Abstract
The transient receptor potential (TRP) superfamily is one of the largest families of cation channels. The metazoan TRP family has been subdivided into major branches: TRPC, TRPA, TRPM, TRPP, TRPV, TRPML, and TRPN, while the TRPY family is found in fungi. They are involved in many physiological processes and in the pathogenesis of various disorders. An efficient high-yield expression system for TRP channels is a necessary step towards biophysical and biochemical characterization and structural analysis of these proteins, and the budding yeast, Saccharomyces cerevisiae has proven to be very useful for this purpose. In addition, genetic screens in this organism can be carried out rapidly to identify amino acid residues important for function and to generate useful mutants. Here we present an overview of current developments towards understanding TRP channel function and structure using Saccharomyces cerevisiae as an expression system. In addition, we will summarize recent progress in understanding gating mechanisms of TRP channels using endogenously expressing TRPY channels in S. cerevisiae, and insights gained from genetic screens for mutants in mammalian channels. The discussion will focus particular attention of the use of cryo-electron microscopy (cryo-EM) to determine TRP channel structure, and outlines a “divide and concur” methodology for combining high resolution structures of TRP channel domains determined by X-ray crystallography with lower resolution techniques including cryo-EM and spectroscopy.
Keywords: cryo-electron microscopy, protein expression, TRP channel, X-ray crystallography
Introduction
Ion channels regulate the flow of ions across the plasma membrane in response to a variety of chemical, electrical, temperature, or mechanical signals. Determining ion channel structure is essential for understanding mechanisms of channel gating, modulation, ion selectivity and permeation. TRP channels display multifunctional and polymodal behavior in their regulation and interactions with proteins, lipids, and other small molecules and ions, and with electric fields. They thus present themselves as intriguing candidates for structural analysis. Detail structural information on TRP channels will allow development of new strategies for drug design targeting these channels. Several research groups have expended considerable effort towards understanding TRP channel structure, and structure-function relationships. In this chapter, we will review the current progress in understanding TRP channel structures through structural analysis of both full-length proteins and channel fragments.
The TRP channel family
The TRP family of channels derive their name from a Drosophila trp mutant with defective vision characterized by a transient receptor potential that was reported forty years ago(Cosens & Manning, 1969; Minke et al, 1975). Twenty years later, molecular cloning and functional analysis led to the discovery that the defect lies in a gene encoding a cation channel, known as TRP in Drosophila (Montell & Rubin, 1989). In the last decade, subsequent investigations identified over 70 homologues to the original TRP channel in invertebrates and vertebrates. To date, a total of 27 mammalian genes belonging to the TRP family have been reported and are subdivided into six major branches (Clapham et al, 2003): TRPC (canonical), TRPA (ankyrin), TRPM (melastatin), TRPP (polycystin), TRPV (vanilloid), and TRPML (mucolipin). The TRPN (NOMP-C homologues) sub-family of proteins are not found in mammals, but they are expressed in invertebrates such as flies and worms (Walker et al, 2000), and in cold-blooded vertebrates(Shin et al, 2005; Sidi et al, 2003). Yeast and other fungi express TRP channels known as the TRPY sub-family(Palmer et al, 2001; Zhou et al, 2005).
Based on sequence comparison and structural prediction algorithms, TRP channels are related to the superfamily of voltage-gated cation channels. Typically, TRP channels are predicted to have 6 transmembrane helices (TM1-6) per subunit, with varying sizes of cytoplasmic amino- and carboxy-termini, and are thought to form tetrameric assemblies (Clapham, 2003; Montell, 2005). Depending on the TRP family branch, the cytoplasmic amino-terminal domain contains different number of ankyrin repeats, ranging from zero to twenty nine, which have been proposed to be involved in a range of interactions (reviewed in (Gaudet, 2008)), including activating ligands, protein-protein interactions, and gating by voltage and temperature (Lepage et al, 2009). Recently it was discovered that mutations in the ankyrin domain of TRPV4 underlie autosomal dominant disorders of the peripheral nervous system (Auer-Grumbach et al,; Landoure et al, 2010), including Charcot-Marie-Tooth disease type 2C, the most common inherited neurological disease(Deng et al, 2010). The carboxy-termini of most contain a signature “TRP box” motive and coiled-coiled regions important in protein assembly. The majority of functionally characterized TRP channels are permeable to Ca2+ with the exception of TRPM4 and TRPM5, which are permeable to monovalent cations (Nilius et al, 2005). Ca2+ selectivity is poor for many TRP channels but TRPV5 and TRPV6 are highly permeable Ca2+ channels (Nilius et al, 2002). These channels function as polymodal sensors and are gated by diverse stimuli that include the binding of intracellular and extracellular messengers; changes in temperature, and chemical or mechanical stress (Venkatachalam & Montell, 2007).
TRP channels are widely distributed through the body, expressed in a vast number of different cell types and have numerous splice variants. TRP channels are particularly abundant in sensory receptor cells, and play a critical role in vision, touch, hearing, taste, pain and temperature sensation. The importance of determining TRP channel structure is highlighted by their emerging roles as major drug targets for the treatment of pain, inflammation, and a range of disorders (Colsoul et al, 2009; Cortright & Szallasi, 2009; Inoue et al, 2009; Lee & Gu, 2009; Watanabe et al, 2009; White et al, 2010; Woudenberg-Vrenken et al, 2009), and by the association of genetic defects in TRP channels with a number of devastating diseases, ranging from the most common single-gene neurological defect to polycystic kidney disease (Gallagher et al), to night blindness (Audo et al, 2009; Koike et al, 2010; Li et al, 2009; Morgans et al, 2009; Shen et al, 2009; van Genderen et al, 2009). The long-term hope is that understanding TRP channel structures, the structural determinants of ligand binding, and the effects of disease mutations on structure, will aid in the development of new therapeutics.
Ion channel structural biology
In the past several years, considerable progress has been made in the field of membrane protein structural biology. High-resolution structures for the pharmaceutically relevant eukaryotic membrane proteins, such as G protein-coupled receptors (Jaakola et al, 2008; Palczewski et al, 2000; Park et al, 2008; Rasmussen et al, 2007), transporters (Shinoda et al, 2009) and ion channels (Gonzales et al, 2009; Kawate et al, 2009; Long et al, 2005a; Long et al, 2005b; Long et al, 2007; Sobolevsky et al, 2009; Tao et al, 2009), have been obtained and provided very valuable information about mechanism of action for these proteins. Still, membrane protein structure determination remains a difficult task. Despite extensive efforts in many laboratories, the number of solved membrane protein structures remains small because of the many challenges presented by membrane proteins. Early success in crystallization of eukaryotic membrane proteins came from the use of the native sources that provide a large amount of protein, for example, bovine retinas providing a milligram of rhodopsin per cow (Palczewski et al, 2000). These problems have been especially challenging for eukaryotic ion channels among which only five high-resolution structures have been determined. Because of the low levels at which ion channels are typically expressed endogenously, structural studies of TRP channels and others require efficient heterologous systems for high level expression and purification. It is important that the cells used for expression are capable of properly folding and assembling the multiple subunits, and of producing them in stable and active form.
