Abstract
Few studies have investigated heterogeneity of selection response in replicate lines subjected to equivalent selection. We developed four replicate lines of mice based on high levels of voluntary wheel running (high runner or HR lines) while also maintaining four nonselected control lines. This led to the unexpected discovery of the HR minimuscle (HRmini) phenotype, recognized by a 50% reduction in hindlimb muscle mass, which became fixed in 1 of the four HR selected lines. Here, we report genome-wide expression profiling describing transcriptome differences between HRnormal and HRmini medial gastrocnemius. Consistent with the known reduction of type IIB fibers in HRmini, Myh4 gene expression was −8.82-fold less (P = 0.0001) in HRmini, which was closely associated with differences in the “calcium signaling” canonical pathway, including structural genes (e.g., Mef2c, twofold greater in HRmini, P = 0.0003) and myogenic factors (e.g., Myog, 3.8-fold greater in HRmini, P = 0.0026) associated with slow-type myofibers. The gene that determines the HRmini phenotype is known to reside in a 2.6335-Mb interval on mouse chromosome 11 and 7 genes (Myh10, Chrnb1, Acadvl, Senp3, Gabarap, Eif5a, and Clec10a) from this region were differentially expressed. Verification by real-time PCR confirmed 1.5-fold greater (P < 0.05) expression of very long chain acyl-CoA dehydrogenase (Acadvl) in HRmini. Ten other genes associated with fatty acid metabolism were also upregulated in HRmini, suggesting differences in the ability to metabolize fatty acids in HRnormal and HRmini muscles. This work provides a resource for understanding differences in muscle phenotypes in populations exhibiting high running capacity.
Keywords: artificial selection, experimental evolution, wheel running
artificial selection experiments allow the study of evolution in real time and may reveal genetic correlations with the trait under selection. For example, selection of mice for enhanced protein content produced a hypermuscular phenotype (42) that subsequently was found to be caused by a deletion in myostatin (40). We (37) initiated artificial selection for increased wheel-running behavior to investigate the evolution of locomotor behavior and physiological capacities for exercise in outbred house mice (Mus domesticus). After 10 generations of selection, individuals from the four replicate high-runner (HR) lines ran, on average, 70% more revolutions/day, and exhibited significantly greater (12%) maximal oxygen uptake (V̇o2max) compared with mice from four nonselected control lines (38). The differential in daily wheel running eventually plateaued at approximately +200% compared with control lines, and at generation 49 the endurance capacity of HR lines during forced treadmill exercise was found to be 35% greater than control lines based on total distance run prior to exhaustion (30). Moreover, the endurance running capacity of HR mice (30) was significantly greater than reported in studies using direct genetic manipulations or pharmacological approaches to alter endurance performance (e.g., Refs. 17, 32, 43).
These comparisons indicate the efficacy of artificial selection as a tool to alter complex, polygenic performance traits (12), but relatively few studies have addressed the issue of heterogeneity in the response to selection (13). That is, when replicate lines are subjected to a selection regimen their response may be dissimilar particularly regarding correlated traits. Our HR mice were developed as four replicate lines each exhibiting elevated levels of voluntary running capacity, while four nonselected lines were maintained as controls. This approach led to the unexpected discovery of the HR minimuscle (HRmini) phenotype, originally recognized by a 50% reduction in triceps surae muscle mass (11), as well as whole limb muscle mass (21) compared with HR normal-sized (HRnormal) mice. Caused by a Mendelian recessive allele, the HRmini phenotype increased in frequency in two of the four selected lines, demonstrating that the selection regimen has favored the phenotype (11), and the HRmini allele has become fixed in one selected line and remains polymorphic in another (Ref. 39 and unpublished results).
Total hind limb muscle mass in the HRmini phenotype is approximately half that of control lines, but despite the smaller muscle mass, there is no statistical difference in total number of wheel revolutions completed per day between HRmini and HRnormal mice (21). The enhanced whole animal aerobic exercise capacity (35, 38) of HRmini mice is in part underpinned by greater muscle capillarity (45), doubled mass-specific content of some metabolic enzymes (14), and pronounced loss of myofibers expressing myosin heavy chain (MyHC) IIb (15). The selective reduction of IIb myofibers, which have relatively large cross-sectional areas, may be largely responsible for the diminutive size of HRmini muscles (14, 15) and likely underpins the observed differences in muscle performance. For example, medial gastrocnemius of HRmini animals produces approximately half the mass-specific isotonic power of HRnormal, and fatigue resistance is significantly enhanced (39).
Diversity in response to selection is useful for testing hypotheses about mechanisms of correlated evolution, “trade-offs,” and “constraints.” Importantly, genetic correlations can differ from expectations based on first-principles knowledge of physiological mechanisms. The gene that determines the HRmini phenotype is known to behave as a Mendelian recessive and to reside in a 2.6335-Mb interval on mouse chromosome 11 (MMU11) (20). This region harbors ∼100 expressed or predicted genes, many with known roles in muscle development and function, and direct sequencing of this region is currently in progress. Microarray analysis of RNA transcripts is a powerful tool for determining potential associations between gene expression and phenotypic outcomes. We used microarray of HRmini and HRnormal to gain a better understanding of the differences and similarities in these two alternate solutions to selection on high running capacity. In addition, differentially expressed genes from the area showing linkage were prioritized to provide insight to the potential causal gene.
METHODS
Animal model.
Experimental procedures were approved by the University of California Riverside Institutional Animal Care and Use Committee.
Male mice were used from an animal model of four closed lines selectively bred for high voluntary wheel-running behavior (HR) and four control lines bred without intentional selection, described in Swallow et al. (37). Animals used in the current work were sampled from generation 37 at a mean age of 84 days (range = 79–86). To determine muscle masses, we sampled HR line #3 (lab designation), which is fixed for the minimuscle phenotype (all individuals express this phenotype) and is here termed HRmini, as well as two other HR lines (line #7 and #8) in which the minimuscle phenotype has never been observed (here, termed HRnormal) (11). Gastrocnemius, plantaris, and soleus from both left and right legs were dissected, and masses (wet weight) of HRnormal and HRmini muscles were compared by analysis of covariance using body mass as the covariate. For microarray analysis, we used gastrocnemius muscles (including both lateral and medial portions) from the mice described above for HRmini line #3 and HRnormal line #8 (n = 6, in each group). To obtain sufficient RNA, gastrocnemius samples of HRmini were pooled from left and right limbs from each animal, whereas HRnormal gastrocnemius was sampled from either left or right limb only.
Microarray analysis.
Total RNA was extracted from whole gastrocnemius muscle (>100 mg) homogenized in TRIzol reagent (Life Technologies, Gaithersburg, MD) following the manufacturer's protocol. The RNA was purified using RNeasy purification columns (Qiagen, Germantown, MD), and then RNA integrity, purity and quantity were assessed by Bioanalyzer (Agilent Technologies, Santa Carla, CA) and Nanodrop (NanoDrop Technologies, Baltimore, MD) spectrometry. The expression profiling procedure was performed following the standard Affymetrix protocol. Briefly, total RNA from each sample (5 μg) was used for cDNA synthesis followed by cRNA synthesis. Biotin-labeled and fragmented cRNAs were hybridized to the Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA) followed by washing, staining and image scanning per manufacturer's protocol. All profiles have been made publically accessible via the National Center for Biotechnology Information Gene Expression Omnibus (GSE41910) at http://wwwncbi.nlm.nih.gov/geo/.
Absolute analyses of the array images were performed using Affymetrix MAS 5.0. Satistically significant expression changes between HRmini and HRnormal mice were determined using GeneSpring version 7. The expression level of each probe set was imported in to GeneSpring for probe-set filtering (10% present calls), and statistical analysis was performed using Welch t-test (corrected by Benjamini and Hochberg multiple testing with 5% false positive rate). Genes that showed significant changes (P < 0.05 after correction) were retained for functional enrichment analyses.
