Abstract
Chronic helminth infections are associated with modulation of host cellular immune responses, presumably to prolong parasite survival within the mammalian host. This phenomenon is attributed, at least in part, to the elaboration of parasite molecules, including orthologs of host cytokines and receptors, at the host–parasite interface. This review describes recent progress in the characterization of macrophage migration inhibitory factor (MIF) orthologs from parasitic nematodes. The roles of these molecules in parasite developmental biology and pathogenesis are discussed. Further knowledge of the species-specific activities and three-dimensional structures of human and parasitic nematode MIF molecules should make them ideal targets for drug- and/or vaccine-based strategies aimed at nematode disease control.
Helminth immunomodulation
At the host–parasite interface parasitic nematodes produce a panoply of molecules, both on the cuticular surface and/or released in excretory–secretory (ES) products, which mediate their ability to survive for long periods of time despite the actions of the host immune system [1]. These parasite mediators drive potentially immune-evasive processes that allow for the parasites’ prolonged survival within their hosts [2]. There is functional diversity in these parasite-derived immune modulators that presumably reflects a long-standing coevolutionary relationship between nematode parasites and their respective hosts. Putative nematode virulence factors that might subvert host immune responses effectively include proteases, protease inhibitors, antioxidant proteins and orthologs of mammalian cytokines and their receptors [3]. Studies by several investigators have provided evidence of the elaboration by parasitic nematodes of macrophage migration inhibitory factor (MIF) orthologs, which modulate the taxis of immune cells, altering their gene expression and the subsequent production of cytokines.
The role of MIF in human disease
MIF is an important mediator of mammalian inflammatory conditions [4]. MIF is a potent effector cytokine that was described in 1966 as a soluble factor (credit for its discovery can be attributed to Miriam George and John Vaughn in the late 1950s) [5] after the observation that supernatants of activated T cells inhibited the random migration of peritoneal exudate macrophages [6–8]. Despite being one of the first cytokines described, the gene responsible for MIF activity was not isolated until 1989 [9]. However, with the availability of recombinant bioactive MIF protein [10] and a neutralizing antibody, by 1993 [11] more than 1000 scientific papers had been published on MIF. These subsequent studies have defined an important proinflammatory role for MIF in immunobiology [12]. Mammalian MIFs are both ubiquitous and constitutive in their expression; they are found in numerous tissues and cell types, including T cells, eosinophils and fibroblasts, as well as monocytes and macrophages [13,14]. Lacking an N-terminal signal sequence sufficient for classical secretion via the ER and Golgi pathway, mammalian MIF is secreted by a leaderless, nonconventional pathway [15].
In the context of the mammalian immune system, MIF has been ascribed multiple functions, including roles in pathogenesis of septic shock [16], rheumatoid arthritis [17], inflammatory bowel disease and tumor metastasis [18]. Although the molecular mechanisms by which MIF mediates its actions at the cellular level remain only partially understood, one pathway that has been defined clearly is the human MIF (hMIF), a ligand for the CD74–CD44 receptor complex at the surface of target cells [8,19]. Intracellular signaling leads to the activation of ERK1 and ERK2 MAPK-specific cascades, which potentiate downstream proinflammatory gene expression. MIF-induced activation of target cells has been demonstrated to modulate cytokine expression (e.g. TNF-α, IL-1β, IL-6, IL-8 and IL-12) and counter-regulate the anti-inflammatory and immunosuppressive effects of glucocorticoid steroids [4]. Recent studies have defined a role for MIF as a ligand for chemokine receptors (CXCR2 and CXCR4), capable of triggering calcium influx and integrin activation and modulating T-cell and monocyte migration [20]. Moreover, MIF stimulates the production of matrix metalloproteases, cycloxygenase 2 and prostaglandin E2 [8,21], emphasizing the importance of MIF in innate immunity. In addition to the inhibition of the random migration of monocytes and its effect on cytokine production, two different catalytic activities have been described for MIFs from mammals: tautomerase [4] and thiol-protein oxidoreductase activities [22].
Nematode MIF orthologs
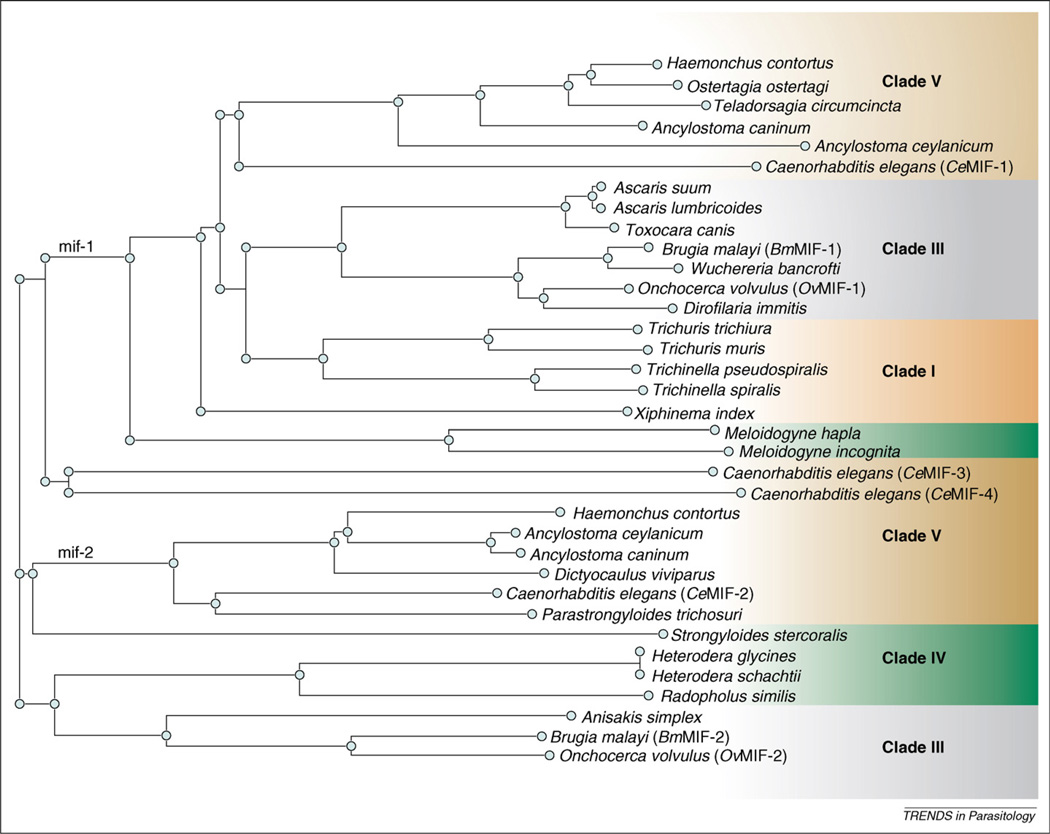