E. coli expression has been widely used for soluble eukaryotic proteins and for bacterial membrane proteins, as well as for soluble domains of transmembrane proteins. In the case of TRP channels, soluble fragments successfully produced in high yields from bacteria include the ankyrin repeat domains of proteins in the TRPV family (Jin et al, 2006; Lishko et al, 2007; McCleverty et al, 2006; Phelps et al, 2008; Phelps et al, 2010), domains of TRPM7 including the C-terminal cytoplasmic coiled-coil assembly domain (Fujiwara & Minor, 2008) and the α-kinase (Yamaguchi et al, 2001), and the coiled-coil region of the TRPP2 C-terminal domain (Yu et al, 2009).
Unfortunately, methods have not been found for routine expression in functional form of multi-pass eukaryotic membrane proteins such as ion channels in bacteria, likely as a result of lack of appropriate chaperones or other components of the folding and assembly machinery associated with the endoplasmic reticulum. No success has been achieved with full length TRP channels or their fragments containing transmembrane domains.
Baculovirus-mediated expression in insect cells offers another useful tool for generating recombinant membrane proteins (Brondyk, 2009). In 2009, several new eukaryotic ion channels structures were solved using insect cell expression (Gonzales et al, 2009; Kawate et al, 2009; Sobolevsky et al, 2009). However, the high cost of this methodology represents a drawback. Expression of some TRP channels using baculovirus has been reported, including overexpression of TRPV1 (Korepanova et al, 2009) and TRPV4 (Shigematsu et al, 2010). In the case of TRPV4 sufficient protein was purified for structure determination by in insect cells showed the possibly of using this system for structural biology of TRP channels, and the TRPV4 structure was determined using electron microscopy (Shigematsu et al, 2010).
Although even more expensive, mammalian tissue culture cells offer a native folding and assembly environment for mammalian membrane proteins. COS-1 cells have been used to express sufficient amounts of a rhodopsin mutant engineered for enhanced thermal stability (Standfuss et al, 2007). Mammalian cells have been used extensively for expression and functional studies of TRP channels, but only in a few cases have TRP channels been purified from them. TRPC3 (Mio et al, 2005; Mio et al, 2007) and TRPM2 (Maruyama et al, 2007) were expressed in mammalian cells, and used for structural studies by electron microscopy and single particle analysis.
Yeast is another traditional and powerful tool for the expression of eukaryotic recombinant proteins (Brondyk, 2009). The advantages include relatively low cost, rapid cell growth and ease of producing large volume cultures. Although distant in evolution from mammals, yeast possess conserved protein folding and assembly machinery that can be exploited to produce mammalian membrane proteins in large amounts. Most commonly, Pichia pastoris and Saccharomyces cerevisiae are used for the overexpression of membrane proteins. P. pastoris can achieve exceptionally high cell densities, that in favorable cases can provide high levels of protein production for structural biology. The first atomic structure of a mammalian potassium channel was possible only after the methodology for the overexpression of functional channel using P. pastoris was published (Parcej & Eckhardt-Strelau, 2003). Since that published work, the P. pastoris system has been used to obtain structures for a number of ion channels, including aquaporin (Nyblom et al, 2007) and potassium channels (Long et al, 2005a; Long et al, 2007; Tao et al, 2009; Tao et al, 2010; Tao & Mackinnon, 2008). Although attempts have been made, there has been no success reported in overexpression in functional form of TRP channels in P. pastoris.
TRP channels expression in Saccharomyces cerevisiae and functional analysis
One of the most characterized methods for the overexpression of recombinant membrane proteins has been budding yeast, S. cerevisiae. As eukaryotic organisms, yeast contain the machinery necessary for overexpression of eukaryotic membrane proteins, including rough endoplasmic reticulum and Golgi apparatus with associated molecules required for translocation, folding, and post-translational modifications, in addition to membrane trafficking machinery (Figler et al, 2000). They also have the advantage of easy plasmid and genetic manipulation for protein expression. A multitude of different strains including protease-deficient strains as well as a variety of expression vectors comprising yeast episomal plasmids (Yeps) and yeast integrating plasmids (Yips) are commercially available and allow genetic manipulation (Hunte et al, 2003). In addition, yeast express an endogenous TRP channel, known as TRPY1 or YVC1 in S. cerevisiae (Denis & Cyert, 2002; Palmer et al, 2001), so they have the necessary factors for folding and assembling channels of this family.
The earliest example of heterologous expression of a mammalian TRP channel in yeast was that of TRPV1 (Moiseenkova et al, 2003). The approach was based on a method that was published for the overexpression of P-glycoprotein (MDR1) from the Al-Shawi laboratory (Figler et al, 2000). The method is simple, and incorporates important steps that help to increase the expression of the protein by several-fold. High levels of expression of the protein were obtained using a maximally active PMA1 promoter. Although high levels of expression of a Ca2+-permeable channel, even under conditions of low activity, might be expected to be toxic to yeast; however, by transient expression produces large amounts of protein. Protein folding and stability are improved by addition of glycerol, which apparently acts as a sort of chemical chaperone. The optimal concentration of glycerol was determined for each construct, and was found to vary.
For heterologously expressed protein to be useful for structural studies, it needs to be functional. Ca2+ flux studies using Fura-2 in yeast confirmed that mammalian TRPV1 expressed in S. cerevisiae conducts Ca2+ in response to its well known agonist, capsaicin. A comparison of different detergents suggested that good solubilization could be obtained using either 1% egg L-α-lysophosphatidylcholine or 50 mM n-dodecyl-β-D-maltoside; subsequent studies (see below, and VYM and TGW unpublished observations) suggest that related detergents may be more suitable. Although TRPV1 expressed with a C-terminal His12 tag was purified to about 80% purity using nickel-chelate affinity chromatography, the yield was only about 1 mg per 16 L of yeast culture.
