Abstract
Megakaryocyte (MK) development is critically informed by plasma membrane-localized receptors that integrate a multiplicity of environmental cues. Given that the current understanding about receptors and ligands involved in megakaryocytopoiesis is based on single targets, we performed a genome-wide search to identify a plasma membrane receptome for developing MKs. We identified 40 transmembrane receptor genes as being upregulated during MK development. Seven of the 40 receptor-associated genes were selected to validate the dataset. These genes included: interleukin-9 receptor (IL9R), transforming growth factor, β receptor II (TGFBR2), interleukin-4 receptor (IL4R), colony stimulating factor-2 receptor-beta (CSFR2B), adiponectin receptor (ADIPOR2), thrombin receptor (F2R), and interleukin-21 receptor (IL21R). RNA and protein analyses confirmed their expression in primary human MKs. Matched ligands to IL9R, TGFBR2, IL4R, CSFR2B, and ADIPOR2 affected megakaryocytopoiesis. IL9 was unique in its ability to increase the number of MKs formed. In contrast, MK colony formation was inhibited by adiponectin, TGF-β, IL4, and GM-CSF. The thrombin-F2R axis affected platelet function, but not MK development, while IL21 had no apparent detectable effects. ADP-induced platelet aggregation was suppressed by IL9, TGF-β, IL4, and adiponectin. Overall, six of seven of the plasma membrane receptors were confirmed to have functional roles in MK and platelet biology. Also, results show for the first time that adiponectin plays a regulatory role in MK development. Together these data support a strong likelihood that the 40 transmembrane genes identified as being upregulated during MK development will be an important resource to the research community for deciphering the complex repertoire of environmental cues regulating megakaryocytopoiesis and/or platelet function.
Keywords: receptors, megakaryocytopoiesis, thrombocytopoiesis, hematopoiesis, microarray
megakaryocytopoiesis involves the proliferation and differentiation of hematopoietic stem cells to form megakaryocyte (MK) progenitors that undergo a maturation process that culminates in a release of ∼1011 platelets per day into the blood circulation (31). This process is tightly regulated by a number of factors, which include extracellular cues such as cytokines, cell-to-cell interactions, and cell-to-extracellular matrix interactions. Among cytokines that are known to act as important extracellular regulators of megakaryocytopoiesis is the main physiological stimulator of this specific cell-lineage commitment, thrombopoietin (TPO) (17). Additionally, interleukin 3 (IL3), interleukin 6 (IL6), interleukin 11 (IL11), stem cell factor (SCF), and fms-like tyrosine kinase 3 ligand act as positive regulators of MK development (17, 31). There are also several cytokines such as transforming growth factor-beta (TGF-β), platelet factor 4, and interleukin 4 (IL4) that are documented as negative regulators of MK development (31).
The importance of megakaryocytopoiesis and platelet biogenesis is apparent; morbidity and mortality from bleeding due to moderate to severe thrombocytopenia are major problems facing a wide range of patients. An effective treatment to combat bleeding disorders is the transfusion of allogeneic platelets into patients with low platelet numbers and/or functionally defective platelets. However, because platelet products have a short shelf-life and because there is also a high demand for their use in transfusion settings, it is not uncommon for medical facilities to experience shortages of transfusable platelets. Consequently, achieving a readily available and constant supply of allogeneic platelets for transfusion purposes continues to be a challenging endeavor for blood banks.
Platelet shortages may one day be alleviated through clinical-scale in vitro production of platelets from stem cells. Efforts to achieve such an endeavor are currently underway, but despite the progress made the number and quality of platelets produced in vitro are inefficient (25). As a result, it is still not feasible to produce large-scale numbers of functional MKs that can generate platelets in quantities that are needed for clinical applications (17). A key to achieve this endeavor will be knowing how to regulate an artificial environment that supports MK development and platelet biogenesis. This will necessitate a comprehensive understanding of the complex nature of the numerous environmental cues that facilitate the proliferation and differentiation of MK progenitors, MK maturation, and release of platelets from mature MKs.
In view of the fact that receptors found on the surface of a cell are responsible for integrating a multiplicity of environmental cues to instruct cells to perform biological responses (i.e., divide, differentiate, die), the objectives of this study are to use a systems biology approach to generate a map of receptors that are upregulated during the development of human MKs and to validate the robustness of the dataset. A list of receptor-associated genes upregulated during MK development is presented and is composed of well-characterized genes previously documented as playing a role in MK development and platelet biology. Also, there are genes in the dataset with assigned functions or unknown functions that have no prior association with the MK lineage of development. The strength of this dataset indicates that it should be useful for identifying novel ligand-receptor systems that play functional roles in MK development and/or in modulating platelet biology.
MATERIALS AND METHODS
Cytokines and antibodies.
Recombinant human cytokines IL3, IL6, SCF, interleukin 21 (IL21), interleukin 9 (IL9), thrombin, TGF-β, IL4, granulocyte/macrophage-colony stimulating factor (GM-CSF) and adiponectin were purchased from R&D Systems (Minneapolis, MN). Phycoerythrin (PE)-conjugated anti-human antibodies CD41a, CD61, and FITC-conjugated anti-human CD61 were purchased from BD Biosciences (San Jose, CA; http://www.bdbiosciences.com). PE-conjugated anti-human antibodies to IL9R, TGFBR2, and CSF2RB, and allophycocyanin-conjugated anti-human antibodies to IL4Ra, and IL21R were purchased from R&D Systems. Anti-human antibodies F2R was obtained from R&D and anti-human ADIPOR2 was purchased from ABBIOTEC (San Diego, CA). Rabbit anti-goat IgG, goat anti-mouse IgG1, and goat IgG isotype controls were purchased from Southern Biotech (Birmingham, AL). Mouse IgG1 isotype control was purchased from eBioscience (San Diego, CA).
Purification of human bone marrow CD34+/CD38lo cells.
Institutional Review Board approval was obtained for the isolation and purification of human progenitor cells. Briefly, after obtaining informed consent from the families of organ donors, we obtained total nucleated cells (TNCs) (42.1 ± 6.80 × 109, mean ± SE) from vertebral bodies procured by Northwest Tissue Services (Seattle, WA) (32). TNCs were applied to an Isolex 300 SA (Nexell, Irvine, CA) column to yield on average of 22.4 ± 2.95 × 107 CD34+ cells with purities of 87 ± 3.4% as determined by flow cytometry. Enriched fractions of CD34+ cells were subsequently frozen and stored in 1 ml aliquots containing 5–10 × 106 cells per vial at −135°C in X-VIVO 10 (Lonza, Walkersville, MD) medium containing 10% dimethyl sulfoxide (Research Industries Corporations, Salt Lake City, UT), 20% HyClone bovine serum (BS) (Invitrogen, Carlsbad, CA) and 2 mM l-glutamine (Invitrogen).
To isolate highly purified populations of CD34+/CD38lo cells, aliquots of frozen CD34+ cells were thawed rapidly at 37°C and costained with antibodies directed against CD34 and CD38 antigens. Adult bone marrow (BM) CD34+ cells were subpopulated based on coexpression of CD38 using a FACSVantage flow cytometer (Becton Dickinson, San Jose, CA) to obtain a CD34+/CD38lo population of cells with >95% purity (26).
Purification of human umbilical cord blood CD34+ cells.
We received donated human umbilical cord blood (UCB) after obtaining informed consent. UCB was processed by addition of 6% (wt/vol) Hetastarch (Hospira, Lake Forest, IL) to a final concentration of 1.2% (28), and gravity sedimentation over 60 min. The leukocyte-enriched supernatant was removed and centrifuged for 10 min at 300 g, and the leukocyte-poor supernatant was removed. The cell pellet was treated with ACK lysing buffer (Invitrogen), followed by a wash with PBS. To obtain highly purified CD34+ cells, leukocyte-enriched fractions were labeled with anti-CD34 antibody conjugated to magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). CD34+ cells were positively selected with an autoMACS separator according to the manufacturer's instruction. Greater than 90% of the enriched cells were CD34+ cells as determined by flow cytometry (FACSCaliber, Becton Dickinson).
Culture-derived megakaryocytes.
Purified populations of marrow CD34+/CD38lo cells or UCB CD34+ cells were seeded at a density of 5 × 104 cells/ml and cultured in serum-free X-VIVO 10 medium supplemented with a cytokine combination consisting of IL3 (10 ng/ml), IL6 (10 ng/ml), SCF (10 ng/ml), and TPO (50 ng/ml) (day 0) (12). The suspension cultures were incubated for 10 days at 37°C in a 5% CO2 humidified chamber.
Isolation of total RNA and cDNA synthesis.
According to manufacturer's instructions (RNAeasy kit; QIAGEN, Valencia, CA), RNA was isolated from uncultured cells, culture-derived MKs and other cell types as indicated. Isolated RNA was stored at −80°C until use. Total RNA (2 mg) from each sample was reverse transcribed into cDNA using Qiagen Omniscript according to the manufacturer's protocol.