Functional enrichment analyses of differentially (P < 0.05, corrected) expressed genes was performed by two different methods. First, clustering of genes in to functional groups and the statistical significance of overrepresentation among the groups was calculated with the Database for Annotation, Visualization and Integrated Discovery [DAVID; http://david.abcc.ncifcrf.gov/home.jsp, (22)]. Genes were clustered according to their association with pathways of the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/), and the statistical significance of clusters was estimated by modified Fisher exact P value. Second, differentially expressed genes were uploaded to Ingenuity Pathways Analysis to identify overrepresented canonical pathways from the Ingenuity Knowledge Base. To examine associations that are considered to occur specifically in skeletal muscles, we narrowed the Ingenuity Knowledge Base database to interactions reported in skeletal muscle only.
Quantitative real-time RT-PCR.
Quantitative real-time RT-PCR was used to verify the difference in acyl-coenzyme A dehydrogenase, very long chain (Acadvl) expression highlighted by microarray analysis. Gastrocnemius muscles (n = 6, in each group) were sampled from HRnormal and HRmini mice (mean age 108 days, range 100–114 days) from generation 62 of selection. Total RNA (1 μg) isolated from gastrocnemius muscles were treated with DNase I before cDNA synthesis. cDNA were synthesized using superscript II RT (200 U/μl) with oligo-DT primers (1,000 pmol/μl). The primers used for Acadvl were 5′ GCGGACGGCGCCATTGACCTCTATG-3′ (forward) and 5′-ATCCGAGTTGCAGCCTCAATGCA-3′ (reverse). Ribosomal RNA 18S was used as the internal control. Primers used for 18S rRNA were 5′-TAGCCTTCGCCATCACTGCCATTA-3′ (forward) and 5′-AACCTGGCTGTACTTCCCATCCTT-3′ (reverse). The ABI 7900HT system (Applied Biosystem) was used for mRNA quantification as previously described (6). Briefly, SYBR Green PCR Master Mix (Applied Biosystems) was used. Samples were amplified in triplicate with cycling conditions of 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All primers were tested for nonspecific amplicons and primer dimers by visualizing PCR products on 2% agarose gels prior to performing qPCR and disassociation curve after the reactions. Delta delta CT method was used to determine the expression changes of the Acadvl and an independent t-test performed to determine statistical significance of the differences.
RESULTS
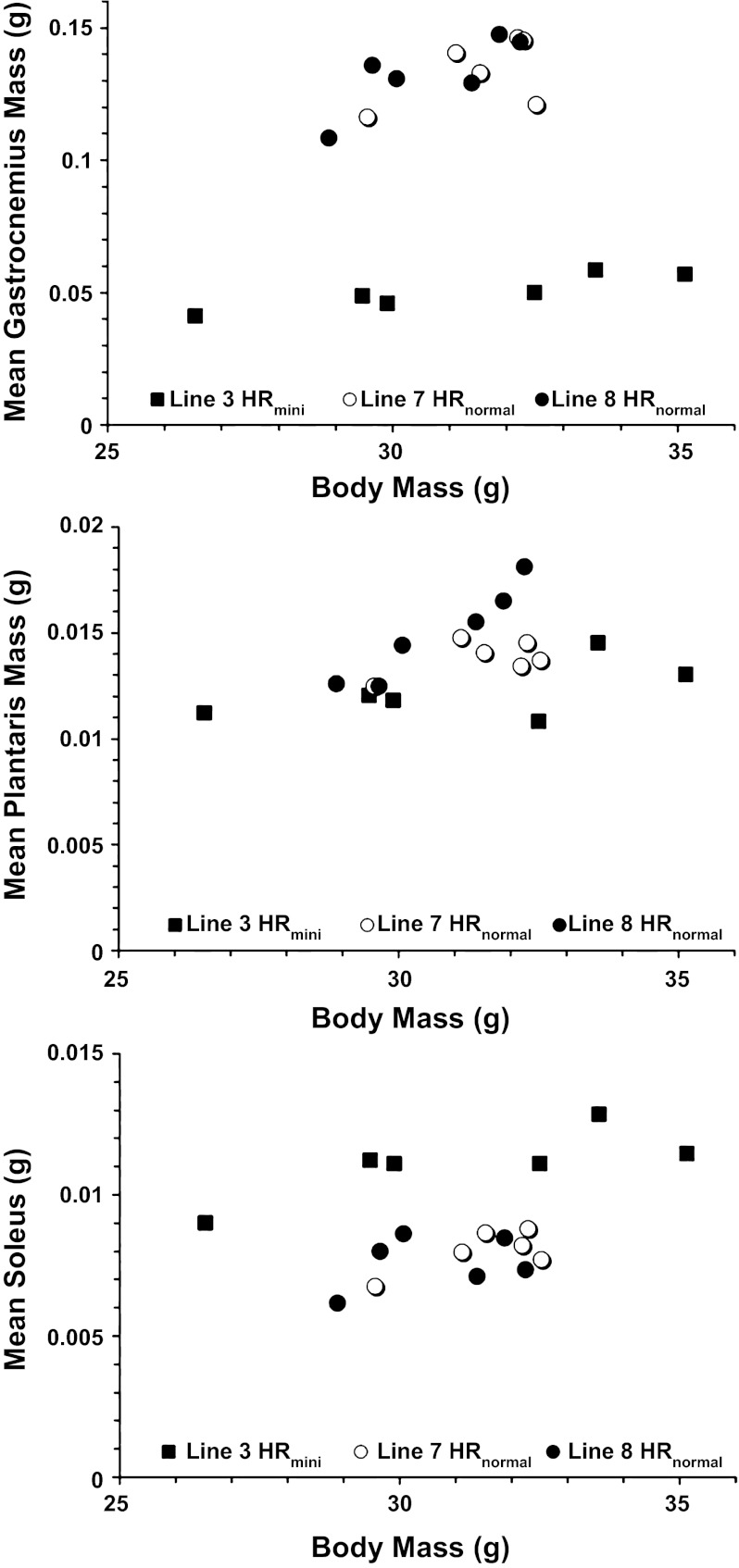
Consistent with our previous findings, gastrocnemius mass was significantly less in HRmini. Muscle mass was related (P = 0.0151) with body mass (Fig. 1), but no interactions (P > 0.07) were found between body mass and experiment group (HRmini and HRnormal) for each of the muscles investigated. After adjusting for a mean body mass of 31.14 g, we found the mass of gastrocnemius (50 mg) in HRmini to be 37.5% of the mass (133 mg) in HRnormal (P < 0.0001). In plantaris, the effect of body mass was also positive and significant (P = 0.0409), yet plantaris mass was significantly (P = 0.0084) less in HRmini. After body mass adjustment, HRmini plantaris (12.2 mg) was 84.9% of the mass of HRnormal (14.4 mg). Body mass was also significantly (P = 0.0039) related with soleus mass but in contrast to the fast-twitch muscles the mass of the slow-twitch soleus was significantly (P < 0.0001) greater in HRmini. That is, after adjusting for body mass, soleus mass (11.1 mg) in HRmini was 142% greater than HRnormal (7.8 mg).
Fig. 1.
Muscle mass in relation to body mass in high-runner normal-sized (HRnormal) and HR minimuscle (HRmini) mice. Muscle mass (g) of gastrocnemius (top), plantaris (middle), and soleus (bottom) relative to body mass (g) of line #3 HRmini (■), line #7 HRnormal (○), and line #8 HRnormal (●) mice. Data represent n = 6 mice in each group.