The MIFs isolated from parasitic nematodes appear to exhibit bioactivities similar to those of their mammalian hosts, although some studies were performed with recombinant fusion proteins that do not share native protein structure [23–27]. The MIF gene family in the free-living nematode Caenorhabditis elegans contains four distinct genes [28]. The four corresponding proteins (CeMIF-1, -2, -3 and -4) bear 15%–32% amino acid sequence identity to each other and 22%–35% identity to hMIF. As a result of genome- and transcriptome-sequencing initiatives [25,27,29,30], MIF homologs have been identified in other parasitic helminths representing four major clades of the phylum Nematoda (Table 1). This list includes MIF homologs from Trichinella and Trichuris spp. (clade I); Ascaris suum, Brugia spp., Wuchereria bancrofti and Onchocerca volvulus (clade III); Strongyloides stercoralis, Meloidogyne and Heterodera spp. (clade IV); and Haemonchus contortus and Ancylostoma spp. (clade V), among others. Alignment of these 35 amino acid sequences reveals two distinct types of MIF proteins, based on homology to either CeMIF-1 or CeMIF-2 (Figure 1). CeMIF-3 and CeMIF-4 cluster separately and are distinct from the other members of the nematode MIF protein family, with CeMIF-3 being significantly larger (146 aa) and the open reading frame of CeMIF-4 predicted to contain a glutamine residue instead of a proline in the crucial second position of the open reading frame. The 20 MIF-1-type sequences display between 18% and 51% (mean = 39% ± 0.08) identity with CeMIF-1 (Table 1) and cluster by clade (Figure 1). Pairwise analysis of the 13 MIF-2-type sequences reveals a greater level of conservation between homologs, with 28%–65% (mean = 47% ± 0.13) identity (Table 1). When included in a multiple-sequence alignment with 35 nematode MIFs, hMIF clusters with the MIF-1-type sequences (not shown), although CeMIF-1 and -2 are 35% and 33% identical to hMIF, respectively.
Table 1.
Distribution of transcribed MIF genes across the phylum Nematoda
| Speciesa | Accession numberb |
Sequence/ DBc |
Typed | %IDe |
|---|---|---|---|---|
| Clade I | ||||
| Trichuris muris | BM277412 | E | 1 | 42 |
| Trichuris trichiura | CAB46355 | P | 1 | 43 |
| Trichinella pseudospiralis | AAL12630 | P | 1 | 42 |
| Trichinella spiralis | CAB46354 | P | 1 | 41 |
| Xiphinema index | CV507928 | E | 1 | 40 |
| Clade III | ||||
| Anisakis simplex | EH005411 | E | 2 | 40 |
| Ascaris lumbricoides | BU586680 | E | 1 | 40 |
| Ascaris suum | BAD24819 | P | 1 | 40 |
| Toxocara canis | BQ458133 | E | 1 | 39 |
| Brugia malayi | AAB60943 | P | 1 | 40 |
| Brugia malayi | AAF91074 | P | 2 | 40 |
| Dirofilaria immitis | BQ482396 | E | 1 | 38 |
| Onchocerca volvulus | AAK66563 | P | 1 | 43 |
| Onchocerca volvulus | AAK66564 | P | 2 | 41 |
| Wuchereria bancrofti | AAC82615 | P | 1 | 40 |
| Clade IV | ||||
| Heterodera schachtii | CF100446 | E | 2 | 35 |
| Heterodera glycines | CD749065 | E | 2 | 35 |
| Meloidogyne hapla | CF804195 | E | 1 | 26 |
| Meloidogyne incognita | CK984698 | E | 1 | 18 |
| Radopholus similis | CO961429 | E | 2 | 36 |
| Parastrongyloides trichosuri | BM513256 | E | 2 | 63 |
| Strongyloides stercoralis | BG224821 | E | 2 | 28 |
| Clade V | ||||
| Ancylostoma caninum | AW626839 | E | 1 | 48 |
| Ancylostoma caninum | ABU68338 | P | 2 | 63 |
| Ancylostoma ceylanicum | CB190146 | E | 1 | 28 |
| Ancylostoma ceylanicum | ABO31935 | P | 2 | 65 |
| Dictyocaulus viviparus | EV854186 | E | 2 | 57 |
| Haemonchus contortus | CB012470 | E | 1 | 51 |
| Haemonchus contortus | CB015598 | E | 2 | 57 |
| Ostertagia ostertagi | BQ457911 | E | 1 | 49 |
| Teladorsagia circumcincta | CB043804 | E | 1 | 41 |
| Caenorhabditis elegans (CeMIF-1) | NP_499536 | P | 1 | 100 |
| Caenorhabditis elegans (CeMIF-2) | NP_506003 | P | 2 | 100 |
| Caenorhabditis elegans (CeMIF-3) | NP_492069 | P | - | - |
| Caenorhabditis elegans (CeMIF-4) | NP_500968 | P | - | - |
Nematodes are classified into five clades. Clade II is not shown because there are no genetic data available for any clade II species.
Accession numbers of all sequences are available on GenBank (http://www.ncbi.nlm.nih.gov).
Sequence/DB (database); E, expressed sequence tag(s) (EST) is known; P, a complete protein open reading frame cDNA is available.
MIF sequence type based on homology to either Caenorhabditis elegans MIF-1 (CeMIF-1; NP_499536) or MIF-2 (CeMIF-2; NP_506003).
Percent identity to either CeMIF-1 or CeMIF-2 based on BLASTP of translated amino acid sequences.
Figure 1.
Dendrogram of type-1 and type-2 MIF sequences from across the phylum Nematoda based on amino acid sequence homology. All sequence information was obtained from NCBI GenBank as either translated expressed sequence tags (ESTs) or primary amino acid sequences. GenBank accession numbers for all sequences are listed in Table 1. The dendrogram was generated with Clustalw, and corresponding species clades are shaded by clusters or branches.