Microarrays.
As we previously published (32), microarray analyses of uncultured CD34+/CD38lo cells and culture derived MKs were performed according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). Briefly, the quantity and purity of total RNA were analyzed by UV spectrophotometry, and the integrity of intact total RNA was verified. Biotin-labeled target cRNA was prepared from 8 μg of total RNA from each sample and hybridized to Human Genome U133A and U133B Affymetrix GeneChips (Affymetrix). The hybridized chips were washed and stained with streptavidin phycoerythrin solution, then scanned at a wavelength of 570 nm using an Affymetrix HP GeneArray Scanner (Santa Clara, CA).
Microarray data were analyzed with the Rosetta Resolver Expression Data Analysis System (Resolver) (Rosetta Biosoftware, Kirkland, WA), which utilizes an error-modeling-based approach verified through self-vs.-self experiments, rather than the nonparametric statistical method of MAS 5.0 (http://www.rosettainpharmatics.com/products/resolver/default.htm, website no longer accessible). The calculation of statistical confidence (P value) was derived from the total calculated error consisting of both random and systematic error. Gene expression profiles of significantly changed transcripts were then generated using a fold change cut-off of ≥1.5 and a P value of ≤0.01. Profiles from each of three biological replicate experiments were intersected to attain a set of reproducible genes.
Megakaryome and genome receptome enrichment gene list.
To define a list of training genes that make up the megakaryome, we used the Toppgene database (http://toppgene.cchmc.org/) (5). To derive this list, genes were identified from mouse and human gene ontology annotations, pathway associations, and those that have been observed in mouse gene knockout models that cause abnormal MK development or abnormal platelet function. Receptors localized to the plasma membrane were identified using the Human Plasma Membrane Receptome database (http://Receptome.Stanford.edu) (2).
Semiquantitative reverse transcriptase-polymerase chain reaction.
Total RNA (100 ng) from uncultured or cultured-derived MKs was reverse transcribed into cDNA using the SuperScript III First-Strand Synthesis Kit (Invitrogen), and PCR was performed using Platinum Taq DNA polymerase (Invitrogen). The thermocycling program consisted of: denaturation at 95°C for 10 s, followed by 40 cycles (35 cycles for Adiponectin Receptor 2) of PCR (95°C for 10 s, 55°C for 5 s, 72°C for 10 s), and an additional incubation for 10 min at 72°C. In each case, product identity was demonstrated by the presence of a single band of the appropriate size when analyzed by electrophoresis on a 1% agarose gel.
Platelet immunophenotyping and aggregation studies.
For immunophenotyping, seven volumes of human whole blood was drawn into one volume of anticoagulant citrate dextrose solution A (Citra Anticoagulant, Braintree, MA) and centrifuged at 120 g for 10 min with the centrifuge brake in the off mode. The top layer or platelet-rich plasma (PRP) was removed and centrifuged at 1,200 g for 10 min. The supernatant was removed, and the platelet pellet was resuspended in citrate-glucose-saline buffer containing 120 mM sodium chloride, 13 mM sodium citrate, and 30 mM glucose buffer prior to labeling with antibodies for flow cytometry or for preparing a cell lysate for Western blot analysis.
For aggregation studies, human whole blood was drawn into 3.8% sodium citrate and centrifuged at 120 g for 10 min, and the PRP was transferred into a fresh tube. The remaining whole blood was again centrifuged at 2,000 g for 10 min to obtain a platelet-poor plasma (PPP) fraction. Using the PPP, we adjusted the platelet count of the PRP to 250,000 platelets/μl. We determined percent aggregation using an AggRAM aggregometer (Helena Laboratories, Beaumont, TX) after adding 50 or 500 ng/ml of a given cytokine as indicated in text. Tyrode's buffer was added as negative control. For adenosine diphosphate (ADP)-induced platelet aggregation, we preincubated PRP samples with 500 ng/ml of a given cytokine for 5 min and then added 20 μM of ADP. The percent aggregation of platelets was determined by an AggRAM aggregometer.
Direct and indirect antigen labeling.
Cells were collected by centrifugation and resuspended into PBS/2% BS. Cells were then stained with FITC- or PE-conjugated antibodies or the corresponding isotype controls and incubated for 15 min. Cells were washed and analyzed on a FACSCalibur flow cytometer. For indirect labeling, after labeling cells with a primary antibody, we stained cells with FITC- or PE-conjugated secondary antibodies. Cells were then analyzed on a FACSCalibur flow cytometer.
MK colony-forming-unit assay.
Megakaryocyte colony-forming-unit (CFU-MK) assays were preformed according to the manufacturer's instructions (Stemcell Technologies, Vancouver, Canada). Briefly, UCB CD 34+ cells were cultured in collagen and serum-free MegaCult-C medium supplemented with 10 ng/ml IL3, 10 ng/ml IL6, 10 ng/ml SCF, 50 ng/ml TPO (36ST) along with an additional cytokine as indicated in the text. After 12 days, cultures were dehydrated, fixed, and then immunostained for CD41, a marker of MK differentiation.
TGF-β detection.
UCB CD34+ cells were cultured in X-VIVO 10 supplemented with 36ST for 10 days. Supernatant was collected and concentrated using Amicon Ultra-15 centrifugal filter units with ultracel-3 membrane (Millipore, Billerica, CA). Culture supernatant was split in half, and one-half of the fraction was treated with HCl for 10 min followed by neutralization with NaOH/HEPES, and the other half was not treated. Detection of TGF-β1 in the treated (i.e., total TGF-β1) and untreated (i.e., activated TGF-β1) fractions were performed using a human TGF-β1 ELISA immunoassay kit (R&D Systems). Concentrations of total and activated TGF-β1 were determined against a standard curve. The concentration of latent TGF-β1 in the culture supernatant was calculated by subtracting the concentration of activated TGF-β1 from total TGF-β1.
Statistical analysis.
Data are presented as means ± standard error of the mean, and significant differences were determined by ANOVA.
RESULTS
Receptor-associated genes upregulated during in vitro megakaryocytopoiesis.
A genome-wide search to identify all receptors whose ligands might have important functional roles in MK development was performed by using a powerful in vitro MK differentiation platform. This involved isolating marrow CD34+/CD38lo cells and culturing the cells for 10 days with a cytokine combination that was previously reported to induce unilineage megakaryocytopoiesis (12). In our hands, this resulted in 72 ± 5% of cells expressing the CD41 antigen at 10 days of culture and ∼90% by 14 days. Using uncultured CD34+/CD38lo cells and culture-derived MKs, we generated transcript expression profiles and compared the profiles with each other to identify differentially expressed genes. A heat map of gene expression levels for replicate samples showed that there was reasonable similarity among the datasets (data not shown). Analyses of the data indicated that a total of 3,035 genes significantly changed. Of the differentially expressed transcripts, expression levels for 1,267 increased (i.e., >1.5-fold, P ≤ 0.01) and expression levels for 1,768 decreased (−1.5-fold, P ≤ 0.01) (Supplements 1, A and B).1
To identify potential paracrine/autocrine signaling systems involved in MK development, we used the Human Plasma Membrane Receptome database (http://receptome.stanford.edu) to interrogate genes contained within the microarray dataset that are classified as membrane receptors. Investigation of the transcriptome database against a liganded receptome revealed that there were 40 upregulated (Table 1) and 37 downregulated ligand-receptor pairs (Table 2). The analyses also revealed that among the 40 upregulated receptors there were 10 classified as orphan receptors and among 37 downregulated genes there were 11 orphan receptors. The receptors were then surveyed against a defined set of “training genes” known to be specifically critical for the development and function of megakaryocytes and platelets (i.e., megakaryome, 519 genes) (Supplement 2). Of the 40 upregulated genes, 11 transcripts were found to be common to the megakaryome dataset. The common upregulated genes included ADAM10, CD55, CD63, F2R, ICAM2, IL6ST, ITGA2B, ITGB1, ITGB3, PTGIR, and SELP. Since it is unlikely that genes downregulated during MK development are involved in the function of MKs and platelets, it was not surprising that only one gene of the 37 downregulated receptor genes, ITGA2, was also found in the megakaryome dataset.
Table 1.