In order to obtain higher yields of functional and highly purified protein, a technique widely used for purification of rhodopsin and related visual pigments (Oprian, 1993; Oprian et al, 1987) was used (Moiseenkova-Bell et al, 2008). An epitope from the C-terminus of rhodopsin, recognized by a monoclonal antibody 1D4 (Molday & MacKenzie, 1983), was engineered into the C-terminus of TRPV1, allowing purification to homogeneity in a single step using an affinity column of immobilized 1D4 antibody, and elution with a peptide corresponding to the epitope. For this purification, n-decyl-β-D-maltoside was found to be the most useful detergent among those tried.
In this preparation, some heterogeneity in subunit molecular weight was revealed by SDS PAGE. A band appearing at a lower molecular weight than full length TRPV1 was consistently observed, but in varying relative amounts. Enzymatic de-glycosylation and stringent disulphide reduction did not eliminate the heterogeneity, suggesting that the smaller fragment results from proteolysis. The smaller fragment is recognized by the C-terminal 1D4 antibody, so presumably a piece of the cytoplasmic N-terminus is removed. A construct lacking part of the N-terminal domain does not display the lower band (VYM and TGW, unpublished observations).
Gel filtration in detergent revealed that purified TRPV1 migrated predominantly as a monodisperse homotetramer. Thus the mammalian protein assembled in yeast appears to have the appropriate stoichiometry for a native TRP channel.
Although the presence of functional protein in the yeast membrane had been demonstrated, it was possible that only a small fraction of the total expressed protein was functional, or that detergent extraction disrupted the native structure and function. To address this question, the purified TRPV1 was reconstituted by dialysis in the presence of phospholipids into phospholipid bilayers under conditions that yielded, on average, less than one tetramer per vesicle. This ratio can be determined accurately by measurement of protein-to-lipid ratios in the final vesicles, and by using electron microscopy to determine the vesicle size distribution. When the reconstitution was carried out in the presence of 5 mM Ca2+, and the external Ca2+ removed by chelate chromatography, the fraction of Ca2+ released by a TRPV1 agonist, resinaferatoxin, provided an estimated lower limit for the fraction of protein that was in functional form. The estimate is a lower limit because of the likelihood that more Ca2+ leaks out of the TRPV1-containing vesicles than out of the protein-free vesicles used as controls. This method provided a lower limit of 72% active protein, consistent with nearly all of the purified protein being in active form. This method does not distinguish among channels with cytoplasmic domains outside vs. inside the vesicles, due to the membrane permeability of the agonist. Future studies using antibodies, lectins, and/or cytoplasmic ligands such as calmodulin, will be needed to determine whether vesicle insertion happens with a preferential orientation, and to optimize conditions for achieving preferential orientation.
Other TRP channels have been expressed at high levels in S. cerevisiae and purified, using the same approaches as for TRPV1. These include TRPV2, TRPY1-4, TRPM8, and TRPA1 (unpublished data). They are all behaving well in this system, allowing purification of sufficient quantities of protein for functional and structural work. Thus the strategy of transient over-expression in budding yeast and epitope affinity chromatography in detergent appears to be one of general utility for members of the TRP family.
Cryo-EM structures of TRP channels
Single-particle EM can provide structural information for a large variety of biological molecules without the need to produce crystals (Chiu et al, 2005). Very little sample is required (Cheng & Walz, 2009). Single particle cryo-EM (Penczek et al, 1992) is a method in which the specimen, typically a protein embedded in vitreous ice, is held at cryogenic temperatures while images are obtained by the electron microscope (Wang & Sigworth, 2006). After completing the imaging, single-particle reconstruction methods are used to align and classify the individual particle images, and solve the structure of the protein (Frank et al, 1996; Hohn et al, 2007; Tang et al, 2007). Because the individual molecules (particles) are randomly oriented in the ice layer, the particle images can be classified into groups representing distinctive views of the original 3D particle. Particle images in each group can then be aligned with each other and a consensus shape determined. Once these averaged views are obtained, a “map” of density throughout the volume of the particle can be calculated to complete the reconstruction process.
Ion channels are excellent candidates for single-particle analysis work (Wang & Sigworth, 2006). Several channel structures have also been determined by cryo-EM, including a the voltage gated sodium channel (Sato et al, 2001), inositol triphosphate receptor (Sato et al, 2004; Serysheva et al, 2003), muscle L-type voltage gated calcium channel (dihydropyridine receptor) (Serysheva et al, 2002; Wolf et al, 2003), muscle calcium release channel/ryanodine receptor (RyR1) (Serysheva et al, 2005; Serysheva et al, 2008; Sharma et al, 2000; Wagenknecht & Samso, 2002), voltage-gated channel KvAP (Jiang et al, 2004), large-conductance calcium- and voltage-activated potassium channel (BK) in a lipid environment(Wang & Sigworth, 2009) and others.
In the last few years, several TRP channel structures have been studied by electron microscopy (Moiseenkova-Bell & Wensel, 2009). These include TRPV1(Moiseenkova-Bell et al, 2008), TRPV4 (Shigematsu et al, 2010), and TRPC3 (Mio et al, 2007), imaged in ice, and TRPC3 (Mio et al, 2005), and TRPM2 (Maruyama et al, 2007) imaged in negative stain.
The structure of TRPV1 using a cryo-EM approach was solved recently using preparation affinity purified from budding yeast and tested for tetrameric structure and ligand-gated ion flux as described above (Moiseenkova-Bell et al, 2008). Cryo-EM images and single particle reconstruction revealed that the structure is four-fold symmetric and consists of two well-defined domains (Figure 1). The more compact has the right dimensions to correspond to the membrane-spanning domain, likely composed of six transmembrane segments per subunit. This domain is 40 Å in the dimension thought to be normal to the membrane surface, and about 60 Å in diameter. This domain fits well with the high-resolution structure of the voltage-gated potassium channel Kv1.2 (Long et al, 2005a). The other domain, which contains the majority of the mass, as expected for the N- and C-terminal cytoplasmic domains, is an open, basket light structure connected by thin densities to the putative transmembrane domain. There is a region near these connecting densities, and therefore relatively near to the proposed membrane surface region, that fits well with the high-resolution structure of the ankyrin repeat domain of TRPV1 (Lishko et al, 2007).
Figure 1.