Structure of nematode MIF proteins
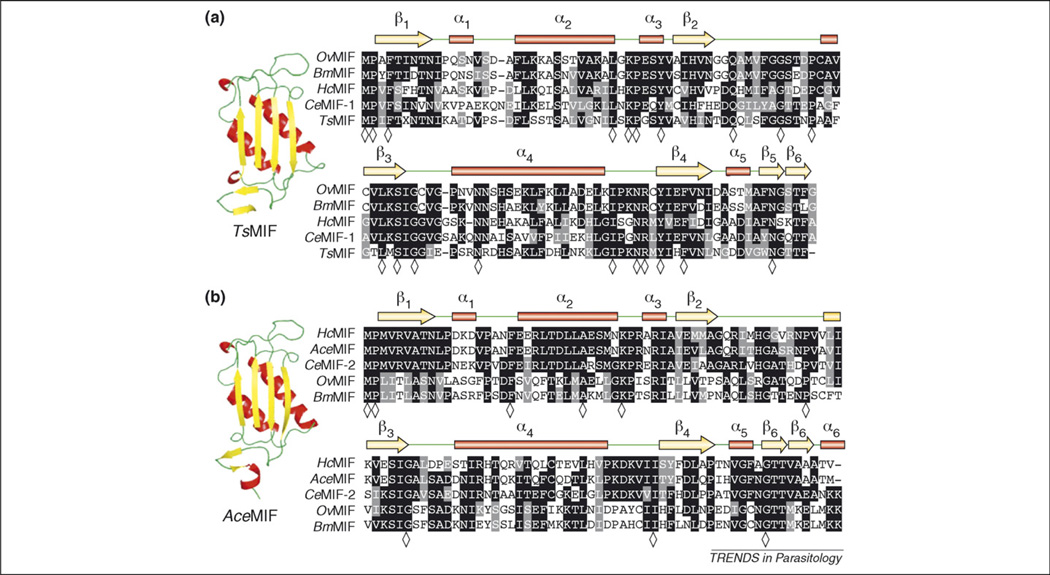
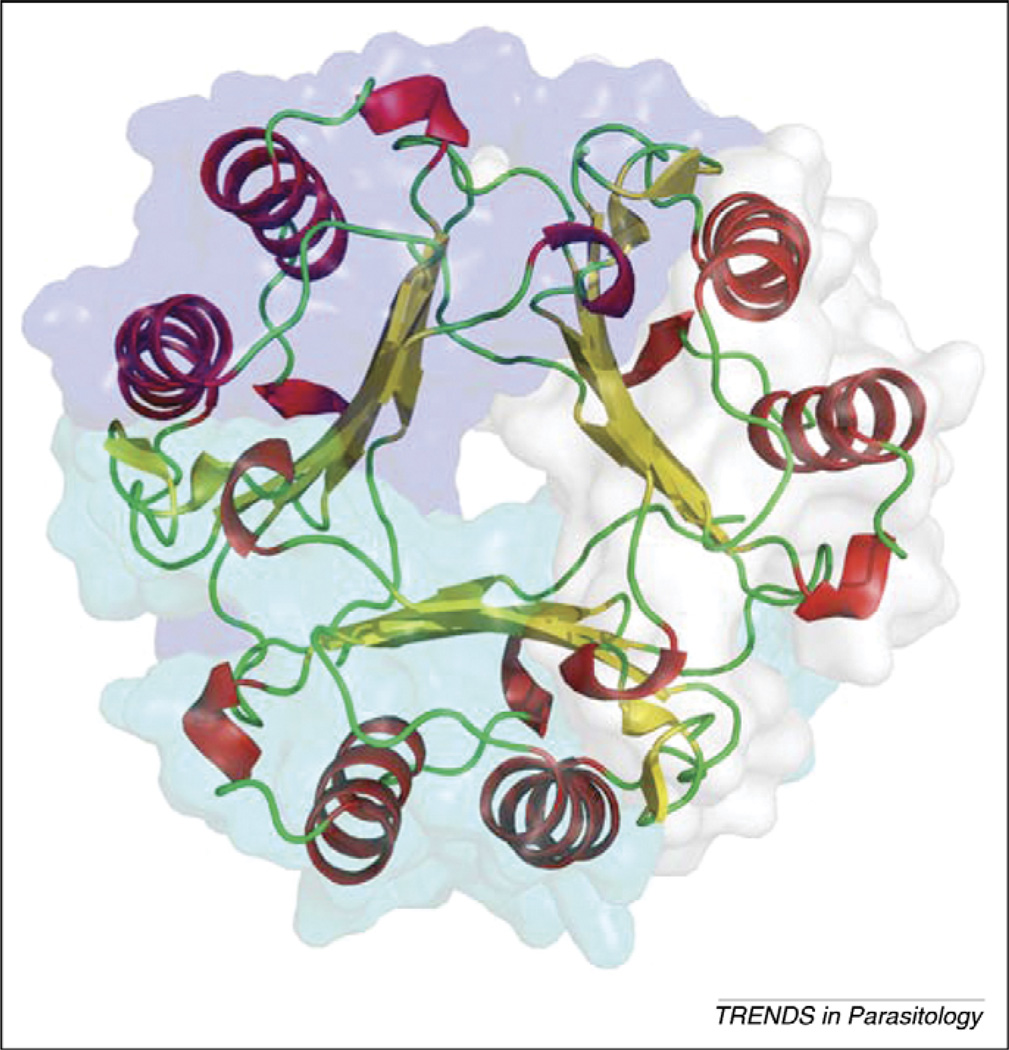
MIF proteins from Brugia malayi (BmMIF-1 and -2), Trichinella spiralis (TsMIF) and Ancylostoma ceylanicum (AceMIF) each have been well characterized, and the three-dimensional crystal structures have been solved [25,27,30]. Although the primary amino acid sequences of these parasite MIF molecules are not highly conserved among one another (<45% identical), secondary, tertiary and quaternary structures of the molecules are very similar [25,27,30] (Figure 2). The MIF monomer core consists of an antiparallel, four-stranded β-sheet. An additional β-strand is present before and after the core β-sheet, which is involved in subunit–subunit interactions in the homotrimer. The last β-strand is part of the C-terminal domain, which is particularly important for stable trimer formation [31]. The two β-strands from adjacent MIF monomers extend the core four-stranded β-sheet from neighboring monomers on either side to create a six-stranded β-sheet for each of the monomers, which pack against each other to form a cylinder surrounded by six α-helices (Figure 3). The formation of the trimer is essential for the protein’s catalytic function because residues from two adjoining subunits are involved in each of the three catalytic sites. Sedimentation velocity and equilibrium experiments show that hMIF is a strongly associated trimeric quaternary structure [32]. To examine whether nematode MIFs could have regulatory activity on hMIF by formation of heterotrimers, the electrostatic potential of the subunit interfaces of hMIF and AceMIF were examined and found to be non-complementary, meaning they would not allow the formation of heterotrimers [30].
Figure 2.