Upregulated plasma membrane receptor genes during megakaryocyte development
| Fold Change | Gene ID | Gene Identifier | Gene Title | Ligand |
|---|---|---|---|---|
| 55.9 | ITGB3 | M35999 | Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | Fibronectin, ADAM15, ADAM23 |
| 17.2 | ITGA2B | AF098114 | Integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) | Fibronectin 1, ICAM 4 |
| 16.8 | IL21R | NM_021798 | Interleukin 21 receptor | IL21 |
| 14.5 | LRP12 | NM_013437 | Low-density lipoprotein-related protein 12 | orphan receptor |
| 10.4 | ITGA6 | AV733308 | Integrin, alpha 6 | ADAM 2, ADAM9, DUSP18 |
| 9.0 | PTGER3 | L27489 | Prostaglandin E receptor 3 (subtype EP3) | Prostglandin E2 |
| 8.7 | CCRL2 | AF015524 | Chemokine (C-C motif) receptor-like 2 | orphan receptor |
| 8.6 | IL9R | NM_002186 | Interleukin 9 receptor | IL9 |
| 7.8 | ADAM10 | AU135154 | ADAM metallopeptidase domain 10 | orphan receptor |
| 7.2 | ITGB5 | NM_002213 | Integrin, beta 5 | ADAM9 |
| 7.1 | F2R | NM_001992 | Coagulation factor II (thrombin) receptor | Thrombin |
| 6.1 | HMMR | NM_012485 | Hyaluronan-mediated motility receptor (RHAMM) | Hyaluronan synthase 2 |
| 5.8 | PTGIR | NM_000960 | Prostaglandin I2 (prostacyclin) receptor (IP) | Prostacyclin |
| 5.2 | SELP | NM_003005 | Selectin P (granule membrane protein 140 kDa, antigen CD62) | Selectin P ligand, CD24 |
| 4.9 | SORT1 | BE742268 | Sortilin 1 | Nerve growth factor, beta subunit |
| 4.8 | THBD | NM_000361 | Thrombomodulin | Platelet factor 4 |
| 4.5 | CD40 | BF664114 | CD40 molecule, TNF receptor superfamily member 5 | CD40 ligand, lymphotoxin beta (TNF superfamily, member 3) |
| 4.0 | CSF2RB | AV756141 | Colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | Granulocyte-macrophage colony stimulating factor, IL3, IL5 |
| 3.9 | C3AR1 | U62027 | Complement component 3a receptor 1 | complement component 3 |
| 3.8 | ICAM2 | NM_000873 | Intercellular adhesion molecule 2 | ITGAL, ITGB2 |
| 3.7 | ADRA2A | AF284095 | Adrenergic, alpha-2A-, receptor | Dopamine beta-hydroxylase, plasma |
| 3.7 | GPR137B | NM_003272 | G protein-coupled receptor 137B | orphan receptor |
| 3.5 | LRP8 | NM_004631 | Low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | orphan receptor |
| 3.1 | HTR2A | NM_000621 | 5-hydroxytryptamine (serotonin) receptor 2A | Tryptophan hydroxylase 1 |
| 3.0 | CD63 | NM_001780 | CD63 molecule | orphan receptor |
| 2.9 | TGFBR2 | D50683 | transforming growth factor, beta receptor II (70/80 kDa) | TGFb |
| 2.8 | CD47 | Z25521 | CD47 molecule | Thrombospondin I & II |
| 2.7 | IL6ST | AL049265 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | IL6, leukemia inhibitory factor, oncostatin M, ciliary neurotrophic factor, IL11, IL6, NP (ortholog of mouse neuropoietin) |
| 2.7 | ACVR1 | NM_001105 | Activin A receptor, type I | Bone morphogenetic protein 6 & 7 |
| 2.7 | ITGB1 | BG500301 | integrin, beta 1 | ADAM 12, 15, 17, 2, 9 |
| 2.5 | IL4R | NM_000418 | Interleukin 4 receptor | IL4, IL13 |
| 2.2 | TSPAN4 | AF054841 | Tetraspanin 4 | orphan receptor |
| 2.1 | CD55 | BC001288 | CD55 molecule, decay accelerating factor for complement (Cromer blood group) | CD97 |
| 2.0 | ADIPOR2 | NM_024551 | Adiponectin receptor 2 | Adiponectin, C1Q and collagen domain containing |
| 1.9 | IL10RB | BC001903 | Interleukin 10 receptor, beta | IL10 |
| 1.8 | SEMA4D | NM_006378 | Sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D | Plexin B1 |
| 1.7 | PTPRA | AL121905 | protein tyrosine phosphatase, receptor type, A | orphan receptor |
| 1.6 | TSPAN31 | NM_005981 | Tetraspanin 31 | orphan receptor |
| 1.6 | TFG | NM_006070 | TRK-fused gene | orphan receptor |
| 1.6 | ATRN | AL132773 | attractin | Agouti signaling protein |
Fold change from 3 independent experiments; P value ≤ 0.01.
Table 2.
Plasma membrane receptor genes downregulated during megakaryocyte development
| Fold Change | Gene ID | Gene Identifier | Gene Title | Ligand |
|---|---|---|---|---|
| 0.63 | ADAM22 | AW242701 | Transcribed locus | Leucin-rich, glioma inactivated 1 (LGI1) |
| 0.62 | TYRO3 | D50479 | TYRO3 protein tyrosine kinase | growth arrest-specific 6 (GAS6), Protein S, alpha |
| 0.61 | GPR35 | AF089087 | G protein-coupled receptor 35 | kynurenic acid |
| 0.57 | EDA2R | NM_021783 | Ectodysplasin A2 receptor | Ectodysplasin A |
| 0.54 | GPR56 | AL554008 | G protein-coupled receptor 56 | Transglutaminase 2 (TGM2) |
| 0.50 | LILRB3 | NM_024318 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | orphan receptor |
| 0.46 | EMR2 | NM_013447 | Egf-like module containing, mucin-like, hormone receptor-like 2 | orphan receptor |
| 0.45 | NOTCH4 | AI743713 | Notch homolog 4 (Drosophila) | Delta-like 4 (DLL4) |
| 0.45 | CNTN1 | U07820 | Contactin 1 | Contactin-associated protein 1 (CNTNAP1), protein-tyrosine phosphatase zeta precursor (PTPRZ1) |
| 0.41 | FLT1 | AA058828 | Fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | Vascular endothelial growth factor (VEGF) |
| 0.39 | IL18R1 | NM_003855 | Interleukin 18 receptor 1 | IL18 |
| 0.39 | FGFR1 | M60485 | Fibroblast growth factor receptor 1 | FGF1, FGF3, FGF4 |
| 0.38 | CD53 | NM_000560 | CD53 molecule | Orphan Receptor |
| 0.36 | LPHN1 | NM_024679 | latrophilin 1 | SH3 and multiple ankyrin repeate domains 1 (SHANK1) |
| 0.29 | SELL | NM_000655 | Selectin L | CD34-molecule (CD34), complement factor H (CFH), ppodocalyxin-like 2 (PODXL2) |
| 0.28 | ITGA9 | NM_002207 | Integrin, alpha 9 | ADAM12, ADAM15 |
| 0.26 | PTGER2 | NM_000956 | Prostaglandin E receptor 2 (subtype EP2), 53 kDa | Prostglandin E2 |
| 0.21 | CD97 | NM_001784 | Leucocyte antigen CD97 | Decay-accelerating factor for complement (CD55) |
| 0.21 | PVRL2 | AI520949 | Poliovirus receptor-related 2 (herpesvirus entry mediator B) | orphan receptor |
| 0.21 | EPHB4 | NM_004444 | EPH receptor B4 | Ephrin B2 (EFNB2), ephrin B1 (EFNB1) |
| 0.20 | TSPAN32 | AF176071 | Tetraspanin 32 | orphan receptor |
| 0.16 | TSPAN18 | AL565381 | Transcribed locus | orphan receptor |
| 0.16 | ITGA2 | N95414 | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | CHAD, COL1A2, COL2A1 |
| 0.15 | IL17RA | BF939473 | Hypothetical protein LOC150166 | DUSP18, FN1 |
| 0.12 | GPRC5B | NM_016235 | G protein-coupled receptor, family C, group 5, member B | orphan receptor |
| 0.11 | TSPAN14 | BF025955 | Tetraspanin 14 | orphan receptor |
| 0.10 | TNFRSF10D | AI738556 | Tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain | TNF, Oligodendrocyte myelin glycoprotein (OMG) |
| 0.10 | ROBO4 | AA156022 | Roundabout homolog 4, magic roundabout (Drosophila) | SLIT, drosophila, homolog of, 2 (SLIT2) |
| 0.09 | GPR160 | BC000181 | G protein-coupled receptor 160 | orphan receptor |
| 0.07 | CD44 | AV700298 | CD44 molecule (Indian blood group) | hyaluronan synthase 2 (HAS2) |
| 0.04 | TSPAN5 | AI928507 | Tetraspanin 5 | orphan receptor |
| 0.03 | GPR161 | AI743151 | G protein-coupled receptor 161 | Dopamine beta-hydroxylase, plasma (DBH) |
| 0.02 | ITGA8 | BF939224 | Integrin, alpha 8 | Nephronectin (NPNT) |
| 0.02 | IFNGR1 | AI458949 | Interferon gamma receptor 1 | Interferon-gamma (IFNG) |
| 0.02 | GPR98 | AW339783 | G protein-coupled receptor 98 | orphan receptor |
| 0.003 | AGR2 | AI922323 | Anterior gradient homolog 2 (Xenopus laevis) | Prod 1 in the salamander model |
| 0.002 | ROR1 | AK000776 | CDNA FLJ20769 fis, clone COL06674 | orphan receptor |
Fold-change from 3 independent experiments; P value ≤ 0.01.