To identify genes differentially expressed between the two HR mouse lines, expression profiling was performed on six animals from line 8 (HRnormal) and six animals from line 3 (HRmini). Of 45,307 probe sets on the Affymetrix mouse genome 430_2.0 chip, 56% of them (25,570 probe sets) detected target expression in at least one of the 12 arrays. These probe sets were further analyzed, and 2,650 transcripts were identified to be significantly different between HRmini and HRnormal (P < 0.05 after multiple testing correction). Among them, 1,241 transcripts were downregulated in HRmini, and 1,409 were upregulated. HRmini exhibited particularly high expression levels (>20-fold greater expression) of keratin 8 and ankyrin repeat domain 1, and low (>20-fold lesser expression) levels of transmembrane protein 232, HtrA serine peptidase 4, serine/threonine kinase 33, and carbonic anhydrase 3 compared with HRnormal mice.
Functional annotation was conducted in DAVID to link genes with KEGG pathways and the top-ranking (P < 0.05) KEGG pathways are shown in Table 1. The most significant association was the KEGG pathway “focal adhesion,” which included 21 genes significantly less expressed in HRmini, such as IGF-1, glycogen synthase kinase 3 beta, and mitogen-activated protein kinase 8; and 29 genes exhibiting greater expression in HRmini, such as integrins, epidermal growth factor, and vascular endothelia growth factor. Other prominent associations included “regulation of actin cytoskeleton” and “ubiquitin-mediated proteolysis.” Expression of two-thirds of the genes (32 of 48) linked to regulation of actin cytoskeleton was greater in HRmini, whereas the majority of genes (22 of 34) associated with the KEGG pathway ubiquitin-mediated proteolysis were significantly less expressed in HRmini. Some associations, such as “hypertrophic/dilated cardiomyopathy” (Table 1), arise due to expression of specific isoforms of myofibrillar proteins, which are upregulated in failing hearts but are part of the normal expression pattern in the context of skeletal muscle. Similarly, associations to “pathways in cancer,” “glioma,” and “renal cell carcinoma” in Table 1 arise due to enrichment of genes associated with growth factor signaling pathways. Evidence exists to link the majority of genes listed within such pathways with the observed differences in muscle phenotype. However, because current ontology databases contain a proportionally greater emphasis on pathophysiological processes, particularly oncological research, this clustering of growth factor-related genes is misinterpreted as tumorigenesis. Therefore, to minimize such erroneous associations the data were further assessed using Ingenuity Pathway Analysis with the Ingenuity Knowledge Base restricted to skeletal muscle only. Under this constraint, significant (P < 0.05) enrichment was reported for 12 canonical pathways, and a cluster of four pathways (calcium signaling, glycolysis/gluconeogenesis, protein kinase A, and cAMP-mediated signaling) exhibited highly significant (P < 0.001) associations with the experiment dataset (Table 2). The validity of the microarray data is demonstrated by comparing known phenotypic differences between HRmini and HRnormal with the most highly significant (P < 0.01) genes that were up- or downregulated in the four top-ranked canonical pathways.
Table 1.
Top-ranking KEGG pathways reported by DAVID
| Name | Genes, n | BH Corrected P Value |
|---|---|---|
| Focal adhesion | 50 | 0.000019 |
| Regulation of actin cytoskeleton | 48 | 0.00099 |
| Ubiqutin-mediated proteolysis | 34 | 0.0012 |
| Hypertrophic cardiomyopathy | 32 | 0.0062 |
| Pathways in cancer | 59 | 0.0098 |
| Pyruvate metabolism | 14 | 0.012 |
| Glycolysis/gluconeogenesis | 19 | 0.012 |
| Dilated cardiomyopathy | 22 | 0.03 |
| Glioma | 17 | 0.035 |
| Renal cell carcinoma | 18 | 0.033 |
| Ribosome | 21 | 0.035 |
| MAPK signaling pathway | 47 | 0.037 |
Top-ranking KEGG pathways reported by the DAVID functional annotation tool applied to all genes exhibiting significant (P < 0.05 corrected) differences in expression between normal high-runner (HRnormal) and minimuscle high-runner (HRmini) mice.
Table 2.
Skeletal muscle genes associated with the top-ranking IPA canonical pathways
| Fold Difference |
||||
|---|---|---|---|---|
| Pathway | Gene | Description | (HRmini) | Campbell |
| Calcium signaling | ||||
| Myl4 | Myosin, light chain 4, alkali; atrial, embryonic | 9.78 | ||
| Myl3 | Myosin, light chain 3, alkali; ventricular, skeletal, slow | 2.82 | ||
| Tpm2 | Tropomyosin 2 (beta) | 2.30 | ||
| Tnni1 | Troponin I type 1 (skeletal, slow) | 2.23 | ||
| Ppkar1a | Protein kinase, cAMP-dependent, regulatory, type I, alpha | 2.05 | ||
| Mef2c | Myocyte enhancer factor 2C | 2.05 | ||
| Myh7 | Myosin, heavy chain 7, cardiac muscle, beta | 1.97 | ||
| Tnnt1 | Troponin T type 1 (skeletal, slow) | 1.93 | ||
| Camk2d | Calcium/calmodulin-dependent protein kinase II delta | 1.88 | ||
| Tpm3 | Tropomyosin 3 | 1.67 | ||
| Myl9 | Myosin, light chain 9, regulatory | 1.62 | ||
| Myh4 | Myosin, heavy chain 4, skeletal muscle | −8.82 | −10.0 | |
| Calm1 | Calmodulin 1 (phosphorylase kinase, delta) | −3.08 | −2.3 | |
| Camk2a | Calcium/calmodulin-dependent protein kinase II alpha | −2.20 | ||
| Calm3 | Calmodulin 3 (phosphorylase kinase, delta) | −2.06 | ||
| Calm2 | Calmodulin 2 (phosphorylase kinase, delta) | −1.85 | −1.6 | |
| Tnni2 | Troponin I type 2 (skeletal, fast) | −1.79 | ||
| Trdn | Triadin | −1.57 | ||
| Ppp3r1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform | −1.53 | ||
| Asph | Aspartate beta-hydroxylase | −1.51 | ||
| Ppp3cb | Protein phosphatase 3, catalytic subunit, beta isozyme | −1.50 | −6 | |
| Atf4 | Activating transcription factor 4 (tax-responsive enhancer element B67) | −1.35 | ||
| Tnnt3 | Troponin T type 3 (skeletal, fast) | −1.22 | ||
| Ppkar2a | Protein kinase, cAMP-dependent, regulatory, type II, alpha | −1.21 | ||
| Glycolysis/Gluconeogenesis | ||||
| Ldhb | Lactate dehydrogenase B | 1.91 | 17.0 | |
| Aldh1a1 | Aldehyde dehydrogenase 1 family, member A1 | 1.83 | ||