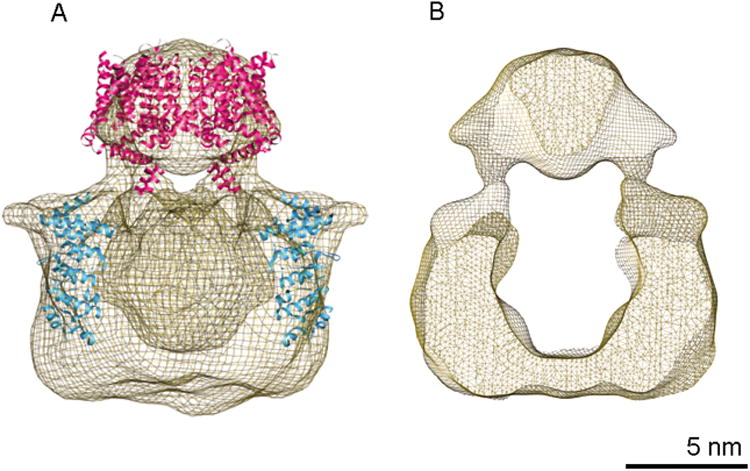
Structure of TRPV1 determined by electron cryo-microscopy and single particle analysis (Moiseenkova-Bell et al, 2008). On the left is an iso-dense surface in transparent mesh representation. Superimposed are ribbon diagrams of the x-ray structures of the transmembrane domain of Kv1.2 (Long et al, 2005a), 2A79 (magenta), and the ankyrin repeats of TRPV1 (Lishko et al, 2007), 2PNN (cyan). On the right is a similar representation cut through a center plane perpendicular to the proposed plane of the membrane, showing the large space within the basket-like cytoplasmic domain of TRPV1.
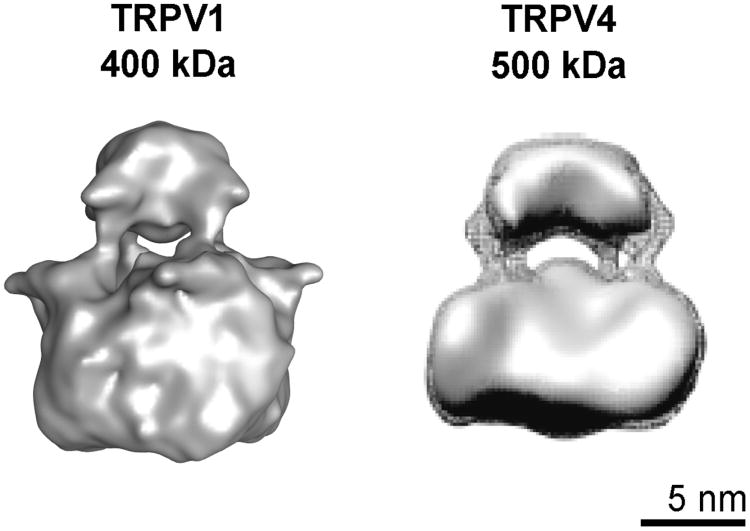
Another structure of the TRPV sub-family has been determined using electron microscopy in ice (Shigematsu et al, 2010). The structure of TRPV4 revealed considerable similarity to the TRPV1 structure, and contains two-distinct regions, likely corresponding to the transmembrane and cytoplasmic domains of the channel respectively (Figure 2). The results from these studies provided insight into structural organization of TRPV sub-family of ion channels and can be used as an initial testable structural template for studying full-length TRPV channels at higher resolution.
Figure 2.
Similarity of structures of TRPV1, left, (Moiseenkova-Bell et al, 2008) and TRPV4, right, (Shigematsu et al, 2010); (figure reproduced with permission) determined by electron cryo-microscopy.
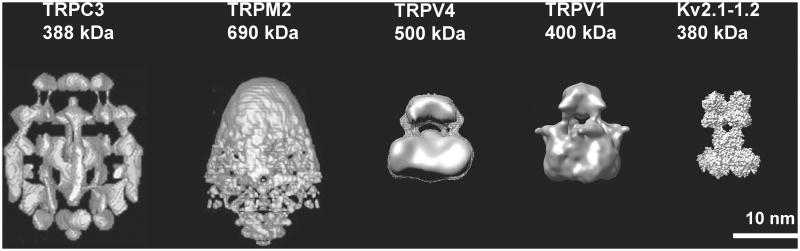
As described above, there is reason for some confidence in the published structures for the TRPV sub-family, given their self-consistency and the extensive characterization of the TRPV1 preparation. From the reported structures for the TRPC and TRPM sub-families, many questions remain (Moiseenkova-Bell & Wensel, 2009) (Figure 3). Structures for TRPC3 (Mio et al, 2005) and TRPM2 (Maruyama et al, 2007) determined in negative stain indicated a bullet-like shape for each, with the dense bullet-head region proposed to be the channel domain with its transmembrane segments, and a more open and larger domain proposed to be the cytoplasmic regions. There are some qualitative features of these structures reminiscent of the TRPV family structures, but the detailed structures are quite distinct. Limitations on the interpretation of these structures arise from the lack of functional characterization, concerns about the presence of lipid aggregates, and the inherent limitations and artifacts associated with negative stain. A very different structure of TRPC3 was reported by the same group, based on images collected in ice (Mio et al, 2007). The ice structure is lace-like and very open, with a very large overall volume. No regions of appropriate size and continuous density for a membrane-spanning domain are obvious.
Figure 3.
A comparison of reported channel structures. Images reproduced by permission from (Mio et al, 2007), TRPC3, (Maruyama et al, 2007), TRPM2, (Shigematsu et al, 2010), (Moiseenkova-Bell et al, 2008), TRPV1. The Kv2.1-1.2 chimera structure is from coordinates of 2R9R (Long et al, 2007), and reproduced by permission from (Moiseenkova-Bell & Wensel, 2009).
The major strength of single-particle reconstruction method is its ability to produce structural information about proteins that are especially challenging for X-ray crystallography. These proteins include ones for which high-yield expression systems are not yet available, as well as large multi-domain proteins, which are too flexible and too large (Frank, 2009), and membrane proteins for which suitable crystallization conditions have not yet been found. Cryo-EM allows proteins to be imaged in their native aqueous environment, and in a variety of functional states, without constrains of crystal contacts. In addition, EM data can be collected and structural information extracted as soon as purified protein is available, whereas for X-ray crystallography, considerable effort must be expended in finding suitable crystallization conditions. The major traditional disadvantage of single-particle electron microscopy has been the limitations on resolution. There have been improvements on this front in recent years, especially for large complexes and those with high symmetry, so that for the most favorable cases, near atomic resolution can be achieved, and some side chains can be visualized as well as accurate peptide backbone traces (Wolf et al, 2010; Zhang et al, 2010a; Zhang et al, 2010b). However, for smaller molecules, such as TRP channels, achieving atomic resolution by this method is not feasible with current methods, so that other methods such as homology modeling, mutagenesis and spectroscopy are needed to answer questions about positions of specific amino acid residues within the low resolution maps. Another limitation, which may come into play with some preparations of TRP channels, is that images of individual particles have low signal-to-noise, so it is possible to select objects other than the protein of interest, such as lipid aggregates or protein-free micelles, or to include degraded, aggregated or denatured forms of the protein in the data set. If care is not taken to avoid such “bad” particles, bizarre results can be obtained. More work will be needed, using more well-characterized TRP proteins and higher resolution cryo-EM data, to resolve the discrepancies among published TRP channel structures.