To validate the robustness of the microarray dataset, a subset of seven genes from the list of 40 upregulated ligand-receptor pairs was chosen for further examination. These seven receptor genes were chosen because their ligands were commercially available. Of the seven receptor genes, only the coagulation factor II (thrombin) receptor (F2R) was common to both the megakaryome and Human Plasma Membrane Receptome. The remaining six genes, IL21R, IL9R, CSF2RB, ADIPOR2, TGFBR2, and IL4R, were not found in the megakaryome database.
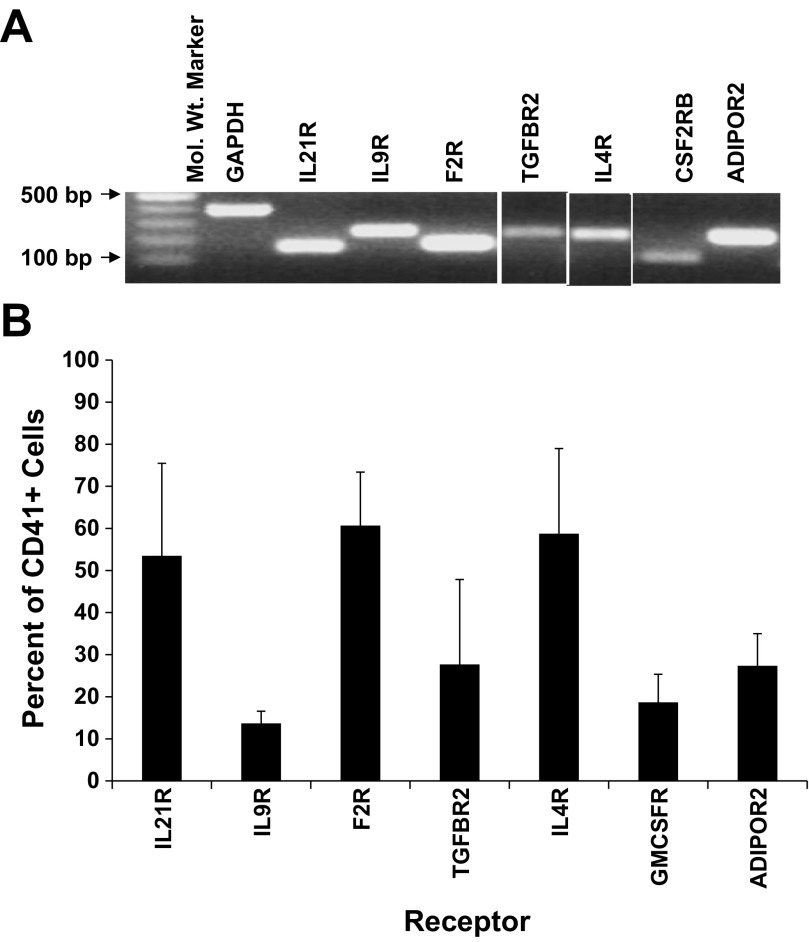
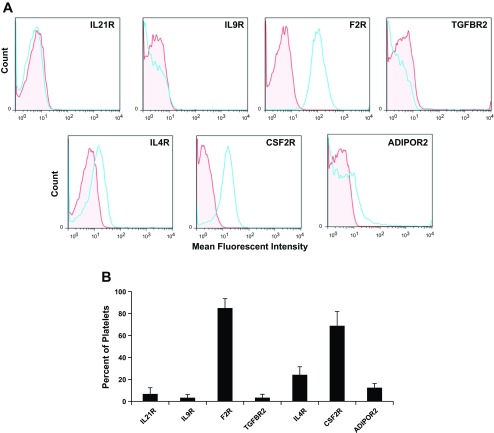
RNA and protein expression levels in MKs were confirmed for each of the seven genes. RT-PCR was used to verify RNA levels in culture-derived MKs (Fig. 1A), and flow cytometry was used to examine protein levels. Greater than 50% of culture-derived CD41+ cells expressed IL21R, F2R, and IL4R (Fig. 1B). Receptors IL9R, TGFBR2, CSF2RB, and ADIPOR2 were expressed on 14, 28, 19, and 27% of MKs, respectively (Fig. 1B).
Fig. 1.
RNA and protein expression for each of 7 receptors is confirmed in megakaryocytes (MKs). Umbilical cord blood (UCB) CD34+ cells were cultured with interleukin (IL)3, IL6, stem cell factor (SCF), and thrombopoietin (TPO). After 10 days of culture, the CD41+ cells were isolated by flow cytometry. A: RT-PCR confirmation of receptor expression in CD41+ cells. B: percent of CD41+ cells that express each receptor as measured by flow cytometry. Data are shown as means ± SE and are representative of 3 independent experiments from 3 biological samples.
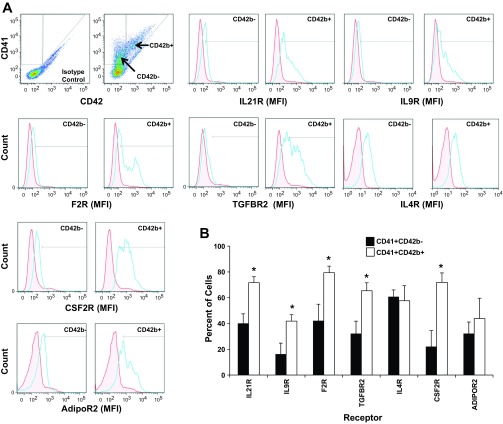
To further investigate receptor expression during MK maturation, we compared receptor expression on a subpopulation of CD41+CD42b− MKs to a more mature subset of MKs defined as CD41+CD42b+ (Fig. 2). Histogram plots indicated that the mean fluorescent intensity (MFI) for six of the seven receptors (i.e., IL21R, IL9R, F2R, TGFBR2, CSF2R, ADIPOR) was greater on CD41+CD42b+ cells than on CD41+CD42b− cells (Fig. 2A). The percent of cells expressing each of the five receptors was significantly higher (P < 0.05) on CD41+CD42b+ cells than on CD41+CD42b− cells (Fig. 2B). The percent of CD41+CD42b+ expressing ADIPOR2 slightly increased but did not reach significance.
Fig. 2.
Protein expression for each of 7 receptors increases as MKs mature. Culture-derived MKs were isolated and immunostained for CD41, CD42b, and for each of the indicated receptors. A: dot plot analysis of CD42 and CD41 expression on culture-derived MKs was used to define gates for the subpopulations, CD41+/CD42b+ and CD41+/CD42b−. Representative single-parameter histogram profiles for gated CD41+CD42b− cells or CD41+CD42b+ cells were generated depicting the mean fluorescent intensity (MFI) for each receptor relative to an isotype control (red, isotype control; blue, receptor). B: the percent of CD41+ CD42b− cells and CD41+CD42b+ cells that express the indicated receptor as measured by flow cytometry. Values are means ± SE and are representative of 3 independent experiments and from 3 biological samples. *P < 0.05.
Proliferation and differentiation responses to each receptor's cognate ligand.
