Abstract
The DNA-binding protein TRF2 is essential for telomere protection and chromosome stability in mammals. We show here that TRF2 expression is activated by the Wnt/β-catenin signalling pathway in human cancer and normal cells as well as in mouse intestinal tissues. Furthermore, β-catenin binds to TRF2 gene regulatory regions that are functional in a luciferase transactivating assay. Reduced β-catenin expression in cancer cells triggers a marked increase in telomere dysfunction, which can be reversed by TRF2 overexpression. We conclude that the Wnt/β-catenin signalling pathway maintains a level of TRF2 critical for telomere protection. This is expected to have an important role during development, adult stem cell function and oncogenesis.
Keywords: telomere, TRF2, Wnt signalling, cancer
INTRODUCTION
Telomeres are of paramount importance in determining cell fate, as they link replicative history to a great diversity of functions regulating tissue homeostasis. If the telomere length maintenance depends on telomerase, which can compensate for replicative erosion [1, 2], telomere capping relies on the organization of chromatin in a way that protects the chromosome ends from aberrant signallization and repair [3, 4].
The shelterin complex is a key protein component of telomeric chromatin in mammals and consists of six polypeptides: TRF1, TRF2, RAP1, TIN2, TPP1 and POT1 [5]. This complex appears to act as a protein hub that protects telomeres from being recognized as a double-strand break, thereby avoiding inappropriate DNA damage response activation and aberrant recombinational repair. Among the shelterin components, TRF2 has a crucial role in regulating the molecular events that maintain telomere integrity [3]. Suppression of TRF2 activates an ATM-dependent DNA damage response pathway that induces apoptosis or senescence, depending on the cell type [6].
Expression of telomerase is tightly regulated during normal development and aging, and telomerase is upregulated in the vast majority of human cancers [7]. However, very little is known on the regulation of the shelterin components expression. The current view is that shelterin is ubiquitously expressed to protect telomeres in all tissues in which the telomeres are not too short to preclude DNA binding [5, 8]. Recent studies challenge this prevailing paradigm by showing that the expression of shelterin subunits, notably TRF2, is increased during malignant transformation, suggesting that shelterin expression might be the target of oncogenic signalling pathways [9,10,11,12,13,14].
RESULTS AND DISCUSSION
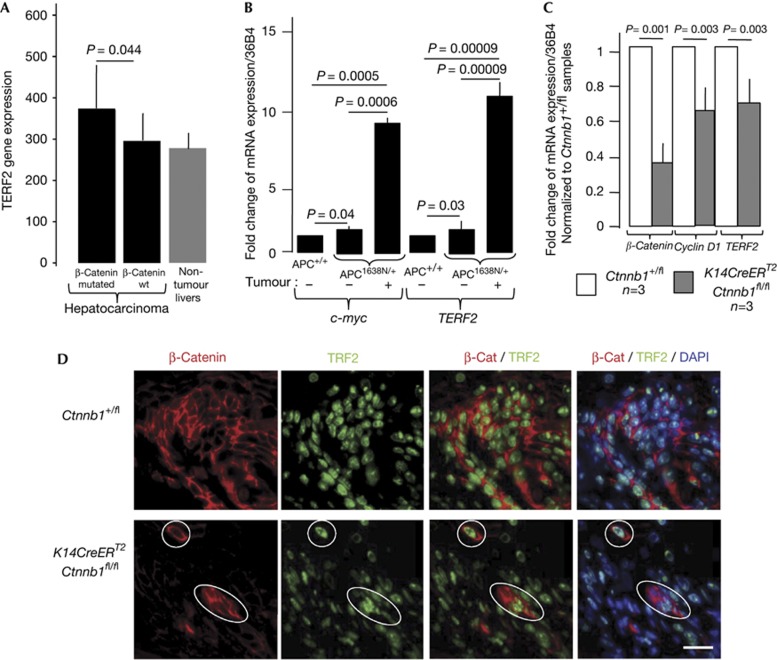
To determine whether TERF2 mRNA expression correlates with particular features of hepatocellular carcinomas (HCCs), we analysed an expression profile done using an Affimetrix microarray on 56 human HCCs with a known mutational status of the Ctnnb1 gene encoding β-catenin (Fig 1A). An increase of TERF2 gene expression, but not of the other shelterin genes, was noticed in HCC with Ctnnb1 mutation when compared to HCC with intact Ctnnb1 gene (P<0.05) or to a set of five non-tumour liver samples (Fig 1A).
Figure 1.