Multiple sequence alignments of nematode MIFs. (a) Type 1 MIFs, including prototype CeMIF-1, with corresponding secondary amino acid structure assignments listed above alignment including α-helices represented as red bars, β-strands as yellow arrows and random coils in green. Representative ribbon backbone diagram of type 1 MIF member Trichinella spiralis MIF (TsMIF). (b) Type 2 MIFs, including prototype CeMIF-2, with corresponding secondary amino acid structure assignments as described above. Representative ribbon backbone diagram of type-2 MIF member Ancylostoma ceylanicum MIF (AceMIF). Sequence alignments were generated with Clustalw and BoxShade programs. Identical residues are shaded in black and conserved residues are shaded in gray; residues identical in alignments of all type 1 (21 sequences) or type 2 (14 sequences) MIFs from Table 1 are indicated in the consensus line with a diamond (◇).
Figure 3.
Secondary structure and surface presentation of hMIF trimer. Helices (red), strands (yellow) and loops (green) are drawn with the surface of each subunit in different color.
Parasitic nematode MIF tautomerase activity: structure and function
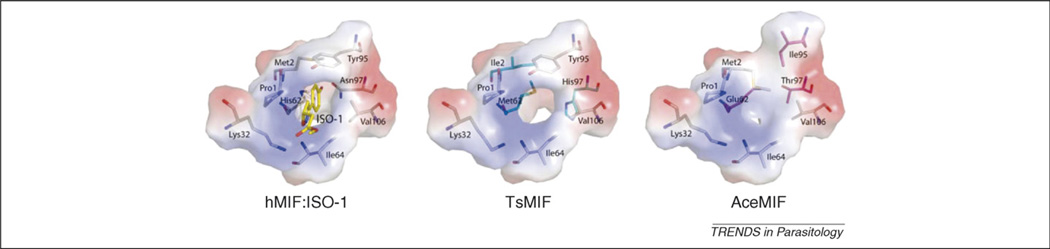
Both mammalian and helminth MIFs catalyze the ketoenol isomerization of small aromatic substrates such as hydroxyphenylpyruvate and L-dopachrome methyl ester (DCME). Post-translational cleavage of the initiating methionine exposes an N-terminal catalytic proline (Pro1) that is essential for MIF tautomerase activity [33]. This amino acid and other invariant residues (including Lys32, Ile64 and Val106) form the MIF active site where substrate molecules interact (Figure 4). The amino acid composition of the active site also modifies the size of the catalytic pocket and subsequent activities [30] because mutation of Pro1 to a glycine significantly reduces both the catalytic and cytokine-stimulatory activities of mammalian and nematode MIF molecules [27,33]. By contrast, the Cys57-Xaa-Xaa-Cys60 motif, responsible for the thiol-protein oxidoreductase activity of the thioredoxin family of proteins, uniformly is present in the mammalian MIF proteins but not in all helminth orthologs [34]. Although the Cys-Xaa-Xaa-Cys motif is present in type-1 MIFs of clade III parasitic nematodes, evidence for oxidoreductase activity in parasitic nematode MIFs currently is lacking.
Figure 4.
Distinct amino acid composition in the active sites of human and nematode MIFs. The active site residues located within non-covalent interaction distances to the hMIF inhibitor ISO-1 are represented in stick model in each MIF with the corresponding electrostatic surface potential. The crystallographically determined structure of ISO-1 is colored in yellow with hMIF (human MIF) on the left. The active site of TsMIF (Trichinella spiralis) is in the middle with the residues distinct from those of hMIF in cyan. The active site of AceMIF is on the right with the residues distinct from those of hMIF in maroon. The electrostatic surface potential of all the MIFs are scaled equally to each other.
Parasitic nematode MIFs as chemoattractants
Leukocyte recruitment is a crucial step in mediating inflammation within tissues. Recombinant BmMIF-1 and -2 (rBmMIF-1 and -2), TsMIF (rTsMIF) and AceMIF (rAceMIF) induce chemotaxis and prevent the random migration of human monocytes similar to recombinant hMIF. The elaboration of MIF by helminths might regulate the number and function of immune cells at the host–parasite interface because sufficient quantities of MIF at the site of infection could theoretically prevent the egress of sensitized antigen-presenting cells to peripheral immune centers. Similarly, parasite MIF molecules might function by delaying immune recognition by lymphocytes during parasite tissue migration, enabling the establishment of a successful infection.
Recently, AceMIF was demonstrated to interact with the hMIF receptor, CD74 [30]. Using an in vitro capture assay, it was demonstrated that excess rAceMIF partially displaces hMIF from its cognate receptor. It remains to be seen whether AceMIF acts as an agonist, driving activation of downstream proinflammatory pathways, or as an antagonist, engaging CD74 in a nonproductive or inhibitory fashion. Both scenarios could directly affect cellular function at the site of hookworm attachment and result in modulation of the host immune response (Box 1).
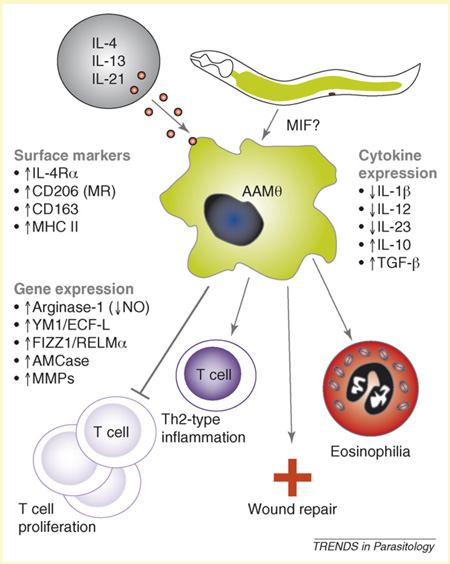
Box 1. Role of MIF in alternative activation of macrophages.
Alternative activation of macrophages in the setting of a variety of helminth infections, including Brugia malayi, Schistosoma mansoni, Nippostrongylus brasiliensis and Ascaris suum [53]. Alternatively activated macrophages (AAMΦs) are characterized by surface expression of IL-4R and the mannose receptor (CD206); upregulation of arginase-1 and subsequent decrease of NO production; and downregulation of IL-1β, IL-12 and IL-23 and increased IL-10 production [54]. AAΦs are associated with polarized Th2-type inflammatory responses, and their induction is stimulated by IL-4 and IL-13 (Figure I).