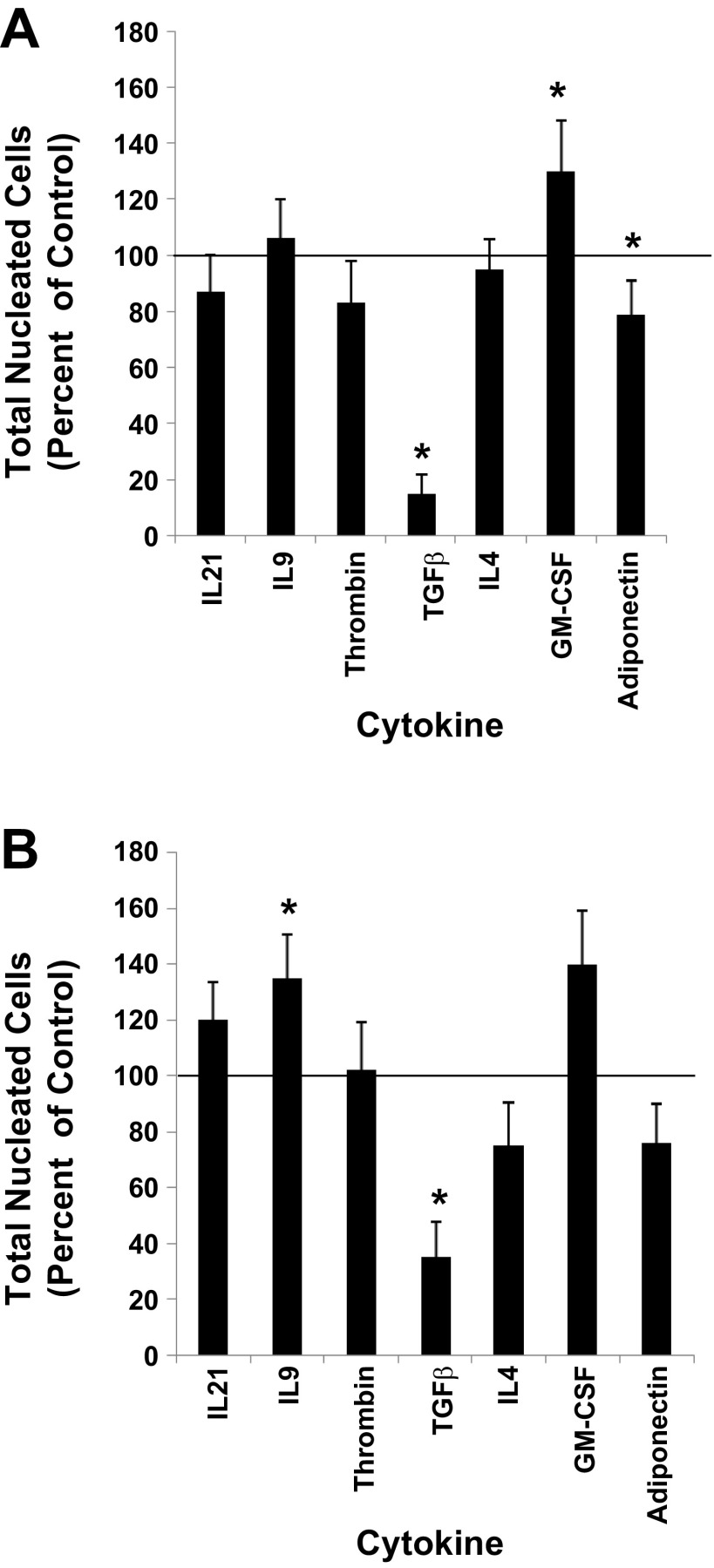
Functional roles for each of the seven receptors during megakaryocytopoiesis were probed by evaluating the effects of their cognate ligands on in vitro MK proliferation and differentiation. The ligands IL21, IL9, Thrombin, TGF-β, IL4, adiponectin, and GM-CSF were used to test whether the plasma membrane receptors IL21R, IL9R, F2R, TGFBR2, IL4R, ADIPOR2, and CSF2RB facilitated MK development, respectively. This was accomplished by initiating cultures of UCB CD34+ cells with an MK induction base media containing the cytokine combination of 36ST (i.e., control) plus one of the following cytokines: IL21, IL9, Thrombin, TGF-β, IL4, adiponectin, or GM-CSF. After 10 days of culture, results showed that relative to control cultures (i.e., 36ST only) the proliferation of UCB CD34+ cells was significantly increased with the addition of GM-CSF and inhibited with TGF-β and adiponectin (Fig. 3A). The remaining cytokines did not have any apparent effect on proliferation (Fig. 3A).
Fig. 3.
Proliferation responses. UCB CD34+ cells were cultured with IL3, IL6, SCF, TPO (36ST) (i.e., control). A: 36ST was added at day 0 along with one of the indicated cytokines. B: 36ST was added at day 0, and after 7 days of culture one of the indicated cytokines was added. The number of total nucleated cells was counted at day 14 of culture. The proliferation response is expressed as a percent of the control group. The line at the 100% level in each graph represents the proliferation response of control cultures. Data are shown as means ± SE in triplicate culture and are representative of 3 independent experiments from 3 biological samples. *P < 0.05.
Since gene expression for each of the seven receptors increased as cells underwent MK development, we rationalized that addition of their matched-ligand might have a greater impact on culture outcome if added late in the cultures (i.e., at day 7 of culture) rather than at the time that the cultures were initiated. To execute these experiments, UCB CD34+ cells were cultured for 7 days with 36ST prior to the addition of each individual test cytokine and cultured for an additional 7 days. The results now showed that IL9 had a positive impact on proliferation and even though not significant GM-CSF continued to promote cell proliferation (Fig. 3B). TGF-β strongly inhibited cell proliferation independently of whether it was added at day 0 or day 7 (Fig. 3B). Decreased cellular outputs were also apparent in the presence of adiponectin and IL4; however, neither reached statistical significance.
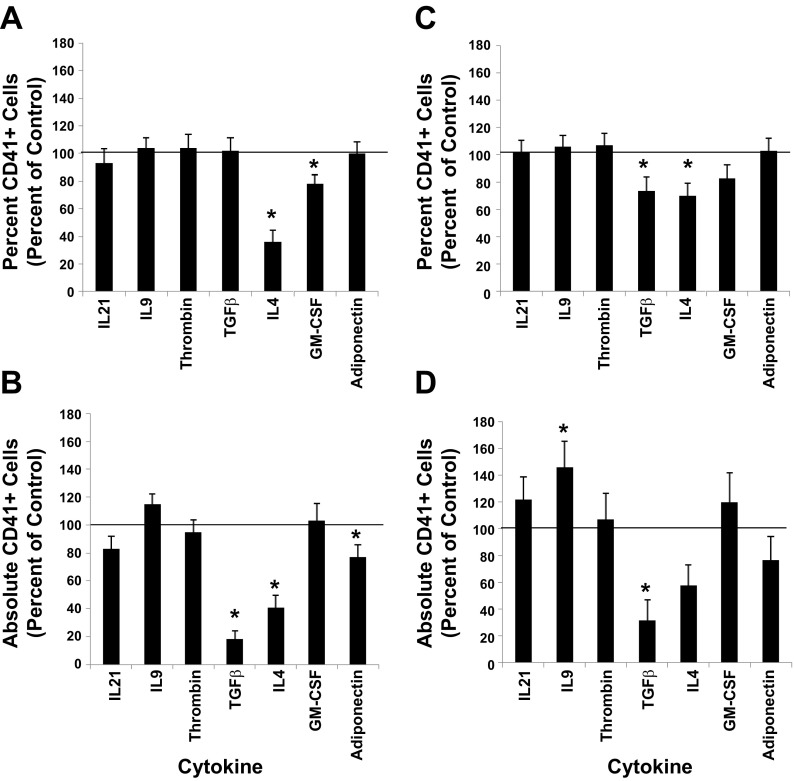
We measured the impact that each of the seven cytokines had on the differentiation response of UCB CD34+ cells as the cells were induced along the MK lineage by monitoring the expression level of the CD41 antigen, as well as a feature unique to megakaryocytopoiesis, polyploidization. When one of the seven cytokines in question was added at the time UCB CD34+ cultures were initiated with base medium containing 36ST, the percent of cells expressing CD41 at day 10 of culture was similar to control cultures except when IL4 and GM-CSF were present in the cultures (Fig. 4A). The absolute counts show that fewer total CD41+ cells were produced relative to control cultures in the presence of TGF-β, IL4, and adiponectin (Fig. 4B). The results also show that, when ligand addition occurred 7 days after culture initiation with 36ST, the percent of cells expressing CD41 was similar to the control except in the presence of TGF-β and IL4 (Fig. 4C), while absolute counts indicate, relative to control cultures, that there were significantly greater numbers of CD41+ cells with IL9 and fewer total CD41+ cells with TGF-β (Fig. 4D). The results of ploidy analyses indicate that no significant changes in polyploidization were observed when each of the seven cytokines were added at the time that UCB CD34+ cultures were initiated with 36ST (data not shown).
Fig. 4.
Differentiation responses. A, B: UCB CD34+ cell cultures were initiated with IL3, IL6, SCF, TPO (36ST) (i.e., control) along with one of the indicated cytokines. After 10 days of culture, the percent of CD41+ cells in culture. C, D: UCB CD34+ cell cultures were initiated with 36ST. After 7 days in culture, one of the indicated cytokines was added. A–D: at day 10 of culture, the percent of CD41+ cells in each culture was measured by flow cytometry. The percent and absolute number of cells expressing CD41+ are normalized to the control group. The line at the 100% level in each graph represents the response of control cultures. Data are shown as means ± SE in triplicate culture and are representative of 3 independent experiments and from 3 biological samples. *P < 0.05.
