibly misleading picture of a 3D organism, a single 3D data stack at one time point gives an incomplete and possibly misleading picture of the 4D evolution that defines that organism. The strength of Bessel plane SR-SIM lies in its ability to balance the requirements for high resolution to visualize this evolution in sufficient 3D spatial detail against the needs to follow this evolution with sufficient temporal resolution and for sufficient time, even in densely fluorescent or multicellular specimens, without adverse effects.
EXPERIMENTAL PROCEDURES
Bessel Beam Structured Plane Illumination Microscope
Two different configurations of the microscope were used to acquire the data herein. The first, identical to that used in our earlier work (Planchon et al., 2011) in terms of optical configuration, control electronics, and control software, used 0.8 NA objectives for both generation of the Bessel beam and detection of the resulting fluorescence within the specimen (Figure S1A, right). The other (FigureS1A, left) employed a custom 0.65 NA objective for excitation (Special Optics), and a 25×, 1.1 NA objective for detection (Nikon, MRD77220). For most samples (Table S1), a holographic diffractive optical element (Holo/OR Ltd. MS-107-Q-Y-A) creating a fan of Gaussian beams was placed immediately before the annular mask (Figure S1C), resulting in seven parallel Bessel beams within the sample (Figure S1D, S1E). This facilitated faster imaging over larger fields of view, and spread the excitation energy across more of the sample, to reduce phototoxicity.
Sample Preparation and Imaging Conditions for Bessel Plane SR-SIM
18 mm diameter coverslips were used in the 0.8 NA system, and either 5 or 8 mm coverslips in the 1.1 NA system. All were cleaned prior to use according to our earlier protocol (Planchon et al., 2011). Mammalian cells were cultured, transfected and grown directly on these coverslips as described previously. Cell fixation and staining were also performed according to our earlier protocol (Planchon et al., 2011).
Fixed cells were imaged in 1× PBS at room temperature. Live mammalian cells were imaged at 37°C in either 1× DMEM medium with 10% serum or L15 medium with 10% serum. Cells of Dictyostelium discoideum strain AX3 (Figure 6) stably expressing mGFP-LimEΔ to label filamentous actin were cultured in HL5 growth medium. This was was replaced with a 20 mM potassium phosphate buffer (PB) (pH 6.6), and cells were allowed to attach to the coverslip for 30–45 minutes in this buffer prior to imaging. PB buffer was also used for imaging at 21°C.
Live imaging of mitotic nuclei in Drosophila syncytial embryos was performed using embryos expressing Enhanced-GFP tagged Histone2Av (His2Av-EGFP/SM6a; Bloomington Stock Center #24163). Flies were placed on apple juice agar plates at room temperature and embryos were collected at 0–3hrs. Embryos were rinsed in 0.1% Triton in water, then dechorionated in 50% bleach for 5 minutes, rinsed three times and directly place on a coverslip covered with double side type for Bessel beam SR-SIM imaging and mounted in PBS and halocarbon oil on PetriPerm dishes with a cover slip for confocal imaging.
Live C. elegans embryos, fixed C. elegans larvae and fixed D. melanogaster brain specimens were placed directly on top of poly-D-lysine coated coverslips for imaging. C. elegans embryos at the desired developmental stage (Figure 5) were dissected from young adult worms, either OD58 ItIs38 [pie-1::GFP::PH domain of PLC1delta1; unc-119(+)] or LP54 ItIs44 [pie-1::mCherry::PH domain of PLC1delta1; unc-119(+)]; zuIs45 [nmy-2::NMY-2::GFP; unc-119(+)], in egg buffer, and imaged in egg buffer at 20°C. The C. elegans L1 stage larva (Figure 7A–D) specimen was prepared by fixing a newly hatched worm, and staining all cell nuclei with DAPI (Long et al., 2009). The D. melanogaster brain specimen (Figure 7E–H) was dissected from the line GAL4-57C10, fixed, and stained with anti-GFP/AF488. Both fixed specimens were imaged in 1× PBS at room temperature.
Imaging parameters for data in all main and supplementary figures are shown in Table S1.
Imaging Conditions for Comparative Microscopy
Spinning disk confocal images of D. discoideum were acquired on an Olympus IX-81 inverted microscope equipped with a CSU-X1 Yokogawa head, an Olympus 60× Plan Apo objective (N.A. 1.42), and a Photometrics QuantEM:512SC EMCCD camera.
Spinning disk confocal images of a C. elegans embryo were acquired on a Nikon Eclipse Ti-E microscope equipped with a CSU-X Yokogawa head, a Nikon Plan Apo VC 100× objective (N.A. 1.49), and a Hamamatsu ImgEM EMCCD camera.
Multibeam confocal images of mitotic nuclei in a live Drosophila embryo were acquired on a Nikon TE2000-E microscope equipped with a Visitech Infinity-Hawk multi-point array scanner, a Nikon Plan APO VC 100× objective (N.A. 1.4), and a Hamamatsu ORCA R2 CCD camera.
All widefield 3D SIM images were acquired by a microscope described elsewhere (Fiolka et al., 2012), using a 63× Zeiss C apochromat water immersion objective (N.A. 1.2), and an Andor iXON 897 EMCCD camera.