Divide and conquer approach: combining X-ray and EM data with computational modeling
Given recent advances in electron microscopy and image processing (Cheng & Walz, 2009; Chiu et al, 2005; Jiang & Ludtke, 2005), it is likely that structures of resolutions approaching 10 Å can be obtained for at least some TRP channels using electron cryo-microscopy. At this resolution, secondary elements, especially α-helices, can be identified to generate sequence-based models for understanding relationships between channel structure and function. However, in the absence of high-quality two-dimensional crystals, it is unlikely that atomic resolution structures will be obtained directly from electron microscopy, and such structures are needed to visualize such features as the conductance pore, selectivity filter, gate, and ligand-binding sites. While determining x-ray structures of full-length TRP channels remains a goal worth pursuing, success has come, and will likely continue to come, more quickly from crystallization of cytoplasmic domains of TRP channels. It should then be possible to fit these high resolution structures into lower resolution structures of complete channels determined by EM, to obtain high resolution models. For this purpose, in addition to x-ray (or perhaps NMR) structures of the domains of interest, homology models based on high resolution structures of homologous proteins (e.g. transmembrane domains of potassium channels) can also be very useful.
High resolution structures of fragments of TRP channels have included the α-kinase domain of TRPM7 (Yamaguchi et al, 2001), ankyrin repeats from TRPV subfamily of proteins (Jin et al, 2006; Lishko et al, 2007; McCleverty et al, 2006; Phelps et al, 2008; Phelps et al, 2010), the C-terminal cytoplasmic coiled-coil domain of TRPM7 (Fujiwara & Minor, 2008), and C-terminal cytoplasmic coiled-coil domain TRPP2 (Yu et al, 2009). As an example of combining these with EM structures, the ankyrin repeat domain of TRPV1 could be readily fit into density near the membrane surface in the cytoplasmic region of the TRPV1 structure determined by cryo-EM (Moiseenkova-Bell et al, 2008). As an example of using structures of homologous proteins for this purpose, the structure of Kv1.2, was readily fit into the transmembrane region of the same TRPV1 map.
Biochemical Studies with TRP channels purified from yeast
In addition to structural analysis, purified channels can be used to measure interactions with other proteins and regulatory small molecules without the confounds arising from studies in cell membranes containing many other proteins. For example, recently, several members of TRP family have been proposed to be regulated by intracellular Ca2+, and/or by calmodulin (CaM). Using calmodulin labeled with a fluorescent dye and measurements of emission anisotropy, it is possible to monitor calmodulin binding to TRPV1, TRPV2, and TRPA1 at nanomolar concentrations. Because the measurements can be carried out in real-time using T-format instrumentation, in which the light intensities for parallel and perpendicular components are detected concurrently using two independent detectors, both kinetics and thermodynamic parameters can be reliably measured (VYM and TGW, unpublished results).
Functional studies and genetic screens of TRP channels in yeast
Once it was found that channels of other species could be expressed in functional form in budding yeast (Moiseenkova et al, 2003) it became possible to use the power and ease of yeast genetic screens to study structure-function relationships in TRP channels. This approach was initially applied to the endogenous channel of S. cerevisiae, TRPY1 (YVC1) (Su et al, 2007; Zhou et al, 2007). Heterologous expression studies revealed that TRP channels from other fungal species are also functional in S. cerevisiae, so they can be studied conveniently in that system (Zhou et al, 2005).
TRPY1 is a large conductance (∼300 pS) channel expressed in vacuolar membranes in yeast. Gain-of-function mutagenesis analysis revealed that aromatic residues on the fifth (phenylalanine) and six (tyrosine) transmembrane segments of the channel control the gating of the TRPY1 channel (Su et al, 2007). Alignment of all the TRP channels revealed that the phenylalanine on the intracellular base of fifth transmembrane segment of the channel is conserved and may be part of a generic gating mechanism for TRP channels. A very similar approach was used to investigate the TRPV4 gaiting mechanism, and revealed not only the importance of the main intracellular gate as reported for TRPY1, but also a new voltage-dependent gaiting mechanism for TRPV4 (Loukin et al, 2010). Genetic screens for functional changes in mammalian TRP channels have been successfully carried out in S. cerevisiae as aids to understanding structure-function relationships (Myers et al, 2008). Results of this study, which used gain-or loss-of-function phenotypes, revealed that pore helix of TRPV1, TRPV2 and TRPV3 play an important role in gaiting mechanisms of these mammalian channels in addition to the intracellular gait (Myers et al, 2008).
In summary, the use of mutagenesis and the robust S. cerevisiae system to study functional and structure-function mechanisms of activation and gating for TRP channels will likely continue to gain popularity and produce very valuable results for understanding TRP channel biology.
Conclusion
As with any eukaryotic ion channels, studies of TRP channels at the molecular level remain challenging. However, the emergence of new tools, such as expression and genetic screens in S. cerevisiae, functional reconstitution protocols, cryo-electron microscopy, and x-ray crystallography of soluble fragments, will likely be combined in the next few years with the huge collection of data from functional studies in vertebrate cell membranes to provide a comprehensive picture of the structures and functional mechanisms of TRP channels.