Surgical implantation of B. malayi adult worms into the peritoneal cavity of mice results in the recruitment of a progression of leukocytes including AAMΦs, a spike in neutrophils and an increase in eosinophils [23]. Transcripts for Ym1/ECF-L (chitinase/eosinophil chemoattractant factor), arginase-1 and other genes whose expression is associated with macrophage alternative activation represented more than 15% of the total mRNA population of macrophages recruited to the site of infection. The investigators further demonstrated that intraperitoneal injection of rBmMIF-1 partially reproduces this recruitment of eosinophils and alternative activation of macrophages as seen in laboratory infections [55]. Mutation of the N-terminal proline residue ablated this effect, suggesting a role of the catalytic center of BmMIF-1 in leukocyte recruitment. Recombinant BmMIF-1 upregulates Ym1/ECF-L expression in macrophages and leads to eosinophil recruitment in the absence of an active filarial infection, pointing to a direct role for BmMIF in the production of AAMΦ and modulation of the immune response to these filarial worms.
Parasitic nematode MIF effects on host cytokine production
An established immunomodulatory property of MIFs produced by parasitic nematodes is the ability to induce cytokine production in human monocytes. Treatment of human monocytes with rBmMIF caused an increase in secreted TNF-α and IL-8 cytokines and, most interestingly, induced the release of endogenous hMIF in vitro [27]. This fact, as Zang et al. point out, suggests that a positive feedback loop might exist in parasite-stimulated hMIF expression. Moreover, although MIF generally acts in a proinflammatory manner, evidence exists that suggests high levels of MIF might actually block AP-1-dependent proinflammatory gene expression by binding the transcription factor Jun activation domain-binding protein 1 (Jab1) [35]. By secreting MIF at the site of infection, helminths also might induce further production of endogenous host MIF, creating a local or possibly systemic anti-inflammatory host environment.
Tissue-specific expression of MIF in parasitic nematodes
The first MIF homologs from parasitic nematodes were described almost simultaneously by two separate groups working with four different parasites [24,36,37]. Pastrana et al. (1998) demonstrated that BmMIF-1 is expressed in the uterine structures of adult female worms and hypodermal muscles of the body wall of males and females [24]. In addition, immunoblots of ES products of microfilariae, L4 and adult-stage parasites were positive for BmMIF-1. These data led the authors to hypothesize a role for BmMIF in modulating cytokine production by antigen-presenting cells and promoting an immunological environment that facilitates parasite growth and survival. Subsequent studies have identified a second homolog (BmMIF-2) [27] from this filarid nematode.
Pennock et al. [37] described the purification of native MIF from soluble protein extracts of several parasitic helminths by using phenyl–agarose chromatography coupled with an evaluation of DCME tautomerase activity. Of the nine species analyzed, column fractions from three of the parasite extracts catalyzed the tautomerization of the DCME substrate. N-terminal amino acid sequencing confirmed that these proteins from T. spiralis, Trichuris muris and Brugia pahangi shared sequence identity with hMIF. Curiously, no tautomerase activity was detectable in any of the platyhelminth parasite extracts, which included the trematodes Schistosoma mansoni, Schistosoma japonicum and Schistosoma haematobium and the cestode Hymenolepis diminuta.
In a separate study [38] western blot analyses identified TsMIF in extracts of adult worms and muscle-encysted larvae. TsMIF-specific antibodies bound to hypodermal muscle cells in the body wall and stichosome (exocrine organ) of encysted larvae. Although TsMIF was not detected in ES products derived from larvae, the authors suggest that TsMIF might alter the host immune response by preventing the accumulation of macrophages around the cysts of T. spiralis-infected cells and cardiac muscle after larval migration through these tissues. This modulation might play a part in the success of long-term infection of host nurse cells by intracellular larvae.
Expression of MIF was shown to be greater in the adult stages of B. malayi and T. spiralis compared to larval stages [26,27]. This is in contrast with an MIF ortholog from AceMIF, which is most highly expressed in infectious L3 larvae and also was present in adult hookworm ES products [30]. Native sequence rAceMIF exhibits tautomerase and chemoattractant activities similar to hMIF and orthologs from other parasitic nematodes. It has been hypothesized that AceMIF is secreted by hookworms to subvert the host immune system during the larval tissue migration and while attached to the small intestine. This might be especially effective during the early phases of infection, when migrating worms are exposed to the host immune system within tissues and locally at the site of attachment after establishment within the gut of the host.
Using reporter gene expression techniques, isoforms of CeMIF-2 and CeMIF-3 protein were localized to the body wall and musculature of C. elegans. This pattern of expression is consistent with that seen in the parasitic nematodes [24,38]. During the entrance into the developmentally arrested dauer stage of C. elegans, CeMIF-2 and CeMIF-3 transcripts were 100-fold more abundant than the preceding L2 stage [28]. Presumably, this increase in transcription indicates that MIF is an important regulator of dauer homeostasis because transcript levels returned to L2 stage after resumption of development to the adult stage (Box 2).
Box 2. MIFs as nematode cytokines?
The possibility that parasitic nematode MIFs might, in fact, be acting in the classical sense as cytokine orthologs exists. They might represent mediators of the molecular crosstalk allowing for proper parasite development, although data supporting such a hypothesis are lacking. Transgenic reporter and immunolocalization studies suggest that C. elegans MIFs have roles in development of the dauer stage [28]. The dauer stage, comparable to the infectious L3 stage in many parasitic nematodes, is developmentally arrested, motile, nonfeeding, environmentally resistant and long-lived. Triggers for the resumption of development are also analogous: increased food availability for C. elegans and the location of a suitable host for parasitic nematodes. In Caenorhabditis spp. the process of dauer formation is mediated, in part, by homologs of the TGF-β pathway. Four TGF-β family members are encoded in the C. elegans genome [36]. One member, DAF-7 (a TGF-β-like ligand), controls entry and exit from the developmental arrest represented by the dauer larvae and acts via the well-characterized TGF-β signaling pathway. The genome of C. elegans also contains the type I and type II receptors that bind DAF-7 [56]. In terms of parasitic nematodes, homologs of DAF-7 have been isolated from B. malayi (tgh-1 and -2) [57] and Ancylostoma caninum [58]. Although only type I TGF-β receptors have been identified thus far, the presence of other members [59] of this pathway suggests that the signal transduction cascade is fully functional. In support of the hypothesis that MIF is involved in parasite development, a TBLASTN search of NCBI EST database (http://www.ncbi.nlm.nih.gov/dbEST/) yields three ESTs with high homology to human CD74 receptor (GenBank accession number NP_001020330) from B. malayi. Investigation in this area might lead to further insight into the potential role of MIF in filarial parasite developmental biology and the possibility that the parasitic MIF-CD74 interaction is a target for drug development.