SR-SIM Data Analysis
Bessel beam structured plane illumination data were analyzed using a custom, lab-developed program. The first step in image reconstruction was the separation of the N mixed signal components generated by the N modulation orders of the excitation profile. To accomplish this, N images were acquired at N different illumination phases at every plane, so that the N unknowns representing the mixed signal components could be solved with N equations. The modulation depth, the starting phase and the precise position of each signal components in reciprocal space were determined thereafter from the data itself by comparing the overlapping regions in reciprocal space between each component and the DC (kx = 0) component. Once these parameters were determined, all signal components were shifted to their original locations in an expanded region of reciprocal space and combined with a generalized Wiener filter (Gustafsson et. al. 2008). To avoid edge artifacts, the filtered results were multiplied by a triangular apodization function. The region of reciprocal space where the apodization function is non-zero has the same shape as the region covered by the reconstructed image. Analyzed data were rendered in 3D or 4D (for movies of cellular dynamics) using Amira 5.3.3.
Supplementary Material
HIGHLIGHTS.
Superresolution microscopy is extended to densely labeled and multicellular specimens
Thin, planar excitation enables multicolor imaging of 3D dynamics over extended times
High axial resolution reveals complex 3D dynamics inaccessible to confocal microscopy
In vivo karyotyping reveals specific chromosome arrangements during cell division
ACKNOWLEDGEMENTS
We thank the following Janelia Farm researchers: Thomas Planchon and Bi-Chang Chen for their help with the instrument; Helen White and Sarah Winfrey for cell culture and transfection; Aliosha Nern and Hulamm Phuson for the D. melanogaster brain sample; and Hanchuan Peng for the C. elegans L1 larva. We also thank Dan Milkie of Coleman Technologies, Chadds Ford, PA for custom instrument control software, Goeh Jung of the NHLBI / NIH for the D. discoideum specimen, and Jennifer Lippincott-Schwartz and John Hammer of the NIH for valuable suggestions. B.G. acknowledges the support of NIH R01-GM083071.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Booth MJ. Adaptive optics in microscopy. Phil. Trans. R. Soc. Lond. Ser. A. 2007;365:2829–2843. doi: 10.1098/rsta.2007.0013. [DOI] [PubMed] [Google Scholar]
- Burnette DT, Manley S, Sengupta P, Sougrat R, Davidson MW, Kachar B, Lippincott-Schwartz J. Nat. Cell Biol. 2011;13:371–381. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella Zanachhi F, Lavagnino Z, Perrone Donnorso M, Del Bue A, Furia L, Faretta M, Diaspro A. Live-cell 3D super-resolution imaging in thick biological samples. Nat. Meth. 2011;8:1047–1049. doi: 10.1038/nmeth.1744. [DOI] [PubMed] [Google Scholar]
- Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, Carvunis A-R, Pulak R, Shingles J, Reece-Hoyes J, Hunt-Newbury R, Viveiros R, Mohler WA, Tasan M, Roth FP, Le Peuch C, Hope IA, Johnsen R, Moerman DG, Barabasi A-L, Baillie D, Vidal M. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotech. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev. Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MGL. Time lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc. Nat. Acad. Sci. 2012 doi: 10.1073/pnas.1119262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL, Shao L, Carlton PM, Wang CJR, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Huisken J, Stainier DYR. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–1975. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N, Milkie DE, Betzig E. Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nat. Meth. 2009;7:141–147. doi: 10.1038/nmeth.1411. [DOI] [PubMed] [Google Scholar]
- Ji N, Sato TR, Betzig E. Characterization and adaptive optical correction of aberrations during in vivo imaging in the mouse cortex. Proc. Nat. Acad. Sci. 2012;109:22–27. doi: 10.1073/pnas.1109202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Imaging the division process in living tissue culture cells. Methods. 2006;38:2–16. doi: 10.1016/j.ymeth.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Gweon D, Lee J-H. Enhancement of fluorescence confocal scanning microscopy lateral resolution by use of structured illumination. Meas. Sci. Technol. 2009;20:1–9. [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- Long F, Peng H, Liu X, Kim SK, Myers E. A 3D digital atlas of C. elegans and its application to single-cell analyses. Nat. Meth. 2009;6:667–671. doi: 10.1038/nmeth.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. The Spatial Arrangement of Chromosomes during Prometaphase Facilitates Spindle Assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching program of mouse lung development. Nature. 2008;453:745–751. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat. Meth. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil MAA, Juskaitis R, Wilson T. Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt. Lett. 1997;22:1905–1907. doi: 10.1364/ol.22.001905. [DOI] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Meth. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C, Bao Z. Chiral forces organize left-right patterning in C. elegans by uncoupling midline and anteroposterior axis. Dev. Cell. 2010;19:402–412. doi: 10.1016/j.devcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder T. Long-term single-cell imaging of mammalian stem cells. Nat. Meth. 2011;8:S30–S35. doi: 10.1038/nmeth.1577. [DOI] [PubMed] [Google Scholar]
- Scott EK, Rabbe T, Luo L. Structure of the vertical and horizontal system neurons of the lobula plate in Drosophila. J. Comp. Neurol. 2002;454:470–481. doi: 10.1002/cne.10467. [DOI] [PubMed] [Google Scholar]
- Shao L, Kner P, Rego EH, Gustafsson MGL. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat. Meth. 2011;8:1044–1046. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- Stephens DJ, Allan VJ. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- York AG, Parekh SH, Nogare DD, Fischer RS, Temprine K, Mione M, Chitnis AB, Combs CA, Shroff H. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat. Meth. 2012 doi: 10.1038/nmeth.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.