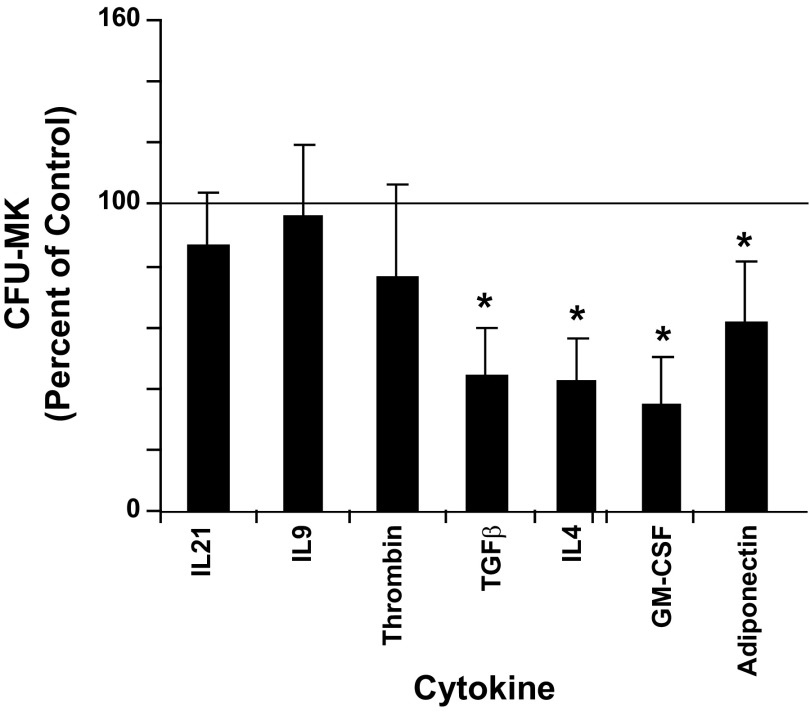
The effect of each cytokine on the formation of CFU-MKs is shown in Fig. 5. Significant reductions (P < 0.5) in colony formation by 56, 57, 31, and 38% relative to control cultures were seen with TGF-β, IL4, GM-CSF, and adiponectin, respectively. The cytokines IL21, IL9, and thrombin did not cause any significant change in CFU-MK formation.
Fig. 5.
Megakaryocyte colony-forming-unit (MK-CFU) formation. Cord blood (UCB) CD34+ cells were cultured for 12 days with IL3, IL6, SCF, TPO (36ST), plus one of the indicated cytokines in collagen containing media. CD34+ cells cultured with only 36ST were designated as controls. The number of colony-forming unit megakaryocytes (CFU-MK) was counted as described in materials and methods and normalized to the control group. The line at the 100% level in each graph represents the response of control cultures. Data are shown as means ± SE from duplicate cultures from 3 independent experiments. *P < 0.05.
Receptor expression on platelets.
As terminal differentiation of MKs culminates in the formation of platelets, we examined the expression level of the seven receptors in platelets. Histogram plots from flow cytometric analyses showed that the MFI for the expression of F2R and CSF2R were substantially greater than for isotype controls (Fig. 6A). The percent of platelets that expressed F2R and CSF2RB was 85 ± 8.5 and 69 ± 13% (means ± SE), respectively (Fig. 6B). IL21R, IL9R, TGFBR2, IL4R, and ADIPOR2 were expressed on 6.8 ± 5.6, 3.4 ± 2.7, 3.4 ± 3.1, 24 ± 7.3, and 12 ± 3.7% of platelets, respectively.
Fig. 6.
Platelet receptor expression. Fresh whole blood platelets were isolated and immunostained with antibodies to CD41 or CD61 and for the indicated receptors. A: representative single parameter histogram profiles for CD41+ or CD61+ gated platelet populations were generated depicting the MFI for each receptor relative to an isotype control (red, isotype control; blue, receptor). B: the percent of platelets that express the indicated receptor as measured by flow cytometry. n = 4.
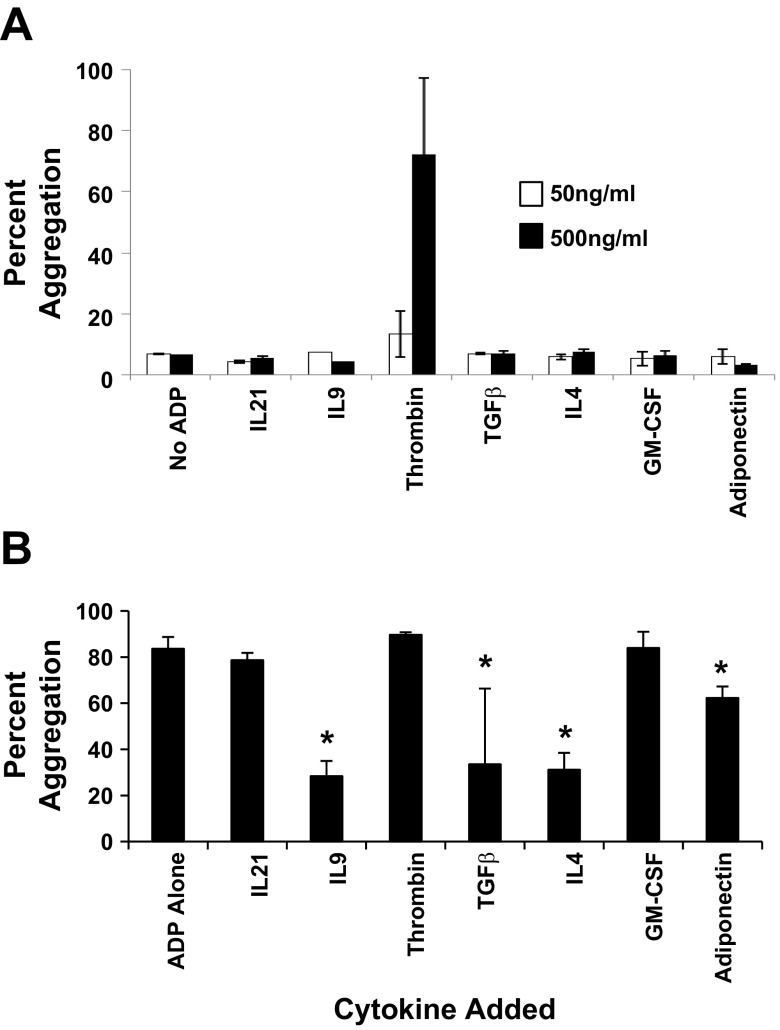
Even though receptors F2R, CSF2RB, and ADIPOR2 were highly expressed on platelets, platelet aggregation studies showed that only thrombin induced platelet aggregation (Fig. 7A). On the other hand, when platelets were independently preincubated with each of the seven cytokines (500 ng/ml) for 5 min, prior to the addition of the platelet agonist ADP, ADP-induced platelet aggregation was significantly inhibited to where only 28 ± 7, 34 ± 31, 32 ± 7, 62 ± 5% of platelet aggregated when IL9, TGF-β1, IL4, or adiponectin was preincubated with platelets prior to ADP addition (Fig. 7B).
Fig. 7.
Platelet aggregation responses. Fresh whole blood platelets were isolated and aggregation of platelets in response to cytokines specific to each of the 7 receptors, IL21R, IL9R, F2R, TGFBR2, IL4R, CSF2RB, and ADIPOR2, was measured. A: maximum percent platelet aggregation in response to 50 or 500 ng/ml of the indicated cytokines. The control was nonstimulated platelets (i.e., no addition of a cytokine or ADP). Data are shown as means ± SE; n = 2. B: platelets were preincubated with 500 ng/ml of the indicated cytokine for 5 min followed by the addition of 20 μM of ADP. Control platelets were preincubated for 5 min followed by the addition of 20 μM of ADP. The percent platelet aggregation was measured with an aggregometer. Data are shown as means ± SE; n = 3. *P < 0.05.
Autoexpression of cognate ligands.
Since autocrine signaling can regulate stem cell development, the microarray data were analyzed to determine whether transcripts for cytokines to each of the seven receptors IL21R, IL9R, F2R, TGFBR2, IL4R, ADIPOR2, and CSF2RB were differentially expressed between uncultured CD34+/CD38lo cells and culture-derived MKs. We found that only the transcript for the cytokine TGF-β increased in expression level. This finding was confirmed by semiquantitative RT-PCR of culture-derived MKs (Fig. 8). To determine whether this translated into a secretion of TGF-β by developing MKs, we examined culture supernatants and found TGF-β levels of 2,228 ± 1,599 pg/ml of media (n = 5). Further analysis indicated that the majority of the secreted TGF-β was in the latent form.
Fig. 8.
TGF-β1 is the only autoexpressed ligand to the 7 receptors. The same microarray data used to identify the upregulation of the receptors IL21R, IL9R, F2R, TGFBR2, IL4R, ADIPOR2, and CSF2RB were searched for the expression of their matched-ligands. A: fold change in relative fluorescent intensities in marrow CD34+/CD38lo cells relative to culture derived MKs. Positive fold changes indicate higher RNA expression levels by culture-derived MKs than day 0 CD34+/CD38lo cells. Data are shown as means ± SD from 3 independent experiments. B: UCB CD34+ cells (day 0) were cultured with IL 3, IL6, SCF, TPO. After 10 days of culture, CD41+ cells were isolated by flow cytometry. RNA was isolated, semiquantitative RT-PCR was performed, and the PCR products are shown on a 1% agarose gel.