MIFs from other parasitic organisms
Orthologs of MIF have been identified in numerous protozoan and metazoan parasite genomes. These other molecules – including DDT (D-dopachrome tautomerase), CHMI (5-carboxymethyl-2-hydroxymuconate isomerase) and 4-OT (oxalocrotonate tautomerase, found only in bacteria) – share similar tautomerization activities with MIF [39]. Miska et al. (2007) constructed a phylogenetic tree containing sequences from members of the tautomerase superfamily of proteins, which produced distinct branch groups that contained bacterial (Yersinia pestis, Escherichia coli, Salmonella enteritica) CHMI sequences, mammalian (Homo sapiens, Mus musculus, Rattus norvegicus) DDT and nematode (C. elegans, O. volvulus, B. malayi) MIF sequences. Apicomplexan (Plasmodium spp., Toxoplasma gondii, Eimeria spp.) MIF and plant (Arabidopsis spp., Oryza sativa) MIF-like sequences clustered in distinct branches, as did other vertebrate and parasitic nematode MIF sequences.
The role of host MIF in macrophage-mediated killing of Leishmania spp. parasites has been investigated extensively. Early studies identified host MIF as a potent activator of macrophages and described its role in cell-mediated immune defenses [40]. MIF−/− mice infected with L. major developed significantly larger lesions and harbored greater parasite burdens than MIF+/+ mice, suggesting a protective role for host MIF in cutaneous leishmaniasis [41]. Interestingly, two MIF orthologs that recently have been isolated from L. major possess structural and functional homology to hMIF, including tautomerase, chemotactic and anti-apoptotic activities [42].
In the mouse model of Taenia crassiceps infection, MIF−/− knockout mice were more susceptible to infection and harbored higher parasite burdens relative to MIF+/+ mice [43]. This phenomenon might be driven by a lack of macrophage activation and higher levels of interferon gamma (IFN-γ) and interleukin-13 (IL-13). Peritoneal macrophages isolated from T. crassiceps-infected, MIF−/− mice also produced significantly less TNF-α, IL-12 and nitric oxide (NO) upon stimulation.
Recently it was reported that coinfection with T. crassiceps is associated with increased susceptibility of mice to Leishmania spp., suggesting an induction of AAMΦ by the cestode despite abundant IFN-γ produced in response to Leishmania infection [44]. Moreover, a search of the NCBI EST database (http://www.ncbi.nlm.nih.gov/dbEST/) yielded no sequences with significant homology to MIF in parasitic platyhelminths, only from the free-living turbellarian flatworms Convoluta pulchra (GenBank accession number EV601112), Macrostomum lignano (GenBank accession number EG957856) and Schmidtea mediterranea (GenBank accession number DN298545). These in silico findings are consistent with the in vitro work of others describing the absence of MIF homologs in parasitic trematodes [37].
Most interestingly, recent characterizations of MIF from Plasmodium spp. [45] and Eimeria spp. [39] describe stage-specific expression patterns and cellular distribution, suggesting that MIF homologs from these parasitic protozoa are expressed throughout development and are secreted into the host environment. Furthermore, MIF−/− knockout mice suffered less severe anemia and exhibited increased survival when infected with Plasmodium chabaudi when compared to age-matched wild-type controls [46]. Data from this same study indicate that MIF might alter normal MAPK signaling, leading to an inhibition of erythroid differentiation and hemoglobin production.
Nematode MIF orthologs as drug targets
Differences in the three-dimensional structures of human and parasitic nematode MIF molecules should enable the successful development of selective inhibitors. In particular, because the catalytic site plays an essential role in the immunomodulatory activity of mammalian and nematode MIFs [27,33], targeting this site of molecular interaction represents a viable strategy for blocking the host and/or parasite cytokine. It is known, for example, that substitution of the first N-terminal proline with alanine reduces both the enzymatic [47] and cytokine activity [33] of MIF. It also has been reported that small molecule tautomerase inhibitors, including (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester (ISO-1) and its phenolic hydrazone or carbonyloxime derivatives, inhibit hMIF activity [48–50]. Recently it was demonstrated that ISO-1, despite its potent inhibitory activity against hMIF, does not block AceMIF in assays of tautomerase and cell migration activity, providing proof of concept for the development of selective parasite-specific MIF inhibitors [30].
Drugs or vaccines that specifically target nematode MIF could have a therapeutic value by either preventing infection or facilitating parasite expulsion from an infected individual. Just as hMIF inhibitors have been developed by using rational drug design [49–51], selective inhibitors of nematode MIFs also could be designed based on the known structures of the active site and substrate(s). De novo lead compound identification or modification of currently available inhibitors also could be performed in silico, as reviewed recently for hMIF inhibitors [52]. For example, a recently completed high-throughput screen of small molecule libraries successfully identified multiple selective inhibitors of AceMIF, and work currently is under way to define the structural basis for their species specificity (E. Lolis and Y. Cho, unpublished). It is anticipated that these inhibitors will not only facilitate studies of the role of AceMIF in the biology of Ancylostoma but also serve as lead compounds for novel chemotherapeutic agents for the treatment of hookworm and possibly other parasitic nematode infections.
Concluding remarks
It is recognized that most helminth infections are associated with a Th2-biased immune response, altering cytokine production and promoting B-cell antibody class switching [53]. However, the mechanisms underlying helminth immune modulation, as well as the degree to which this phenomenon represents a parasite survival strategy (versus a host defense response), remain to be fully elucidated. Thus, the exact reason(s) why parasitic nematodes produce inflammatory cytokine orthologs like MIF, which are capable of stimulating host immune responses against the pathogen, remains unknown. However, the fact that nematodes produce these highly active cytokines in a stage- and species-dependent manner strongly suggests a crucial role in parasite biology, as well as pathogenesis. In fact, many questions about MIF remain, including:
Do nematode MIFs function specifically to subvert host immune responses to suit the parasite?
Are nematode MIF molecules required for parasite growth and development or do they also regulate responses to host defenses?
Do multiple forms of MIF in a single species of nematode represent individual isoforms or does each play a unique role?
Why are there no MIF genes in parasitic flatworms?
It is hoped that future research will provide answers to these and other important questions, ultimately leading to a better understanding of nematode biology and providing new strategies through which the myriad diseases caused by these globally important parasites might eventually be controlled.
Figure I.