Upregulation of TERF2 mRNA when activation of the canonical Wnt/β-catenin signalling pathway is more pronounced. (A) TERF2 mRNA expression in 56 hepatocellular carcinomas with known β-catenin mutations. P-value determined according to a Mann–Whitney U-test. Error bar represents mean±s.e.m. (B) Relative TERF2 mRNA levels in normal intestine and adenocarcinoma samples from APC+/+ and APC1638N/+ mice (n=6 for each type of mouse). The values are normalized to tissues of APC+/+ mice. The P-value was determined using Bonferroni multiple comparison test. Error bar represents mean±s.e.m. for six independent experiments. (C) Reverse transcription–quantitative PCR analysis of the expression of Ctnnb1, c-Myc and TERF2 genes in skin samples from the indicated newborn mice treated topically with Tamoxifen between days 0.5 and 1.5 and euthanized at day 3. The P-value was determined using Wilcoxon matched-pairs signed rank test. Error bar represents mean±s.e.m. (D) Representative images of immunofluorescence analysis using antibodies directed against TRF2 or β-catenin of skin samples of the indicated mice treated with Tamoxifen. The white circles show examples of cells that still express a high level of β-catenin and of TRF2. The bar represents 15 μm. DAPI, 4′-6-diamidino-2-phenylindole.
We next investigated whether TERF2 gene expression is upregulated in APC1638N/+ mice, a model of familial adenomatous polyposis, which is caused by a germline mutation in the APC gene, whose product targets β-catenin for degradation. We found a modest but significant increase in c-Myc mRNA level, a direct target of β-catenin, in the normal intestine mucosa of APC1638N/+ mice, and a marked overexpression of c-Myc in APC1638N/+ tumours (Fig 1B). Notably, TERF2 mRNA mirrored c-Myc expression pattern. These results are in agreement with the overexpression of TRF2 observed in colorectal carcinoma [15] (J.Y. Scoazec and E.G., unpublished observations).
Next, we assessed whether the Wnt/β-catenin signalling pathway regulates the expression of TERF2 in vivo by the means of a conditional knockout mouse model that allows acute deletion of β-catenin encoding gene, Ctnnb1. The skin of K14CreERT2;Ctnnb1fl/fl newborn mice were topically treated with Tamoxifen at postnatal days 0.5 and 1.5 and killed at day 3. By this time, as expected, the expression of Ctnnb1 and Cyclin D1, a target of the Wnt/β-catenin signalling pathway, were decreased in skin samples from K14CreERT2;Ctnnb1fl/fl mice when compared with similarly treated samples from Ctnnb1+/fl control mice (Fig 1C,D). Concomitantly, TERF2 expression decreased, both at the mRNA and protein levels (Fig 1C,D). It is worth noting that the level of TRF2 remains high in the few keratinocytes from K14CreERT2;Ctnnb1fl/fl Tamoxifen-treated mice that were still expressing β-catenin after Tamoxifen treatment (encircled in Fig 1D), ruling out a problem of TRF2 immunostaining. Together, those results show that β-catenin regulates TRF2 expression in both human and mouse cells.
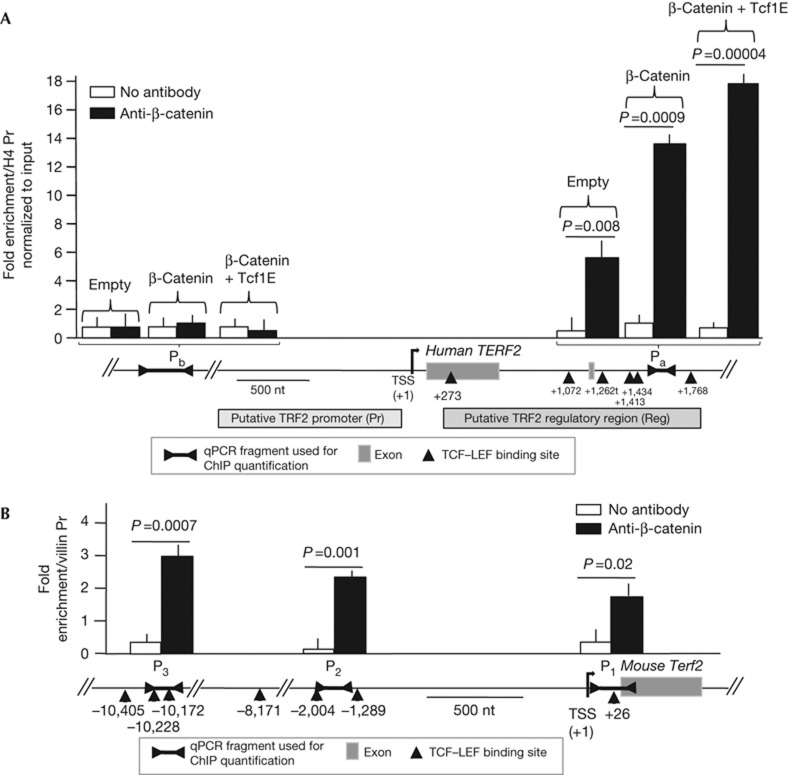