References
- Audo I, Kohl S, Leroy BP, Munier FL, Guillonneau X, Mohand-Said S, Bujakowska K, Nandrot EF, Lorenz B, Preising M, Kellner U, Renner AB, Bernd A, Antonio A, Moskova-Doumanova V, Lancelot ME, Poloschek CM, Drumare I, Defoort-Dhellemmes S, Wissinger B, Leveillard T, Hamel CP, Schorderet DF, De Baere E, Berger W, Jacobson SG, Zrenner E, Sahel JA, Bhattacharya SS, Zeitz C. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2009;85:720–729. doi: 10.1016/j.ajhg.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer-Grumbach M, Olschewski A, Papic L, Kremer H, McEntagart ME, Uhrig S, Fischer C, Frohlich E, Balint Z, Tang B, Strohmaier H, Lochmuller H, Schlotter-Weigel B, Senderek J, Krebs A, Dick KJ, Petty R, Longman C, Anderson NE, Padberg GW, Schelhaas HJ, van Ravenswaaij-Arts CM, Pieber TR, Crosby AH, Guelly C. Alterations in the ankyrin domain of TRPV4 cause congenital distal SMA, scapuloperoneal SMA and HMSN2C. Nat Genet. 42:160–164. doi: 10.1038/ng.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondyk WH. Selecting an appropriate method for expressing a recombinant protein. Methods Enzymol. 2009;463:131–147. doi: 10.1016/S0076-6879(09)63011-1. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Walz T. The advent of near-atomic resolution in single-particle electron microscopy. Annu Rev Biochem. 2009;78:723–742. doi: 10.1146/annurev.biochem.78.070507.140543. [DOI] [PubMed] [Google Scholar]
- Chiu W, Baker ML, Jiang W, Dougherty M, Schmid MF. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure (Camb) 2005;13:363–372. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- Clapham DE, Montell C, Schultz G, Julius D. International Union of Pharmacology XLIII Compendium of voltage-gated ion channels: transient receptor potential channels. Pharmacol Rev. 2003;55:591–596. doi: 10.1124/pr.55.4.6. [DOI] [PubMed] [Google Scholar]
- Colsoul B, Nilius B, Vennekens R. On the putative role of transient receptor potential cation channels in asthma. Clin Exp Allergy. 2009;39:1456–1466. doi: 10.1111/j.1365-2222.2009.03315.x. [DOI] [PubMed] [Google Scholar]
- Cortright DN, Szallasi A. TRP channels and pain. Curr Pharm Des. 2009;15:1736–1749. doi: 10.2174/138161209788186308. [DOI] [PubMed] [Google Scholar]
- Cosens DJ, Manning A. Abnormal electroretinogram from a Drosophila mutant. Nature. 1969;224:285–287. doi: 10.1038/224285a0. [DOI] [PubMed] [Google Scholar]
- Deng HX, Klein CJ, Yan J, Shi Y, Wu Y, Fecto F, Yau HJ, Yang Y, Zhai H, Siddique N, Hedley-Whyte ET, Delong R, Martina M, Dyck PJ, Siddique T. Scapuloperoneal spinal muscular atrophy and CMT2C are allelic disorders caused by alterations in TRPV4. Nat Genet. 2010;42:165–169. doi: 10.1038/ng.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis V, Cyert MS. Internal Ca(2+) release in yeast is triggered by hypertonic shock and mediated by a TRP channel homologue. J Cell Biol. 2002;156:29–34. doi: 10.1083/jcb.200111004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figler RA, Omote H, Nakamoto RK, Al-Shawi MK. Use of chemical chaperones in the yeast Saccharomyces cerevisiae to enhance heterologous membrane protein expression: high-yield expression and purification of human P-glycoprotein. Arch Biochem Biophys. 2000;376:34–46. doi: 10.1006/abbi.2000.1712. [DOI] [PubMed] [Google Scholar]
- Frank J. Single-particle reconstruction of biological macromolecules in electron microscopy--30 years. Q Rev Biophys. 2009;42:139–158. doi: 10.1017/S0033583509990059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Minor DL., Jr X-ray crystal structure of a TRPM assembly domain reveals an antiparallel four-stranded coiled-coil. J Mol Biol. 2008;383:854–870. doi: 10.1016/j.jmb.2008.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher AR, Germino GG, Somlo S. Molecular advances in autosomal dominant polycystic kidney disease. Adv Chronic Kidney Dis. 17:118–130. doi: 10.1053/j.ackd.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R. A primer on ankyrin repeat function in TRP channels and beyond. Mol Biosyst. 2008;4:372–379. doi: 10.1039/b801481g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Hunte C, von Jagow G, Schagger H, editors. Membrane Protein Purification and Crystallization: A Practical Guide. San Diego: Academic Press; 2003. [Google Scholar]
- Inoue R, Jian Z, Kawarabayashi Y. Mechanosensitive TRP channels in cardiovascular pathophysiology. Pharmacol Ther. 2009;123:371–385. doi: 10.1016/j.pharmthera.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, Ijzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang QX, Wang DN, MacKinnon R. Electron microscopic analysis of KvAP voltage-dependent K+ channels in an open conformation. Nature. 2004;430:806–810. doi: 10.1038/nature02735. [DOI] [PubMed] [Google Scholar]
- Jiang W, Ludtke SJ. Electron cryomicroscopy of single particles at subnanometer resolution. Curr Opin Struct Biol. 2005;15:571–577. doi: 10.1016/j.sbi.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Jin X, Touhey J, Gaudet R. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J Biol Chem. 2006;281:25006–25010. doi: 10.1074/jbc.C600153200. [DOI] [PubMed] [Google Scholar]
- Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009;460:592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C, Obara T, Uriu Y, Numata T, Sanuki R, Miyata K, Koyasu T, Ueno S, Funabiki K, Tani A, Ueda H, Kondo M, Mori Y, Tachibana M, Furukawa T. TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc Natl Acad Sci U S A. 2010;107:332–337. doi: 10.1073/pnas.0912730107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korepanova A, Pereda-Lopez A, Solomon LR, Walter KA, Lake MR, Bianchi BR, McDonald HA, Neelands TR, Shen J, Matayoshi ED, Moreland RB, Chiu ML. Expression and purification of human TRPV1 in baculovirus-infected insect cells for structural studies. Protein Expr Purif. 2009;65:38–50. doi: 10.1016/j.pep.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Landoure G, Zdebik AA, Martinez TL, Burnett BG, Stanescu HC, Inada H, Shi Y, Taye AA, Kong L, Munns CH, Choo SS, Phelps CB, Paudel R, Houlden H, Ludlow CL, Caterina MJ, Gaudet R, Kleta R, Fischbeck KH, Sumner CJ. Mutations in TRPV4 cause Charcot-Marie-Tooth disease type 2C. Nat Genet. 2010;42:170–174. doi: 10.1038/ng.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Gu Q. Role of TRPV1 in inflammation-induced airway hypersensitivity. Curr Opin Pharmacol. 2009;9:243–249. doi: 10.1016/j.coph.