| Aldh2 | Aldehyde dehydrogenase 2 family (mitochondrial) | 1.78 | ||
| Gpi | Glucose phosphate isomerase | 1.67 | −1.9 | |
| Dlat | Dihydrolipoamide S-acetyltransferase | 1.22 | ||
| Bpgm | 2,3-bisphosphoglycerate mutase | −2.77 | ||
| Acyp1 | Acylphosphatase 1, erythrocyte (common) type | −2.60 | ||
| Pgm1 | Phosphoglucomutase 1 | −1.97 | ||
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | −1.31 | −12 | |
| Pgk1 | Phosphoglycerate kinase 1 | −1.30 | ||
| Pgam2 | Phosphoglycerate mutase 2 (muscle) | −1.24 | ||
| Tpi1 | Triosephosphate isomerase 1 | −1.22 | −2.3 | |
| Protein kinase A signaling | ||||
| Myl4 | Myosin, light chain 4, alkali; atrial, embryonic | 9.78 | ||
| Flnc | Filamin C, gamma | 3.05 | ||
| Myl3 | Myosin, light chain 3, alkali; ventricular, skeletal, slow | 2.82 | ||
| Gys1 | Glycogen synthase 1 (muscle) | 2.57 | ||
| Tnni1 | Troponin I type 1 (skeletal, slow) | 2.23 | ||
| Prkar1a* | Protein kinase, cAMP-dependent, regulatory, type I, alpha | 2.05 | ||
| Camk2d* | Calcium/calmodulin-dependent protein kinase II delta | 1.88 | ||
| Pde8a* | Phosphodiesterase 8A | 1.66 | ||
| Myl9 | Myosin, light chain 9, regulatory | 1.62 | ||
| Akap1 | A kinase (PRKA) anchor protein 1 | 1.59 | ||
| H3f3c | H3 histone, family 3C | 1.44 | ||
| Akap6 | A kinase (PRKA) anchor protein 6 | 1.40 | ||
| Akap9 | A kinase (PRKA) anchor protein (yotiao) 9 | 1.38 | ||
| Hist1 h1c | Histone cluster 1, H1c | 1.34 | ||
| Add1 | Adducin 1 (alpha) | 1.27 | ||
| Myh4 | Myosin, heavy chain 4, skeletal muscle | −8.82 | −10 | |
| Calm1 | Calmodulin 1 (phosphorylase kinase, delta) | −3.08 | −2.3 | |
| Pde4b | Phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4) | −2.79 | ||
| Camk2a | Calcium/calmodulin-dependent protein kinase II alpha | −2.20 | ||
| Calm3 | Calmodulin 3 (phosphorylase kinase, delta) | −2.06 | ||
| Phkb | Phosphorylase kinase, beta | −1.92 | ||
| Calm2 | Calmodulin 2 (phosphorylase kinase, delta) | −1.85 | −1.6 | |
| Tnni2 | Troponin I type 2 (skeletal, fast) | −1.79 | ||
| Ppp1r14c | Protein phosphatase 1, regulatory (inhibitor) subunit 14C | −1.68 | ||
| Anapc5 | Anaphase promoting complex subunit 5 | −1.61 | ||
| Ppp3r1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform | −1.53 | ||
| Ppp3cb | Protein phosphatase 3, catalytic subunit, beta isozyme | −1.50 | −6 | |
| Ywhae | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | −1.47 | ||
| Gsk3b | Glycogen synthase kinase 3 beta | −1.41 | ||
| Atf4 | Activating transcription factor 4 (tax-responsive enhancer element B67) | −1.35 | ||
| Gnas | GNAS complex locus | −1.27 | ||
| Mylpf | Myosin light chain, phosphorylatable, fast skeletal muscle | −1.26 | ||
| Ppkar2a | Protein kinase, cAMP-dependent, regulatory, type II, alpha | −1.21 | ||
| cAMP-mediated signaling | ||||
| Rgs2 | Regulator of G-protein signaling 2, 24 kDa | 2.06 | ||
| Prkar1a | Protein kinase, cAMP-dependent, regulatory, type I, alpha | 2.05 | ||
| Camk2d | Calcium/calmodulin-dependent protein kinase II delta | 1.88 | ||
| Pde8a | Phosphodiesterase 8A | 1.66 | ||
| Akap1 | A kinase (PRKA) anchor protein 1 | 1.59 | ||
| Akap6 | A kinase (PRKA) anchor protein 6 | 1.40 | ||
| Akap9 | A kinase (PRKA) anchor protein (yotiao) 9 | 1.38 | ||
| Calm1 | Calmodulin 1 (phosphorylase kinase, delta) | −3.08 | −2.3 | |
| Pde4b | Phosphodiesterase 4B, cAMP-specific (phosphodiesterase E4) | −2.79 | ||
| Camk2a | Calcium/calmodulin-dependent protein kinase II alpha | −2.20 | ||
| Calm3 | Calmodulin 3 (phosphorylase kinase, delta) | −2.06 | ||
| Calm2 | Calmodulin 2 (phosphorylase kinase, delta) | −1.85 | −1.6 | |
| Ppp3r1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform | −1.53 | ||
| Ppp3cb | Protein phosphatase 3, catalytic subunit, beta isozyme | −1.50 | −6 | |
| Atf4 | Activating transcription factor 4 (tax-responsive enhancer element B67) | −1.35 | ||
| Gnas | GNAS complex locus | −1.27 | ||
| Prkar2a | Protein kinase, cAMP-dependent, regulatory, type II, alpha | −1.21 | ||
Fold difference (HRmini) represents statistically significant (P < 0.05 after correction for multiple testing) differences in expression in HRmini compared with HRnormal gastrocnemius reported in the current work. Fold difference (Campbell) represents significant differences in expression in soleus compared with white gastrocnemius of ICR mice reported in Campbell et al. (5).
Of the 24 genes associated with “calcium signaling” 11 encode myofibrillar proteins. HRmini are characterized by selective loss of type IIB fibers (15), and we found lesser expression of type IIb MyHC (Myh4; −8.82-fold, P = 0.0001), and attendant differences in phosphorylatable fast myosin light chain (Mylpf; −1.26-fold, P = 0.006) and fast isoforms of troponin I (Tnni2; −1.79-fold, P = 0.0003) and troponin T (Tnnt3; −1.22-fold, P = 0.004) in HRmini. Conversely, expression of type I MyHC (Myh7; 1.97-fold, P = 0.002) and myofibril protein isoforms associated with slow-twitch muscle, including β-tropomyosin (Tpm2; 2.3-fold, P = 0.0001), slow α-tropomyosin (Tpm3; 1.67-fold, P = 0.0006), slow troponin I (Tnni1; 2.23-fold, P = 0.005), slow troponin T (Tnnt1; 1.93-fold, P = 0.0005), and ventricular/slow skeletal myosin light chain (Myl3; 2.82-fold, P = 0.006) was greater in HRmini (Table 2).
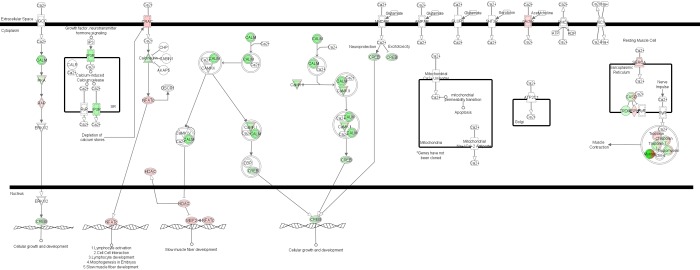
The majority of nonmyofibrillar genes listed in calcium signaling were also associated with the protein kinase A canonical pathway. In addition, the calcium signaling network (Fig. 2) also included triadin (Trdn) and asparate β-hydroxylase (Asph), which are implicated in sarcoplasmic reticulum calcium handling, and myocyte enhancer factor 2C (Mef2c), which is associated with the expression of slow-twitch myofibril genes in adult skeletal muscle.
Fig. 2.
Differential expression of genes associated with calcium signaling. Prominent genes of the Ingenuity Pathway Analysis network “calcium signaling” are shown relative to their cellular location. The expression of genes colored red was significantly greater in HRmini, whereas the expression of genes colored green was greater in HRnormal (P < 0.05, after correction for multiple testing).