Illustration of alternative activation of macrophages and potential mechanism for parasitic nematode-derived, MIF-induced production of AAMΦs [23]. Alternative activation of macrophages is mediated by interleukin-4 (IL-4), IL-13 and possibly IL-21 acting through surface receptors. Characteristic features of AAMΦs include: upregulation of surface receptors including IL-4 receptor (IL-4R), CD-206 (mannose receptor), CD163 (group B scavenger receptor), and MHC II (major histocompatibility class II), among others [54,60]; increased transcription of arginase-1, Ym1/ECF-L (eosinophil chemotactic factor-L), FIZZ1/RELMα (found in inflammatory zone/resistin-like molecule), AMCase (acidic mammalian chitinase) and MMPs (matrix metalloproteinases), among others [53]; modulated expression of cytokines IL-1β, IL-12, IL-10 and transforming growth factor (TGF)- β [4,54]; suppression of T-cell proliferation Th2-type inflammation; and wound repair and eosinophilia [61].
Acknowledgements
The authors would like to thank Richard Bungiro and Blaise Dondji for helpful discussions and Lisa Harrison for critical review of this manuscript. This work was supported by National Institutes of Health Grants AI065029 (to E.L.), AI42310 (to R.B.), AI51306 (to R.B.) and AI058980 (to M.C.) and a Yale University James H. Brown – Alexander B. Coxe Fellowship in the Medical Sciences (to J.J.V.).
References
- 1.Else KJ. Have gastrointestinal nematodes outwitted the immune system? Parasite Immunol. 2005;27:407–415. doi: 10.1111/j.1365-3024.2005.00788.x. [DOI] [PubMed] [Google Scholar]
- 2.van Riet E, et al. Chronic helminth infections induce immunomodulation: consequences and mechanisms. Immunobiology. 2007;212:475–490. doi: 10.1016/j.imbio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Bungiro R, Cappello M. Hookworm infection: new developments and prospects for control. Curr. Opin. Infect. Dis. 2004;17:421–426. doi: 10.1097/00001432-200410000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Flaster H, et al. The macrophage migration inhibitory factor-glucocorticoid dyad: regulation of inflammation and immunity. Mol. Endocrinol. 2007;21:1267–1280. doi: 10.1210/me.2007-0065. [DOI] [PubMed] [Google Scholar]
- 5.Bucala R. MIF: Most Interesting Factor. World Scientific; 2007. [Google Scholar]
- 6.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153:80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 7.David JR. Delayed hypersensitivity in vitro: its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc. Natl. Acad. Sci. U. S. A. 1966;56:72–77. doi: 10.1073/pnas.56.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leng L, Bucala R. Insight into the biology of macrophage migration inhibitory factor (MIF) revealed by the cloning of its cell surface receptor. Cell Res. 2006;16:162–168. doi: 10.1038/sj.cr.7310022. [DOI] [PubMed] [Google Scholar]
- 9.Weiser WY, et al. Molecular cloning of a cDNA encoding a human macrophage migration inhibitory factor. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7522–7526. doi: 10.1073/pnas.86.19.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernhagen J, et al. The emerging role of MIF in septic shock and infection. Biotherapy. 1994;8:123–127. doi: 10.1007/BF01878495. [DOI] [PubMed] [Google Scholar]
- 11.Bernhagen J, et al. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature. 1993;365:756–759. doi: 10.1038/365756a0. [DOI] [PubMed] [Google Scholar]
- 12.Bernhagen J, et al. Purification, bioactivity, and secondary structure analysis of mouse and human macrophage migration inhibitory factor (MIF) Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 13.Lue H, et al. Rapid and transient activation of the ERK MAPK signalling pathway by macrophage migration inhibitory factor (MIF) and dependence on JAB1/CSN5 and Src kinase activity. Cell. Signal. 2006;18:688–703. doi: 10.1016/j.cellsig.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat. Rev. Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flieger O, et al. Regulated secretion of macrophage migration inhibitory factor is mediated by a non-classical pathway involving an ABC transporter. FEBS Lett. 2003;551:78–86. doi: 10.1016/s0014-5793(03)00900-1. [DOI] [PubMed] [Google Scholar]
- 16.Calandra T, et al. Protection from septic shock by neutralization of macrophage migration inhibitory factor. Nat. Med. 2000;6:164–170. doi: 10.1038/72262. [DOI] [PubMed] [Google Scholar]
- 17.Leech M, et al. Regulation of p53 by macrophage migration inhibitory factor in inflammatory arthritis. Arthritis Rheum. 2003;48:1881–1889. doi: 10.1002/art.11165. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa H, et al. An antibody for macrophage migration inhibitory factor suppresses tumour growth and inhibits tumour-associated angiogenesis. Cytokine. 2000;12:309–314. doi: 10.1006/cyto.1999.0562. [DOI] [PubMed] [Google Scholar]
- 19.Shi X, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 2007;13:587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell RA, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 2002;99:345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleemann R, et al. Disulfide analysis reveals a role for macrophage migration inhibitory factor (MIF) as thiol-protein oxidoreductase. J. Mol. Biol. 1998;280:85–102. doi: 10.1006/jmbi.1998.1864. [DOI] [PubMed] [Google Scholar]
- 23.Falcone FH, et al. A Brugia malayi homolog of macrophage migration inhibitory factor reveals an important link between macrophages and eosinophil recruitment during nematode infection. J. Immunol. 2001;167:5348–5354. doi: 10.4049/jimmunol.167.9.5348. [DOI] [PubMed] [Google Scholar]
- 24.Pastrana DV, et al. Filarial nematode parasites secrete a homologue of the human cytokine macrophage migration inhibitory factor. Infect. Immun. 1998;66:5955–5963. doi: 10.1128/iai.66.12.5955-5963.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan TH, et al. Macrophage migration inhibitory factor of the parasitic nematode Trichinella spiralis. Biochem. J. 2001;357:373–383. doi: 10.1042/0264-6021:3570373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Z, et al. Expression of excretory and secretory protein genes of Trichinella at muscle stage differs before and after cyst formation. Parasitol. Int. 2002;51:155–161. doi: 10.1016/s1383-5769(02)00003-x. [DOI] [PubMed] [Google Scholar]
- 27.Zang X, et al. Homologues of human macrophage migration inhibitory factor from a parasitic nematode. Gene cloning, protein activity, and crystal structure. J. Biol. Chem. 2002;277:44261–44267. doi: 10.1074/jbc.M204655200. [DOI] [PubMed] [Google Scholar]
- 28.Marson AL, et al. Macrophage migration inhibitory factor (mif) transcription is significantly elevated in Caenorhabditis elegans dauer larvae. Gene. 2001;278:53–62. doi: 10.1016/s0378-1119(01)00706-5. [DOI] [PubMed] [Google Scholar]