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage PK, Lussier MP, McDuff FO, Lavigne P, Boulay G. The self-association of two N-terminal interaction domains plays an important role in the tetramerization of TRPC4. Cell Calcium. 2009;45:251–259. doi: 10.1016/j.ceca.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Li Z, Sergouniotis PI, Michaelides M, Mackay DS, Wright GA, Devery S, Moore AT, Holder GE, Robson AG, Webster AR. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am J Hum Genet. 2009;85:711–719. doi: 10.1016/j.ajhg.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko PV, Procko E, Jin X, Phelps CB, Gaudet R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron. 2007;54:905–918. doi: 10.1016/j.neuron.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005a;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- Long SB, Campbell EB, Mackinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005b;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- Loukin S, Su Z, Zhou X, Kung C. Forward-genetic analysis reveals multiple gating mechanisms of Trpv4. J Biol Chem. 2010 doi: 10.1074/jbc.M110.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y, Ogura T, Mio K, Kiyonaka S, Kato K, Mori Y, Sato C. Three-dimensional reconstruction using transmission electron microscopy reveals a swollen, bell-shaped structure of transient receptor potential melastatin type 2 cation channel. J Biol Chem. 2007;282:36961–36970. doi: 10.1074/jbc.M705694200. [DOI] [PubMed] [Google Scholar]
- McCleverty CJ, Koesema E, Patapoutian A, Lesley SA, Kreusch A. Crystal structure of the human TRPV2 channel ankyrin repeat domain. Protein Sci. 2006;15:2201–2206. doi: 10.1110/ps.062357206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke B, Wu C, Pak WL. Induction of photoreceptor voltage noise in the dark in Drosophila mutant. Nature. 1975;258:84–87. doi: 10.1038/258084a0. [DOI] [PubMed] [Google Scholar]
- Mio K, Ogura T, Hara Y, Mori Y, Sato C. The non-selective cation-permeable channel TRPC3 is a tetrahedron with a cap on the large cytoplasmic end. Biochem Biophys Res Commun. 2005;333:768–777. doi: 10.1016/j.bbrc.2005.05.181. [DOI] [PubMed] [Google Scholar]
- Mio K, Ogura T, Kiyonaka S, Hiroaki Y, Tanimura Y, Fujiyoshi Y, Mori Y, Sato C. The TRPC3 channel has a large internal chamber surrounded by signal sensing antennas. J Mol Biol. 2007;367:373–383. doi: 10.1016/j.jmb.2006.12.043. [DOI] [PubMed] [Google Scholar]
- Moiseenkova-Bell VY, Stanciu LA, Serysheva II, Tobe BJ, Wensel TG. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc Natl Acad Sci U S A. 2008;105:7451–7455. doi: 10.1073/pnas.0711835105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenkova-Bell VY, Wensel TG. Hot on the trail of TRP channel structure. J Gen Physiol. 2009;133:239–244. doi: 10.1085/jgp.200810123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenkova VY, Hellmich HL, Christensen BN. Overexpression and purification of the vanilloid receptor in yeast (Saccharomyces cerevisiae) Biochem Biophys Res Commun. 2003;310:196–201. doi: 10.1016/j.bbrc.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- Montell C. The TRP superfamily of cation channels. Sci STKE. 2005;2005:re3. doi: 10.1126/stke.2722005re3. [DOI] [PubMed] [Google Scholar]
- Montell C, Rubin GM. Molecular characterization of the Drosophila trp locus: a putative integral membrane protein required for phototransduction. Neuron. 1989;2:1313–1323. doi: 10.1016/0896-6273(89)90069-x. [DOI] [PubMed] [Google Scholar]
- Morgans CW, Zhang J, Jeffrey BG, Nelson SM, Burke NS, Duvoisin RM, Brown RL. TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proc Natl Acad Sci U S A. 2009;106:19174–19178. doi: 10.1073/pnas.0908711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers BR, Bohlen CJ, Julius D. A yeast genetic screen reveals a critical role for the pore helix domain in TRP channel gating. Neuron. 2008;58:362–373. doi: 10.1016/j.neuron.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B, Prenen J, Hoenderop JG, Vennekens R, Hoefs S, Weidema AF, Droogmans G, Bindels RJ. Fast and slow inactivation kinetics of the Ca2+ channels ECaC1 and ECaC2 (TRPV5 and TRPV6) Role of the intracellular loop located between transmembrane segments 2 and 3. J Biol Chem. 2002;277:30852–30858. doi: 10.1074/jbc.M202418200. [DOI] [PubMed] [Google Scholar]
- Nilius B, Prenen J, Janssens A, Owsianik G, Wang C, Zhu MX, Voets T. The selectivity filter of the cation channel TRPM4. J Biol Chem. 2005;280:22899–22906. doi: 10.1074/jbc.M501686200. [DOI] [PubMed] [Google Scholar]
- Nyblom M, Oberg F, Lindkvist-Petersson K, Hallgren K, Findlay H, Wikstrom J, Karlsson A, Hansson O, Booth PJ, Bill RM, Neutze R, Hedfalk K. Exceptional overproduction of a functional human membrane protein. Protein Expr Purif. 2007;56:110–120. doi: 10.1016/j.pep.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Oprian DD. Expression of opsin genes in COS cells. Methods Neuro. 1993;15:301–306. [Google Scholar]
- Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc Natl Acad Sci U S A. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Palmer CP, Zhou XL, Lin J, Loukin SH, Kung C, Saimi Y. A TRP homolog in Saccharomyces cerevisiae forms an intracellular Ca(2+)-permeable channel in the yeast vacuolar membrane. Proc Natl Acad Sci U S A. 2001;98:7801–7805. doi: 10.1073/pnas.141036198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcej DN, Eckhardt-Strelau L. Structural characterisation of neuronal voltage-sensitive K+ channels heterologously expressed in Pichia pastoris. J Mol Biol. 2003;333:103–116. doi: 10.1016/j.jmb.2003.07.009. [DOI] [PubMed] [Google Scholar]
- Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- Phelps CB, Huang RJ, Lishko PV, Wang RR, Gaudet R. Structural analyses of the ankyrin repeat domain of TRPV6 and related TRPV ion channels. Biochemistry. 2008;47:2476–2484. doi: 10.1021/bi702109w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps CB, Wang RR, Choo SS, Gaudet R. Differential regulation of TRPV1, TRPV3, and TRPV4 sensitivity through a conserved binding site on the ankyrin repeat domain. J Biol Chem. 2010;285:731–740. doi: 10.1074/jbc.M109.052548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Sato C, Hamada K, Ogura T, Miyazawa A, Iwasaki K, Hiroaki Y, Tani K, Terauchi A, Fujiyoshi Y, Mikoshiba K. Inositol 1,4,5-trisphosphate receptor contains multiple cavities and L-shaped ligand-binding domains. J Mol Biol. 2004;336:155–164. doi: 10.1016/j.jmb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Sato C, Ueno Y, Asai K, Takahashi K, Sato M, Engel A, Fujiyoshi Y. The voltage-sensitive sodium channel is a bell-shaped molecule with several cavities. Nature. 2001;409:1047–1051. doi: 10.1038/35059098. [DOI] [PubMed] [Google Scholar]