Differential expression of genes associated with “glycolysis/gluconeogenesis” may also link with the low proportion of fast glycolytic fibers in HRmini. Expression of the heart isoform of lactate dehydrogenase (Ldhb), which is more prominent in slow-oxidative muscle, was greater in HRmini, but there were mixed differences in the expression of enzymes of the glycolytic pathway; phosphoglucomutase (−1.50-fold, P = 0.0006), triosephosphate isomerase (−1.22-fold, P = 0.004), and phosphoglycerate kinase (−1.30-fold, P = 0.006) were less expressed in HRmini, whereas expression of hexokinase (1.61-fold, P = 0.001), glucose phosphate isomerase (1.67-fold, P = 0.001), and enolase (1.56-fold, P = 0.001) was greater in HRmini.
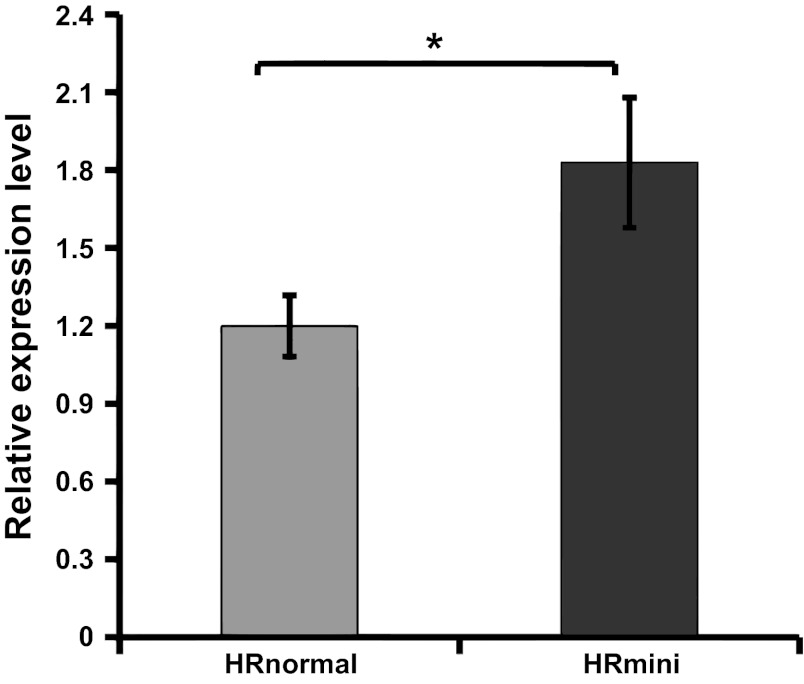
Previous work mapped the region of the HRmini gene to a 2.635-Mb interval on MMU11, which harbors ∼100 expressed or predicted genes (20); seven genes (Table 3) from this region were differentially expressed between HRnormal and HRmini. Among these, very long-chain specific acyl-CoA dehydrogenase (Acadvl) expression exhibited the greatest statistical difference (P = 0.002) and was 1.60-fold greater in HRmini. Furthermore, functional annotation of genes enriched in HRmini highlighted the KEGG pathway “fatty acid metabolism,” which contained 11 genes (Table 4). To validate differential expression of Acadvl, we performed real-time quantitative RT-PCR and showed that the expression level of Acadvl was 1.5-fold (P < 0.05) greater in HRmini compared with HRnormal mice (Fig. 3).
Table 3.
Differentially expressed genes from HRmini region mapped to MU11
| Gene | Affymetrix ID | Description | Fold Difference | P Value |
|---|---|---|---|---|
| Myh10 | 1452740_at | Myosin heavy chain 10, nonmuscle | 1.92 | 0.043 |
| Chrnb1 | 1420682_at | Cholinergic receptor, nicotinic, beta polypeptide 1 (muscle) | 1.76 | 0.0108 |
| Acadv1 | 1424184_at | Acyl-Coenzyme A dehydrogenase, very long chain | 1.60 | 0.00242 |
| Senp3 | 1448866_at | SUMO/sentrin specific protease 3 | 1.27 | 0.0343 |
| Gabarap | 1416937_at | Gamma-aminobutyric acid receptor-associated protein | 1.15 | 0.0425 |
| Eif5a | 1451470_s_at | Eukaryotic translation initiation factor 5A | −1.27 | 0.0254 |
| Clec10a | 1419605_at | Macrophage galactose N-acetyl-galactosamine-specific lectin | −3.12 | 0.0205 |
Fold difference represents differences in expression in HRmini compared with HRnormal. P values are from GeneSpring microarray analysis corrected by Benjamini and Hochberg multiple testing with 5% false positive rate.
Table 4.
Differentially expressed genes associated with KEGG pathway “fatty acid metabolism”
| Gene | Affymetrix ID | Description | Delta | P Value |
|---|---|---|---|---|
| ACAA1A | 1416947_s_at | Aacetyl-Coenzyme A acyltransferase 1A | 1.57 | 0.0299 |
| ACAA2 | 1428145_at, 1428146_s_at | Acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 1.68 | 0.0376 |
| ACSL1 | 1423883_at, 1422526_at | Acyl-CoA synthetase long-chain family member 1 | 1.42 | 0.0146 |
| ACADL | 1448987_at | Acyl-Coenzyme A dehydrogenase, long-chain | 1.84 | 0.00306 |
| ACADS | 1460216_at | Acyl-Coenzyme A dehydrogenase, short chain | 1.67 | 0.01 |
| ACADVL | 1424184_at | Acyl-Coenzyme A dehydrogenase, very long chain | 1.60 | 0.00242 |
| ALDH2 | 1448143_at | Aldehyde dehydrogenase 2, mitochondrial | 1.78 | 0.000667 |
| ALDH7A1 | 1460167_at | Aldehyde dehydrogenase family 7, member A1 | 1.76 | 0.000674 |
| HADH | 1455972_x_at, 1460184_at | Hydroxyacyl-Coenzyme A dehydrogenase | 1.41 | 0.0385 |
| HADHA | 1452173_at | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A-thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 1.74 | 0.00383 |
| PECI | 1431012_a_at | Peroxisomal delta3, delta2-enoyl-Coenzyme A isomerase | 1.41 | 0.0472 |
Delta represents fold difference in expression in HRmini compared with HRnormal. P values from GeneSpring microarray analysis corrected by Benjamini and Hochberg multiple testing with 5% false positive rate.
Fig. 3.
Acadvl expression in HRnormal and HRmini muscle. Relative mRNA expression of Acadvl in HRnormal (line #8) and HRmini (line #3) gastrocnemius muscle. Data are presented as means ± SD (n = 6, in each group). *Statistical significance (P < 0.05) of the difference in expression was determined by 2-tailed t-test for independent samples.
DISCUSSION
The current work provides comprehensive descriptions of two alternative muscle transcriptome “solutions” to a single type of selection. These models of artificial selection involve naturally occurring genetic variants (mutant alleles) rather than chemical or genetic manipulations that are often associated with deleterious correlated traits. HRmini and HRnormal each exhibit enhanced physical capacity and as such might also occur, and be viable, in natural populations of house mice. In contrast, man-made genetic manipulations often induce phenotypes that are unlikely to be viable in nature. Despite their similar running capacities, gastrocnemius of adult HRnormal and HRmini exhibited broad differences in gene expression. Genetic similarities in these two independent lines may encompass genes that are either essential or entirely unrelated to exercise capacity. Whereas gene networks exhibiting significant differences between HRnormal and HRmini highlight aspects of genomic “flexibility” that have resulted in alternative genotypes capable of accomplishing the same task at the biochemical/ physiological level.