- 29.Mitreva M, et al. Comparative genomics of nematodes. Trends Genet. 2005;21:573–581. doi: 10.1016/j.tig.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Cho Y, et al. Structural and functional characterization of a secreted hookworm macrophage migration inhibitory factor (MIF) that interacts with the human MIF receptor CD74. J. Biol. Chem. 2007;282:23447–23456. doi: 10.1074/jbc.M702950200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mischke R, et al. Structure activity studies of the cytokine macrophage migration inhibitory factor (MIF) reveal a critical role for its carboxy terminus. FEBS Lett. 1997;414:226–232. doi: 10.1016/s0014-5793(97)01039-9. [DOI] [PubMed] [Google Scholar]
- 32.Philo JS, et al. Re-examining the oligomerization state of macrophage migration inhibitory factor (MIF) in solution. Biophys. Chem. 2004;108:77–87. doi: 10.1016/j.bpc.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Swope M, et al. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. EMBO J. 1998;17:3534–3541. doi: 10.1093/emboj/17.13.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiele M, Bernhagen J. Link between macrophage migration inhibitory factor and cellular redox regulation. Antioxid. Redox Signal. 2005;7:1234–1248. doi: 10.1089/ars.2005.7.1234. [DOI] [PubMed] [Google Scholar]
- 35.Kleemann R, et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408:211–216. doi: 10.1038/35041591. [DOI] [PubMed] [Google Scholar]
- 36.Patterson GI, Padgett RW. TGF beta-related pathways. Roles in Caenorhabditis elegans development. Trends Genet. 2000;16:27–33. doi: 10.1016/s0168-9525(99)01916-2. [DOI] [PubMed] [Google Scholar]
- 37.Pennock JL, et al. Rapid purification and characterization of L-dopachrome-methyl ester tautomerase (macrophage-migration-inhibitory factor) from Trichinella spiralis, Trichuris muris and Brugia pahangi. Biochem. J. 1998;335:495–498. doi: 10.1042/bj3350495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Z, et al. Molecular expression and characterization of a homologue of host cytokine macrophage migration inhibitory factor from Trichinella spp. J. Parasitol. 2003;89:507–515. doi: 10.1645/0022-3395(2003)089[0507:MEACOA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 39.Miska KB, et al. Characterisation of macrophage migration inhibitory factor from Eimeria species infectious to chickens. Mol. Biochem. Parasitol. 2007;151:173–183. doi: 10.1016/j.molbiopara.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 40.Weiser WY, et al. Human recombinant migration inhibitory factor activates human macrophages to kill Leishmania donovani. J. Immunol. 1991;147:2006–2011. [PubMed] [Google Scholar]
- 41.Satoskar AR, et al. Migration-inhibitory factor gene-deficient mice are susceptible to cutaneous Leishmania major infection. Infect. Immun. 2001;69:906–911. doi: 10.1128/IAI.69.2.906-911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamir D, et al. A Leishmania ortholog of macrophage migration inhibitory factor modulates host macrophage responses. J. Immunol. 2008;180:8250–8261. doi: 10.4049/jimmunol.180.12.8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Sosa M, et al. Macrophage migration inhibitory factor plays a critical role in mediating protection against the helminth parasite Taenia crassiceps. Infect. Immun. 2003;71:1247–1254. doi: 10.1128/IAI.71.3.1247-1254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez-Sosa M, et al. Acute cysticercosis favours rapid and more severe lesions caused by Leishmania major and Leishmania mexicana infection, a role for alternatively activated macrophages. Cell. Immunol. 2006;242:61–71. doi: 10.1016/j.cellimm.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Augustijn KD, et al. Functional characterization of the Plasmodium falciparum and P. berghei homologues of macrophage migration inhibitory factor. Infect. Immun. 2007;75:1116–1128. doi: 10.1128/IAI.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDevitt MA, et al. A critical role for the host mediator macrophage migration inhibitory factor in the pathogenesis of malarial anemia. J. Exp. Med. 2006;203:1185–1196. doi: 10.1084/jem.20052398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lubetsky JB, et al. Pro-1 of macrophage migration inhibitory factor functions as a catalytic base in the phenylpyruvate tautomerase activity. Biochemistry. 1999;38:7346–7354. doi: 10.1021/bi990306m. [DOI] [PubMed] [Google Scholar]
- 48.Al-Abed Y, et al. ISO-1 binding to the tautomerase active site of MIF inhibits its pro-inflammatory activity and increases survival in severe sepsis. J. Biol. Chem. 2005;280:36541–36544. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- 49.Crichlow GV, et al. Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor. J. Biol. Chem. 2007;282:23089–23095. doi: 10.1074/jbc.M701825200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dabideen DR, et al. Phenolic hydrazones are potent inhibitors of macrophage migration inhibitory factor proinflammatory activity and survival improving agents in sepsis. J. Med. Chem. 2007;50:1993–1997. doi: 10.1021/jm061477+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dios A, et al. Inhibition of MIF bioactivity by rational design of pharmacological inhibitors of MIF tautomerase activity. J. Med. Chem. 2002;45:2410–2416. doi: 10.1021/jm010534q. [DOI] [PubMed] [Google Scholar]
- 52.Orita M, et al. Macrophage migration inhibitory factor and the discovery of tautomerase inhibitors. Curr. Pharm. Des. 2002;8:1297–1317. doi: 10.2174/1381612023394674. [DOI] [PubMed] [Google Scholar]
- 53.Kreider T, et al. Alternatively activated macrophages in helminth infections. Curr. Opin. Immunol. 2007;19:448–453. doi: 10.1016/j.coi.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 55.Nair MG, et al. Chitinase and Fizz family members are a generalized feature of nematode infection with selective upregulation of Ym1 and Fizz1 by antigen-presenting cells. Infect. Immun. 2005;73:385–394. doi: 10.1128/IAI.73.1.385-394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw WM, et al. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr. Biol. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maizels RM, et al. Immune evasion genes from filarial nematodes. Int. J. Parasitol. 2001;31:889–898. doi: 10.1016/s0020-7519(01)00213-2. [DOI] [PubMed] [Google Scholar]
- 58.Brand AM, et al. Identification of a DAF-7 ortholog from the hookworm Ancylostoma caninum. Int. J. Parasitol. 2005;35:1489–1498. doi: 10.1016/j.ijpara.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Freitas TC, Arasu P. Cloning and characterisation of genes encoding two transforming growth factor-β-like ligands from the hookworm, Ancylostoma caninum. Int. J. Parasitol. 2005;35:1477–1487. doi: 10.1016/j.ijpara.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 60.Mosser DM. The many faces of macrophage activation. J. Leukoc. Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 61.Maizels RM, Yazdanbakhsh M. Immune regulation by helminth parasites: cellular and molecular mechanisms. Nat. Rev. Immunol. 2003;3:733–744. doi: 10.1038/nri1183. [DOI] [PubMed] [Google Scholar]