- Serysheva II, Bare DJ, Ludtke SJ, Kettlun CS, Chiu W, Mignery GA. Structure of the type 1 inositol 1,4,5-trisphosphate receptor revealed by electron cryomicroscopy. J Biol Chem. 2003;278:21319–21322. doi: 10.1074/jbc.C300148200. [DOI] [PubMed] [Google Scholar]
- Serysheva II, Hamilton SL, Chiu W, Ludtke SJ. Structure of Ca2+ release channel at 14 A resolution. J Mol Biol. 2005;345:427–431. doi: 10.1016/j.jmb.2004.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serysheva II, Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, Sali A, Hamilton SL, Chiu W. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc Natl Acad Sci U S A. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serysheva II, Ludtke SJ, Baker MR, Chiu W, Hamilton SL. Structure of the voltage-gated L-type Ca2+ channel by electron cryomicroscopy. Proc Natl Acad Sci U S A. 2002;99:10370–10375. doi: 10.1073/pnas.162363499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Jeyakumar LH, Fleischer S, Wagenknecht T. Three-dimensional structure of ryanodine receptor isoform three in two conformational states as visualized by cryo-electron microscopy. J Biol Chem. 2000;275:9485–9491. doi: 10.1074/jbc.275.13.9485. [DOI] [PubMed] [Google Scholar]
- Shen Y, Heimel JA, Kamermans M, Peachey NS, Gregg RG, Nawy S. A transient receptor potential-like channel mediates synaptic transmission in rod bipolar cells. J Neurosci. 2009;29:6088–6093. doi: 10.1523/JNEUROSCI.0132-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H, Sokabe T, Danev R, Tominaga M, Nagayama K. A 3.5-nm structure of rat TRPV4 cation channel revealed by Zernike phase-contrast cryoelectron microscopy. J Biol Chem. 2010;285:11210–11218. doi: 10.1074/jbc.M109.090712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JB, Adams D, Paukert M, Siba M, Sidi S, Levin M, Gillespie PG, Grunder S. Xenopus TRPN1 (NOMPC) localizes to microtubule-based cilia in epithelial cells, including inner-ear hair cells. Proc Natl Acad Sci U S A. 2005;102:12572–12577. doi: 10.1073/pnas.0502403102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- Sidi S, Friedrich RW, Nicolson T. NompC TRP channel required for vertebrate sensory hair cell mechanotransduction. Science. 2003;301:96–99. doi: 10.1126/science.1084370. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GF. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhou X, Haynes WJ, Loukin SH, Anishkin A, Saimi Y, Kung C. Yeast gain-of-function mutations reveal structure-function relationships conserved among different subfamilies of transient receptor potential channels. Proc Natl Acad Sci U S A. 2007;104:19607–19612. doi: 10.1073/pnas.0708584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tao X, Avalos JL, Chen J, MacKinnon R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science. 2009;326:1668–1674. doi: 10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Mackinnon R. Functional Analysis of Kv1.2 and Paddle Chimera Kv Channels in Planar Lipid Bilayers. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Genderen MM, Bijveld MM, Claassen YB, Florijn RJ, Pearring JN, Meire FM, McCall MA, Riemslag FC, Gregg RG, Bergen AA, Kamermans M. Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet. 2009;85:730–736. doi: 10.1016/j.ajhg.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam K, Montell C. TRP channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenknecht T, Samso M. Three-dimensional reconstruction of ryanodine receptors. Front Biosci. 2002;7:d1464–1474. doi: 10.2741/A853. [DOI] [PubMed] [Google Scholar]
- Walker RG, Willingham AT, Zuker CS. A Drosophila mechanosensory transduction channel. Science. 2000;287:2229–2234. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- Wang L, Sigworth FJ. Cryo-EM and single particles. Physiology (Bethesda) 2006;21:13–18. doi: 10.1152/physiol.00045.2005. [DOI] [PubMed] [Google Scholar]
- Wang L, Sigworth FJ. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature. 2009;461:292–295. doi: 10.1038/nature08291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Murakami M, Ohba T, Ono K, Ito H. The pathological role of transient receptor potential channels in heart disease. Circ J. 2009;73:419–427. doi: 10.1253/circj.cj-08-1153. [DOI] [PubMed] [Google Scholar]
- White JP, Cibelli M, Rei Fidalgo A, Paule CC, Noormohamed F, Urban L, Maze M, Nagy I. Role of transient receptor potential and acid-sensing ion channels in peripheral inflammatory pain. Anesthesiology. 2010;112:729–741. doi: 10.1097/ALN.0b013e3181ca3179. [DOI] [PubMed] [Google Scholar]
- Wolf M, Eberhart A, Glossmann H, Striessnig J, Grigorieff N. Visualization of the domain structure of an L-type Ca2+ channel using electron cryo-microscopy. J Mol Biol. 2003;332:171–182. doi: 10.1016/s0022-2836(03)00899-4. [DOI] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg-Vrenken TE, Bindels RJ, Hoenderop JG. The role of transient receptor potential channels in kidney disease. Nat Rev Nephrol. 2009;5:441–449. doi: 10.1038/nrneph.2009.100. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Matsushita M, Nairn AC, Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol Cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- Yu Y, Ulbrich MH, Li MH, Buraei Z, Chen XZ, Ong AC, Tong L, Isacoff EY, Yang J. Structural and molecular basis of the assembly of the TRPP2/PKD1 complex. Proc Natl Acad Sci U S A. 2009;106:11558–11563. doi: 10.1073/pnas.0903684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Baker ML, Schroder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, Frydman J, Chiu W. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010a;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010b;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Su Z, Anishkin A, Haynes WJ, Friske EM, Loukin SH, Kung C, Saimi Y. Yeast screens show aromatic residues at the end of the sixth helix anchor transient receptor potential channel gate. Proc Natl Acad Sci U S A. 2007;104:15555–15559. doi: 10.1073/pnas.0704039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XL, Loukin SH, Coria R, Kung C, Saimi Y. Heterologously expressed fungal transient receptor potential channels retain mechanosensitivity in vitro and osmotic response in vivo. Eur Biophys J. 2005;34:413–422. doi: 10.1007/s00249-005-0465-0. [DOI] [PubMed] [Google Scholar]