Previous work (20) mapped the region of the HRmini gene to a 2.635-Mb interval on MMU11 that harbors ∼100 expressed or predicted genes. We discovered seven genes (Table 3) from this region that were differentially expressed between HRnormal and HRmini. Published literature exists to potentially implicate each of these genes in the origin of the HRmini phenotype. For example, nonmuscle myosin (Myh10) is associated with neurite growth cone motility (4) and hepatic mitochondrial DNA number (34). Small ubiquitin-like modifier (SUMO) demodification, mediated by Senp3, is specifically associated with regulation of ribosome biogenesis (16). Gabarap is a known atrophy gene (24) and is upregulated in cisplatin-induced atrophy of C2C12 myotubes (9). Eukaryotic initiation factor 5a (Eif5a) undergoes a unique posttranslational modification (hypusination) that correlates with embryonic myogenic differentiation, and inhibition of EIF5A hypusination impairs C2C12 differentiation (33). Clec10a (MGL1) regulates inflammatory processes, and MGL1−/− mice are protected from glucose intolerance and insulin resistance associated with diet-induced obesity due to diminished adipose tissue macrophage response (44).
Two genes, Chrnb1 and Acadvl, from the 2.635-Mb interval on MMU11 (20) that exhibited the most robust statistical differences between HRmini and HRnormal (Table 3) also link closely with the outcomes of the functional enrichment analyses. Chrnb1 encodes the beta subunit of the muscle acetylcholine receptor, and defects in this receptor are associated with fetal akinesia deformation sequence disorders (31). The greater expression of Chrnb1 in HRmini gastrocnemius could be linked to the differences observed in Ca2+ signaling (Fig. 2). Acadvl encodes a mitochondrial enzyme that performs the first step of beta-oxidation of long-chain and very long-chain fatty acids. Acadvl deficiency is associated with mild cardiac hypertrophy (8) and altered muscle metabolism that protects Acadvl-deficient mice from high fat diet-induced obesity (46). HRmini also exhibit cardiac hypertrophy (11, 41), but the expression of Acadvl was significantly greater in HRmini than HRnormal muscle. Therefore, the difference in expression of Acadvl in HRmini may be associated with the coincident changes in mitochondrial fatty acid metabolism (Table 4).
The concentration of mitochondrial enzymes, including citrate synthase, carnitine palmitoyltransferase 1, and cytochrome c oxidase, is approximately twofold greater in HRmini than HRnormal gastrocnemius (14, 21). However, the size of triceps surae muscles in HRmini is approximately half that of HRnormal, so, at the whole animal level, these factors counterbalance each other, and HRnormal and HRmini muscles afford similar total energetic capacities for exercise (21). Nevertheless, differences in substrate utilization may exist, and whole animal fatty acid oxidation during submaximal exercise is significantly greater in HRmini, which is associated with greater β-hydroxacyl-CoA dehydrogenase activity (41). The current data extend these findings and show the expression of 11 genes (including Hadh) encoding enzymes of the KEGG fatty acid metabolism pathway (Table 4) is greater in HRmini gastrocnemius. In addition, genes associated with the transport of fatty acids to within muscle [solute carrier family 27 (fatty acid transporter member 1), Slc27a1; 1.84-fold, P = 0.0012] and mitochondria (mitochondrial carnitine/acylcarnitine translocase, Cpt1a; 1.76-fold, P = 0.0062) were also expressed at greater levels in HRmini. These findings suggest HRmini skeletal muscle has a greater capacity for fatty acid utilization, which may be consistent with the greater retroperitoneal fat pad mass of HRmini animals (29).
The current genome-wide profiling data reveals the differences in the expression of fatty acid metabolism genes occur in the absence of widespread differences in regulators of mitochondrial biogenesis and genes encoding other mitochondrial enzymes. That is, few components of electron transport chain holoenzymes (e.g., cytochrome c oxidase, subunit VIIIa; 1.5-fold greater in HRmini, P < 0.05) were differentially expressed in HRmini, and the KEGG “oxidative phosphorylation” pathway was not significantly enriched in the current dataset. Peroxisome proliferator-activated receptor alpha (PPARα) is a key transcription factor associated with the expression of enzymes involved in fatty acid utilization (10), and Templeman et al. (41) reports muscle mRNA expression of PPARα, but not sirtuin 1, PPARβ/δ, or PPARγ coactivator-1α (PGC-1α), is significantly greater in HRmini gastrocnemius. Six of the differentially expressed genes in the KEGG fatty acid metabolism pathway contain PPAR elements in their promoter regions, but in the current work Ppara expression was not statistically greater (1.8-fold, P = 0.41) in HRmini. The promoter region of Acadvl contains regulatory elements for activating enhancer binding protein 2 alpha (AP-2α), AP-2α isoform 2, and AP-2α isoform 3 only. Ppara, and genes differentially expressed in HRmini (Table 4) also have regulatory elements in their promoter regions for AP-2 isoforms, but these also were not differentially expressed. Therefore, the differences between HRmini and HRnormal appear to be due to novel mechanisms discrete from current mainstream hypotheses regarding transcriptional regulation of muscle fatty acid metabolism.
Approximately 10% of the total number of genes detected in the current work were differentially expressed between HRmini and HRnormal muscle. This proportion closely resembles analysis (5) comparing white gastrocnemius (100% IIb fibers) and soleus (70% type I, 30% type IIa fibers) from ICR mice, which have similar differences in type IIB myofiber content to HRnormal and HRmini gastrocnemius (note that ICR mice formed the base population for the selection experiment that produced the HRnormal and HRmini mice). Campbell et al. (5) report differences between white gastrocnemius and soleus in the expression of ∼12% of detectable genes including eight (Myh4, Calm1, Calm2, Ppp3cb, Ldhb, Gpi, Gapdh, and Tpi1; Table 2) that are also differentially expressed between gastrocnemius of HRmini and HRnormal. The differences in Myh4, Calm1, Calm2, and Tpi1 expression reported here are similar to those described by Campbell et al. (5) (Table 2), whereas the magnitude of difference in Ppp3cb, Ldhb, and Gapdh expression is relatively less than expected, and HRmini exhibit elevated expression of Gpi, which contrasts with the expected lesser expression of Gpi in slow- compared with fast-twitch muscle.
MyHC IIb is the predominant MyHC isoform expressed in mouse gastrocnemius muscle (18), and homogenous knockout of MyHC IIb results in a significant decrease in the average total number of fibers in hind limb muscles, partial compensatory hypertrophy of IId fibers (1), and a shift to greater expression of IIx/d fibers, consistent with the nearest-neighbor rule (27). Allen et al. (1) comment “We did not see any discernible difference in locomotor behavior between wild-type and IIb null mice. The pattern of gait and posture was unchanged, and there were not any overt differences in the quantity of cage locomotion (data not shown).” Harrison et al. (19), on the other hand, report lower voluntary wheel running in IIb knockout mice and signs of pathology, including fiber degeneration/regeneration. Similar to MyHC IIb knockout mice, HRmini gastrocnemius contains small fibers in the superficial areas of the muscle (15), which normally would be exclusively occupied by IIb fibers. Due to signs of pathology and expression of embMyHC in some regenerating fibers, it is thought that the small muscle phenotype of IIb null mice is caused by atrophy and degeneration of IIb fibers (1). Interestingly, myofibrillar genes upregulated in HRmini include embryonic isoforms of MyHC and myosin light chain along with cardiac isoforms of MyHC and troponin T, which, typically, are only expressed in regenerating adult skeletal muscle (2, 3, 28).