DISCUSSION
This is the first report in which a set of plasma membrane genes is collated to constitute a developing MK receptome. (Table 1). This list is composed of 40 plasma membrane receptor genes that are significantly upregulated as adult hematopoietic CD34+/CD38lo cells are induced to become MKs. Among this list are receptors that are well known (e.g., TGFBR2, F2R) and others that are less well known (e.g., CCRL2, SEMA4D; ADIPOR2) or unknown (e.g., IL21R) to have a role in MK and/or platelet biology.
Of the seven of the 40 differentially expressed transcripts selected to validate the robustness of the dataset, protein expression levels for all seven are observed in MKs. Furthermore, the robustness of the dataset is illustrated by our findings that six of seven ligand-receptor pairs, IL9/IL9R, thrombin/F2R, TGF-β/TGFBR2, IL4/IL4R, GM-CSF/CSF2RB, and adiponectin/ADIPOR2, have significant biological effects on the MK lineage. Of the seven cytokine receptors expressed by MKs, F2R, IL4R, and CSF2RB are readily detected on platelets by flow cytometry, while levels of ADIPOR2 are low and IL21R, IL9R, TGFBR2 are negligible. Of the seven matched-receptor ligands, the IL21 cytokine is the only one with no apparent biological effect on MK development. However, because assays are limited in this study, this does not preclude a role for the IL21/IL21R ligand-receptor axis in the biology of MKs/platelets.
This is also the first study to show that MK colony formation is significantly inhibited by adiponectin (P = 0.037). Thus, suggesting that levels of adiponectin are relevant for regulating megakaryocytopoiesis. Adiponectin is a well-documented regulator of a number of metabolic processes (19); it is expressed in adult BM (34) and in 2007 identified as a novel hematopoietic stem cell growth factor (9). Adiponectin levels inversely correlate with body fat content in adults (8, 33), and in individuals with hematological disorders (1, 23). Adiponectin suppresses BM granulocyte/macrophage colony formation (7) and is secreted by adipocytes, as well as by a number of other cell types (3, 7, 16, 21, 24, 35). Adipocytes are the most abundant stromal cells in adult BM (10) and are a production site for TNF-α (27) and TGF-β (14), which inhibit both fat formation and MK maturation. By use of a human cytokine antibody array, it is also recently shown that adiponectin is among a group of cytokines with the highest levels of detection in platelet lysates (13). On the basis of our findings along with findings from other investigators, it is intriguing to speculate that MK progenitor production may be regulated in part by adiponectin levels. Given the inverse correlation of body fat content to adiponectin levels (1, 24) and our finding that adiponectin inhibits MK colony formation. The prediction is that the production of MK progenitors will increase as adiponectin levels decrease or as body fat content increases. This prediction is partially supported by the observation that obese females have lower adiponectin levels and significantly elevated platelet counts compared with normal-weight females (30). In addition, our observation that adiponectin inhibits ADP-induced platelet aggregation supports a role for adiponectin in modulating platelet function. This observation is in agreement with a previous report that indicates that high concentration of adiponectin (>25 μg/ml) diminishes epinephrine- and ADP-induced platelet aggregation (27).
Our finding that TGF-β strongly inhibits MK development is in agreement with work by other investigators (11, 18, 29). Examination of our microarray dataset for gene expression of cognate ligands to each of the seven selected receptors showed that only the transcript for the cytokine, TGF-β, is upregulated. This is confirmed by findings that TGF-β is secreted into the supernatant of MK cultures. Given that TGF-β when exogenously added to cultures inhibited the proliferation of MK progenitors, it is interesting that autocrine TGF-β has apparently no inhibitory effect on in vitro megakaryocytopoiesis. This is because autocrine TGF-β is predominantly secreted into the culture supernatant in a latent form. The observation that there is an autocrine secretion of latent TGF-β into culture supernatant is important, especially when one considers the design of stem cell culture systems with feeder cells, such as endothelial and/or stromal cells, to support an in vitro MK production. As feeder cells produce TSP-1, MMP-9, and MMP-2, which are known to cleave latent TGF-β (30, 36). Consequently, a bioreactor design with such a coculture design may require an addition of neutralizing antibodies and/or antagonists to TGF-β to enhance MK development.
This is the first report that CSFR2B is expressed on MKs and platelets. CSFR2B is the common beta chain for not only the GM-CSF receptor, but also the IL-3 and IL5 receptors (22). Although other studies have reported that GM-CSF modestly stimulates CFU-MK formation (20), in our study GM-CSF inhibited MK colony formation and MK differentiation. This contradiction in findings may be due to the concentration of GM-CSF, as Chen et al. (6) indicate that low concentrations of GM-CSF stimulate MK colony formation and high doses are inhibitory. On the other hand, we suspect that the high proliferative responses (i.e., increased TNC counts) in the presence of GM-CSF even while CFU-MK progenitor growth is inhibited is most likely due to GM-CSF supporting the growth of other hematopoietic cell lineages such as granulocytes and monocytes (4). As noted by D'Atri et al. (8), the inhibitory effect of GM-CSF on CD41+ cells is a result of a switch of CD34+ cells differentiation toward the monocytic lineage. The finding that CSFR2B is also expressed on platelets suggests that GM-CSF might have a biological role in modulating platelet function. However, despite the presence of CSF2RB on platelets and in agreement with the findings of other investigators (15), GM-CSF alone did not induce platelet aggregation.
Not surprisingly, different proliferation and differentiation responses are dependent upon the timing of when some of the seven cytokines are added to UCB CD34+ cultures that are initiated with a base medium containing 36ST. This is especially evident when comparing the proliferation response of cells supplemented with IL9 at day 0 vs. day 7 of culture. When IL9 is added at day 7 of culture, IL9 significantly increases the proliferative response of cells (Fig. 2B). However, IL9 has no apparent effect on the number of TNCs produced when added at the time that cultures are initiated (Fig. 2A). IL9 stimulatory effects on MK development are in agreement with a previous report from the literature (16).
These studies validate that differentially expressed plasma-membrane receptors identified by microarray analyses of developing MKs have functional roles in megakaryocytopoiesis and platelet biology, thus suggesting that there is a strong likelihood that the remaining 33 of the 40 upregulated plasma membrane-localized receptors have important biological roles in the function and/or development of MKs and platelets (Table 1). Among the remaining 33 genes are ligand-receptor pairs that are implicated in MK development, but receptor expression is not definitively reported in MKs. There are also orphan receptors in the microarray dataset that may help to identify new ligands that participate in megakaryocytopoiesis (Table 1). The creation of a developing MK receptome provides an opportunity for users to identify functional roles for previously uncharacterized matched receptor-ligand systems that integrate a multiplicity of environmental cues that are critical for MK development and platelet biology. Moreover, this work provides a foundation for subsequent studies to characterize the role of the plasma membrane-localized receptors that are downregulated as stem cells are induced to differentiate (Table 2).