Microarray analysis also detected differences in myogenic regulatory factors, including myogenin (3.83-fold, P = 0.0026), alongside robust (21.35-fold greater in HRmini) difference in the expression of cardiac ankyrin repeat domain 1 (CARP), which is a member of the muscle ankyrin repeat transcription factor family (25). Previously we (6) reported elevated expression of CARP in skeletal muscle after eccentric exercise. However, more recent evidence (26) demonstrates CARP expression is also upregulated during muscle atrophy, including denervation-induced atrophy and various models of muscular dystrophy. Interestingly, overexpression of CARP in adult skeletal muscle does not significantly alter muscle size (26). Therefore, CARP may represent a molecular marker of muscle transformation rather than specific regulator of either atrophic or hypertrophic processes. Other prominent features of the microarray data can also be linked to differences in muscle size associated with catabolic and anabolic hormone signaling (36). Keratin 8 (24.9-fold upregulated in HRmini) and Keratin 18 (6.4-fold upregulated in HRmini) are major components in intermediate filaments of the cytoskeleton and modulate tumor necrosis factor-α signaling in simple epithelial cells (23), whereas the ATP-independent protease serine peptidase 4 (HtrA), which regulates the availability of insulin-like growth factors (IGF) by cleaving IGF-binding proteins (7), was 40.5-fold less expressed in HRmini.
In summary, microarray analysis of adult HRnormal and HRmini gastrocnemius revealed the significant decrease in type IIb MyHC content (Myh4 expression) reported in previous studies is associated with attendant differences in the expression of numerous “fast” and “slow” isoforms of myofibrillar genes. The co-occurrence of prominent differences in genes of the calcium, protein kinase A, and cAMP pathways implicate differences in calcium signaling (Fig. 2) and activation of the “slow gene program” via nuclear factor of activated T-cells, cytoplasmic calcineurin-dependent 2 (Nfatc2), and myocyte-specific enhancer factor 2c (Mef2c) as possible contributing mechanisms. However, this cross-sectional study is not able to determine whether these differences in gene expression represent causes or consequences of the minimuscle phenotype.
Genes exhibiting the greatest differences in expression between HRnormal and HRmini point to differences in anabolic/catabolic signaling and indicate the processes of muscle remodeling are ongoing in adult HRmini gastrocnemius, which is further supported by the significant association with the KEGG focal adhesion pathway. In addition, the microarray data extend recent findings regarding the enhanced utilization of fatty acids by HRmini mice (41) and suggest this does not correspond with prototypical transcriptional regulation of muscle fatty acid metabolism. This finding may be related to greater expression of Acadvl, which resides in the 2.6 Mb interval of MMU11 highlighted by linkage mapping as the causal region of the HRmini phenotype.
GRANTS
Supported by National Science Foundation Grant IOS-1121273 to T. Garland. Y-W. Chen was partially supported by National Institutes of Health (NIH) Grants 1RO1AR-052027 and 1R24HD-050846. S. N. Pandey was supported by NIH Grant 1RO1AR-052027.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.G.B., T.G.J., and Y.-W.C. conception and design of research; J.G.B., T.H.M., S.N.P., G.B.-M., and M.F.M. performed experiments; J.G.B., T.H.M., S.N.P., G.B.-M., M.F.M., A.M.B., and Y.-W.C. analyzed data; J.G.B., T.H.M., S.N.P., G.B.-M., M.F.M., A.M.B., T.G.J., and Y.-W.C. interpreted results of experiments; J.G.B., T.H.M., S.N.P., G.B.-M., M.F.M., T.G.J., and Y.-W.C. prepared figures; J.G.B., A.M.B., T.G.J., and Y.-W.C. drafted manuscript; J.G.B., T.H.M., A.M.B., T.G.J., and Y.-W.C. edited and revised manuscript; J.G.B., T.H.M., S.N.P., G.B.-M., M.F.M., A.M.B., T.G.J., and Y.-W.C. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank F. R. Gomes and J. L. Malisch for assistance with tissue collection and preparation, B. Walter for running the Affymetrix microarrays at the UCR genomics center, and Vishal Sharma for assistance with qRT-PCR.
REFERENCES
- 1. Allen DL, Harrison BC, Sartorius C, Byrnes WC, Leinwand LA. Mutation of the IIB myosin heavy chain gene results in muscle fiber loss and compensatory hypertrophy. Am J Physiol Cell Physiol 280: C637–C645, 2001 [DOI] [PubMed] [Google Scholar]
- 2. Barton PJ, Cohen A, Robert B, Fiszman MY, Bonhomme F, Guénet JL, Leader DP, Buckingham ME. The myosin alkali light chains of mouse ventricular and slow skeletal muscle are indistinguishable and are encoded by the same gene. J Biol Chem 260: 8578–8584, 1985 [PubMed] [Google Scholar]
- 3. Bodor GS, Survant L, Voss EM, Smith S, Porterfield D, Apple FS. Cardiac troponin T composition in normal and regenerating human skeletal muscle. Clin Chem 43: 476–484, 1997 [PubMed] [Google Scholar]
- 4. Bridgman PC, Dave S, Asnes CF, Tullio AN, Adelstein RS. Myosin IIB is required for growth cone motility. J Neurosci 21: 6159–6169, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell WG, Gordon SE, Carlson CJ, Pattison JS, Hamilton MT, Booth FW. Differential global gene expression in red and white skeletal muscle. Am J Physiol Cell Physiol 280: C763–C768, 2001 [DOI] [PubMed] [Google Scholar]
- 6. Chen YW, Hubal MJ, Hoffman EP, Thompson PD, Clarkson PM. Molecular responses of human muscle to eccentric exercise. J Appl Physiol 95: 2485–2494, 2003 [DOI] [PubMed] [Google Scholar]
- 7. Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell 10: 443–455, 2002 [DOI] [PubMed] [Google Scholar]
- 8. Cox KB, Liu J, Tian L, Barnes S, Yang Q, Wood PA. Cardiac hypertrophy in mice with long-chain acyl-CoA dehydrogenase or very long-chain acyl-CoA dehydrogenase deficiency. Lab Invest 89: 1348–1354, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fanzani A, Zanola A, Rovetta F, Rossi S, Aleo MF. Cisplatin triggers atrophy of skeletal C2C12 myotubes via impairment of Akt signalling pathway and subsequent increment activity of proteasome and autophagy systems. Toxicol Appl Pharmacol 250: 312–321, 2011 [DOI] [PubMed] [Google Scholar]
- 10. Finck BN. The PPAR regulatory system in cardiac physiology and disease. Cardiovasc Res 73: 269–277, 2007 [DOI] [PubMed] [Google Scholar]
- 11. Garland T, Jr, Morgan MT, Swallow JG, Rhodes JS, Girard I, Belter JG, Carter PA. Evolution of a small-muscle polymorphism in lines of house mice selected for high activity levels. Evolution 56: 1267–1275, 2002 [DOI] [PubMed] [Google Scholar]
- 12. Garland T, Rose M. Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments. Berkeley, CA: University of California Press, 2009 [Google Scholar]
- 13. Garland T, Kelly SA, Malisch JL, Kolb EM, Hannon RM, Keeney BK, Van Cleave SL, Middleton KM. How to run far: multiple solutions and sex-specific responses to selective breeding for high voluntary activity levels. Proc Biol Sci 278: 574–581, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guderley H, Houle-Leroy P, Diffee GM, Camp DM, Garland T., Jr Morphometry, ultrastructure, myosin isoforms, and metabolic capacities of the “mini muscles” favoured by selection for high activity in house mice. Comp Biochem Physiol B Biochem Mol Biol 144: 271–282, 2006 [DOI] [PubMed] [Google Scholar]