GRANTS
This work was supported in part by funding from the Puget Sound Blood Center and National Heart, Lung, and Blood Institute Grant HL-081015 (J.-A. Reems).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: S.S. and J.-A.R. conception and design of research; S.S. and W.W. performed experiments; S.S., B.J.A., and J.-A.R. analyzed data; S.S., B.J.A., and J.-A.R. interpreted results of experiments; S.S. and J.-A.R. prepared figures; Y.L., D.G., B.J.A., and J.-A.R. edited and revised manuscript; J.-A.R. drafted manuscript; J.-A.R. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Najla Abdurraham for technical help and Doug Bolgiano for statistical help.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Avcu F, Ural AU, Yilmaz MI, Bingol N, Nevruz O, Caglar K. Association of plasma adiponectin concentrations with chronic lymphocytic leukemia and myeloproliferative diseases. Int J Hematol 83: 254–258, 2006. [DOI] [PubMed] [Google Scholar]
- 2. Ben-Shlomo I, Yu Hsu S, Rauch R, Kowalski HW, Hsueh AJ. Signaling receptome: a genomic and evolutionary perspective of plasma membrane receptors involved in signal transduction. Sci STKE 2003: RE9, 2003. [DOI] [PubMed] [Google Scholar]
- 3. Berner HS, Lyngstadaas SP, Spahr A, Monjo M, Thommesen L, Drevon CA, Syversen U, Reseland JE. Adiponectin and its receptors are expressed in bone-forming cells. Bone 35: 842–849, 2004. [DOI] [PubMed] [Google Scholar]
- 4. Burgess AW, Camakaris J, Metcalf D. Purification and properties of colony-stimulating factor from mouse lung-conditioned medium. J Biol Chem 252: 1998–2003, 1977. [PubMed] [Google Scholar]
- 5. Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37: W305–W311, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen S, Zhu FM, He J, Liu JH, Qin F, Yan LX. [Effect of GM-CSF on expansion and differentiation of CD34+ megakaryocyte progenitor cells from cord blood in vitro]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 13: 1041–1043, 2005. [PubMed] [Google Scholar]
- 7. Crawford LJ, Peake R, Price S, Morris TC, Irvine AE. Adiponectin is produced by lymphocytes and is a negative regulator of granulopoiesis. J Leukoc Biol 88: 807–811, 2010. [DOI] [PubMed] [Google Scholar]
- 8. D'Atri LP, Pozner RG, Nahmod KA, Landoni VI, Isturiz M, Negrotto S, Schattner M. Paracrine regulation of megakaryo/thrombopoiesis by macrophages. Exp Hematol 39: 763–772, 2011. [DOI] [PubMed] [Google Scholar]
- 9. Diez JJ, Iglesias P. The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur J Endocrinol 148: 293–300, 2003. [DOI] [PubMed] [Google Scholar]
- 10. DiMascio L, Voermans C, Uqoezwa M, Duncan A, Lu D, Wu J, Sankar U, Reya T. Identification of adiponectin as a novel hemopoietic stem cell growth factor. J Immunol 178: 3511–3520, 2007. [DOI] [PubMed] [Google Scholar]
- 11. Gimble JM, Robinson CE, Wu X, Kelly KA. The function of adipocytes in the bone marrow stroma: an update. Bone 19: 421–428, 1996. [DOI] [PubMed] [Google Scholar]
- 12. Greenberg SM, Chandrasekhar C, Golan DE, Handin RI. Transforming growth factor beta inhibits endomitosis in the Dami human megakaryocytic cell line. Blood 76: 533–537, 1990. [PubMed] [Google Scholar]
- 13. Guerriero R, Testa U, Gabbianelli M, Mattia G, Montesoro E, Macioce G, Pace A, Ziegler B, Hassan HJ, Peschle C. Unilineage megakaryocytic proliferation and differentiation of purified hematopoietic progenitors in serum-free liquid culture. Blood 86: 3725–3736, 1995. [PubMed] [Google Scholar]
- 14. Horn P, Bokermann G, Cholewa D, Bork S, Walenda T, Koch C, Drescher W, Hutschenreuther G, Zenke M, Ho AD, Wagner W. Impact of individual platelet lysates on isolation and growth of human mesenchymal stromal cells. Cytotherapy 12: 888–898, 2010. [DOI] [PubMed] [Google Scholar]
- 15. Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 259: 87–91, 1993. [DOI] [PubMed] [Google Scholar]
- 16. Kaplan A, Kaplan S, Marcoe K, Sauvage L. The effect of hematopoietic growth factors on platelet aggregability. Clin Appl Thromb Hemost 4: 238–242, 1998. [Google Scholar]
- 17. Katsiougiannis S, Kapsogeorgou EK, Manoussakis MN, Skopouli FN. Salivary gland epithelial cells: a new source of the immunoregulatory hormone adiponectin. Arthritis Rheum 54: 2295–2299, 2006. [DOI] [PubMed] [Google Scholar]
- 18. Kaushansky K. Historical review: megakaryopoiesis and thrombopoiesis. Blood 111: 981–986, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuter DJ, Gminski DM, Rosenberg RD. Transforming growth factor beta inhibits megakaryocyte growth and endomitosis. Blood 79: 619–626, 1992. [PubMed] [Google Scholar]
- 20. Lara-Castro C, Fu Y, Chung BH, Garvey WT. Adiponectin and the metabolic syndrome: mechanisms mediating risk for metabolic and cardiovascular disease. Curr Opin Lipidol 18: 263–270, 2007. [DOI] [PubMed] [Google Scholar]
- 21. Mazur EM, Cohen JL, Wong GG, Clark SC. Modest stimulatory effect of recombinant human GM-CSF on colony growth from peripheral blood human megakaryocyte progenitor cells. Exp Hematol 15: 1128–1133, 1987. [PubMed] [Google Scholar]
- 22. Miller M, Cho JY, Pham A, Ramsdell J, Broide DH. Adiponectin and functional adiponectin receptor 1 are expressed by airway epithelial cells in chronic obstructive pulmonary disease. J Immunol 182: 684–691, 2009. [DOI] [PubMed] [Google Scholar]
- 23. Miyajima A, Mui AL, Ogorochi T, Sakamaki K. Receptors for granulocyte-macrophage colony-stimulating factor, interleukin-3, and interleukin-5. Blood 82: 1960–1974, 1993. [PubMed] [Google Scholar]
- 24. Molica S, Vitelli G, Cutrona G, Todoerti K, Mirabelli R, Digiesi G, Giannarelli D, Sperduti I, Molica M, Gentile M, Morabito F, Neri A, Ferrarini M. Prognostic relevance of serum levels and cellular expression of adiponectin in B-cell chronic lymphocytic leukemia. Int J Hematol 88: 374–380, 2008. [DOI] [PubMed] [Google Scholar]
- 25. Pineiro R, Iglesias MJ, Gallego R, Raghay K, Eiras S, Rubio J, Dieguez C, Gualillo O, Gonzalez-Juanatey JR, Lago F. Adiponectin is synthesized and secreted by human and murine cardiomyocytes. FEBS Lett 579: 5163–5169, 2005. [DOI] [PubMed] [Google Scholar]
- 26. Reems JA, Pineault N, Sun S. In vitro megakaryocyte production and platelet biogenesis: state of the art. Transf Med Rev 24: 33–43, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reems JA, Torok-Storb B. Cell cycle and functional differences between CD34+/CD38hi and CD34+/38lo human marrow cells after in vitro cytokine exposure. Blood 85: 1480–1487, 1995. [PubMed] [Google Scholar]
- 28. Restituto P, Colina I, Varo JJ, Varo N. Adiponectin diminishes platelet aggregation and sCD40L release. Potential role in the metabolic syndrome. Am J Physiol Endocrinol Metab 298: E1072–E1077, 2010. [DOI] [PubMed] [Google Scholar]
- 29. Rubinstein P, Dobrila L, Rosenfield RE, Adamson JW, Migliaccio G, Migliaccio AR, Taylor PE, Stevens CE. Processing and cryopreservation of placental/umbilical cord blood for unrelated bone marrow reconstitution. Proc Natl Acad Sci USA 92: 10119–10122, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakamaki S, Hirayama Y, Matsunaga T, Kuroda H, Kusakabe T, Akiyama T, Konuma Y, Sasaki K, Tsuji N, Okamoto T, Kobune M, Kogawa K, Kato J, Takimoto R, Koyama R, Niitsu Y. Transforming growth factor-beta1 (TGF-beta1) induces thrombopoietin from bone marrow stromal cells, which stimulates the expression of TGF-beta receptor on megakaryocytes and, in turn, renders them susceptible to suppression by TGF-beta itself with high specificity. Blood 94: 1961–1970, 1999. [PubMed] [Google Scholar]
- 31. Samocha-Bonet D, Justo D, Rogowski O, Saar N, Abu-Abeid S, Shenkerman G, Shapira I, Berliner S, Tomer A. Platelet counts and platelet activation markers in obese subjects. Med Inf (Lond) 2008: 834153, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schultz-Cherry S, Murphy-Ullrich JE. Thrombospondin causes activation of latent transforming growth factor-beta secreted by endothelial cells by a novel mechanism. J Cell Biol 122: 923–932, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schulze H, Shivdasani RA. Mechanisms of thrombopoiesis. J Thromb Haemost 3: 1717–1724, 2005. [DOI] [PubMed] [Google Scholar]
- 34. Shim MH, Hoover A, Blake N, Drachman JG, Reems JA. Gene expression profile of primary human CD34+CD38lo cells differentiating along the megakaryocyte lineage. Exp Hematol 32: 638–648, 2004. [DOI] [PubMed] [Google Scholar]
- 35. Ukkola O, Santaniemi M. Adiponectin: a link between excess adiposity and associated comorbidities? J Mol Med 80: 696–702, 2002. [DOI] [PubMed] [Google Scholar]
- 36. Yokota T, Meka CS, Medina KL, Igarashi H, Comp PC, Takahashi M, Nishida M, Oritani K, Miyagawa J, Funahashi T, Tomiyama Y, Matsuzawa Y, Kincade PW. Paracrine regulation of fat cell formation in bone marrow cultures via adiponectin and prostaglandins. J Clin Invest 109: 1303–1310, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yokota T, Oritani K, Takahashi I, Ishikawa J, Matsuyama A, Ouchi N, Kihara S, Funahashi T, Tenner AJ, Tomiyama Y, Matsuzawa Y. Adiponectin, a new member of the family of soluble defense collagens, negatively regulates the growth of myelomonocytic progenitors and the functions of macrophages. Blood 96: 1723–1732, 2000. [PubMed] [Google Scholar]
- 38. Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev 14: 163–176, 2000. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.