- 15. Guderley H, Joanisse DR, Mokas S, Bilodeau GM, Garland T., Jr Altered fibre types in gastrocnemius muscle of high wheel-running selected mice with mini-muscle phenotypes. Comp Biochem Physiol B Biochem Mol Biol 149: 490–500, 2008 [DOI] [PubMed] [Google Scholar]
- 16. Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep 9: 273–279, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hakimi P, Yang J, Casadesus G, Massillon D, Tolentino-Silva F, Nye CK, Cabrera ME, Hagen DR, Utter CB, Baghdy Y, Johnson DH, Wilson DL, Kirwan JP, Kalhan SC, Hanson RW. Overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) in skeletal muscle repatterns energy metabolism in the mouse. J Biol Chem 282: 32844–32855, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamalainen N, Pette D. The histochemical profiles of fast fibre types IIB, IID and IIA in skeletal muscle of mouse, rat and rabbit. J Histochem Cytochem 41: 733–743, 1993 [DOI] [PubMed] [Google Scholar]
- 19. Harrison BC, Bell ML, Allen DL, Byrnes WC, Leinwand LA. Skeletal muscle adaptations in response to voluntary wheel running in myosin heavy chain null mice. J Appl Physiol 92: 313–322, 2002 [DOI] [PubMed] [Google Scholar]
- 20. Hartmann J, Garland T, Jr, Hannon RM, Kelly SA, Muñoz G, Pomp D. Fine mapping of “mini-muscle,” a recessive mutation causing reduced hindlimb muscle mass in mice. J Hered 99: 679–687, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Houle-Leroy P, Guderley H, Swallow JG, Garland T., Jr Artificial selection for high activity favors mighty mini-muscles in house mice. Am J Physiol Regul Integr Comp Physiol 284: R433–R443, 2003 [DOI] [PubMed] [Google Scholar]
- 22. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Inada H, Izawa I, Nishizawa M, Fujita E, Kiyono T, Takahashi T, Momoi T, Inagaki M. Keratin attenuates tumor necrosis factor-induced cytotoxicity through association with TRADD. J Cell Biol 155: 415–426, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117: 2805–2812, 2004 [DOI] [PubMed] [Google Scholar]
- 25. Kojic S, Medeot E, Guccione E, Krmac H, Zara I, Martinelli V, Valle G, Faulkner G. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. J Mol Biol 339: 313–325, 2004 [DOI] [PubMed] [Google Scholar]
- 26. Laure L, Suel L, Roudaut C, Bourg N, Ouali A, Bartoli M, Richard I, Danièle N. Cardiac ankyrin repeat protein is a marker of skeletal muscle pathological remodelling. FEBS J 276: 669–684, 2009 [DOI] [PubMed] [Google Scholar]
- 27. Leeuw T, Pette D. Coordinate changes in the expression of troponin subunit and myosin heavy-chain isoforms during fast-to-slow transition of low-frequency-stimulated rabbit muscle. Eur J Biochem 213: 1039–1046, 1993 [DOI] [PubMed] [Google Scholar]
- 28. Lefaucheur L, Hoffman R, Okamura C, Gerrard D, Leger JJ, Rubinstein N, Kelly A. Transitory expression of alpha cardiac myosin heavy chain in a subpopulation of secondary generation muscle fibers in the pig. Dev Dyn 210: 106–116, 1997 [DOI] [PubMed] [Google Scholar]
- 29. Meek TH, Eisenmann JC, Garland T. Western diet increases wheel running in mice selectively bred for high voluntary wheel running. Int J Obes (Lond) 34: 960–969, 2010 [DOI] [PubMed] [Google Scholar]
- 30. Meek TH, Lonquich BP, Hannon RM, Garland T. Endurance capacity of mice selectively bred for high voluntary wheel running. J Exp Biol 212: 2908–2917, 2009 [DOI] [PubMed] [Google Scholar]
- 31. Michalk A, Stricker S, Becker J, Rupps R, Pantzar T, Miertus J, Botta G, Naretto VG, Janetzki C, Yaqoob N, Ott CE, Seelow D, Wieczorek D, Fiebig B, Wirth B, Hoopmann M, Walther M, Körber F, Blankenburg M, Mundlos S, Heller R, Hoffmann K. Acetylcholine receptor pathway mutations explain various fetal akinesia deformation sequence disorders. Am J Hum Genet 82: 464–476, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, Kang H, Shaw RJ, Evans RM. AMPK and PPARdelta agonists are exercise mimetics. Cell 134: 405–415, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parreiras-e-Silva LT, Luchessi AD, Reis RI, Oliver C, Jamur MC, Ramos RG, Oliveira EB, Curi R, Costa-Neto CM. Evidences of a role for eukaryotic translation initiation factor 5A (eIF5A) in mouse embryogenesis and cell differentiation. J Cell Physiol 225: 500–505, 2010 [DOI] [PubMed] [Google Scholar]
- 34. Reyes A, He J, Mao CC, Bailey LJ, Di Re M, Sembongi H, Kazak L, Dzionek K, Holmes JB, Cluett TJ, Harbour ME, Fearnley IM, Crouch RJ, Conti MA, Adelstein RS, Walker JE, Holt IJ. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucl Acids Res 39: 5098–5108, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rezende EL, Gomes FR, Malisch JL, Chappell MA, Garland T., Jr Maximal oxygen consumption in relation to subordinate traits in lines of house mice selectively bred for high voluntary wheel running. J Appl Physiol 101: 477–485, 2006 [DOI] [PubMed] [Google Scholar]
- 36. Saini A, Al-Shanti N, Faulkner SH, Stewart CE. Pro- and anti-apoptotic roles for IGF-I in TNF-alpha-induced apoptosis: a MAP kinase mediated mechanism. Growth Factors 26: 239–253, 2008 [DOI] [PubMed] [Google Scholar]
- 37. Swallow JG, Carter PA, Garland T. Artificial selection for increased wheel-running behavior in house mice. Behav Genet 28: 227–237, 1998 [DOI] [PubMed] [Google Scholar]
- 38. Swallow JG, Garland T, Jr, Carter PA, Zhan WZ, Sieck GC. Effects of voluntary activity and genetic selection on aerobic capacity in house mice (Mus domesticus). J Appl Physiol 84: 69–76, 1998 [DOI] [PubMed] [Google Scholar]
- 39. Syme DA, Evashuk K, Grintuch B, Rezende EL, Garland T., Jr Contractile abilities of normal and “mini” triceps surae muscles from mice (Mus domesticus) selectively bred for high voluntary wheel running. J Appl Physiol 99: 1308–1316, 2005 [DOI] [PubMed] [Google Scholar]
- 40. Szabó G, Dallmann G, Müller G, Patthy L, Soller M, Varga L. A deletion in the myostatin gene causes the compact (Cmpt) hypermuscular mutation in mice. Mamm Genome 9: 671–672, 1998 [DOI] [PubMed] [Google Scholar]
- 41. Templeman NM, Schutz H, Garland T, Jr, McClelland GB. Do mice selectively bred for high locomotor activity have a greater reliance on lipids to power submaximal aerobic exercise? Am J Physiol Regul Integr Comp Physiol 303: R101–R111, 2012 [DOI] [PubMed] [Google Scholar]
- 42. Varga L, Szabo G, Darvasi A, Muller G, Sass M, Soller M. Inheritance and mapping of Compact (Cmpt), a new mutation causing hypermuscularity in mice. Genetics 147: 755–764, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol 2: e294, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Westcott DJ, Delproposto JB, Geletka LM, Wang T, Singer K, Saltiel AR, Lumeng CN. MGL1 promotes adipose tissue inflammation and insulin resistance by regulating 7/4hi monocytes in obesity. J Exp Med 206: 3143–3156, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wong LE, Garland T, Jr, Rowan SL, Hepple RT. Anatomic capillarization is elevated in the medial gastrocnemius muscle of mighty mini mice. J Appl Physiol 106: 1660–1667, 2009 [DOI] [PubMed] [Google Scholar]
- 46. Zhang D, Christianson J, Liu ZX, Tian L, Choi CS, Neschen S, Dong J, Wood PA, Shulman GI. Resistance to high-fat diet-induced obesity and insulin resistance in mice with very long-chain acyl-CoA dehydrogenase deficiency. Cell Metab 11: 402–411, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]