Analysis of the human TERF2 gene using the Ensembl genome database (http://www.ensembl.org) revealed a potential regulatory region located downstream of the transcription start site (named Reg in Fig 2A). Activation of the canonical Wnt signalling pathway triggers the stabilization and translocation of β-catenin to the nucleus that subsequently binds to members of the TCF–LEF family of transcription factors to activate gene transcription [16]. Notably, the TERF2 Reg sequence contains six putative TCF–LEF transcription factor binding sites between position +273 and+1768 (Fig 2A). Three clusters of TCF–LEF binding sites were also found in the promoter region of the mouse Terf2 gene (Fig 2B). Chromatin immunoprecipitation (ChIP) experiments with antibodies raised against β-catenin in colorectal cancer cells with a mutant Ctnnb1 gene (HCT116) revealed an enrichment of a DNA fragment localized inside the TCF–LEF site cluster (Pa) relative to a fragment of the histone H4 promoter (H4 Pr), which does not contain TCF–LEF sites (Fig 2A; Supplementary Table S1 online). Importantly, this enrichment was further increased on β-catenin and Tcf1E overexpression and not observed with a DNA fragment located roughly 18-kb upstream of the Reg sequence (Pb in Fig 2A). Similar ChIP experiments performed on normal intestinal tissue from APC1638N/+ mice revealed an enrichment of DNA sequences corresponding to the three clusters of TCF–LEF binding sites of the mouse Terf2 gene relative to a DNA sequence of the Villin promoter, which does not contain TCF–LEF sites (P1, P2 and P3 in Fig. 2B).
Figure 2.
β-Catenin binds to TERF2 regulatory regions containing TCF–LEF sites (marked as triangles). The coordinates of the centres of the DNA sequences of these sites are given relative to the transcription start site (+1). Each quantitative PCR (qPCR) value represents the average of at least three independent chromatin immunoprecipitation (ChIP) experiments. P-value was determined using Mann–Whitney test. (A) Genomic map of the human TERF2 gene and quantification by qPCR of the enrichment of the Pa and Pb DNA fragments relative to a sequence of the histone H4 promoter (H4 Pr). Empty, β-catenin and Tcf1E indicate that the HCT116 cells were transfected with an empty plasmid or with a plasmid expressing a mutated form of β-catenin or with a plasmid expressing Tcf1E. (B) Genomic map of the mouse TERF2 gene and quantification of the enrichment of the P1, P2 and P3 DNA fragments relative to a sequence of the Villin promoter in immunoprecipitates from ChIP experiments of normal intestinal tissues from APC1638N/+ mice using a β-catenin antibody. For both panels, error bar represents the mean±s.e.m. for four independent experiments.
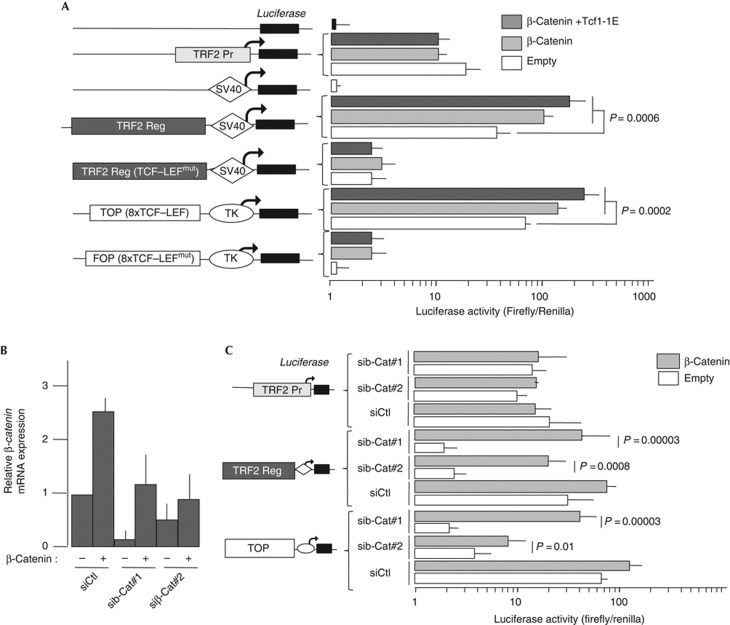
The Reg sequence boosts the transactivating activity of the weak SV40 promoter in a luciferase assay. Importantly, mutations of the TCF–LEF binding sites abrogated the ability of the Reg region to drive transcription of the reporter gene suggesting that those TCF–LEF binding sites mediated the responsiveness of TERF2 expression to Wnt/β-catenin signalling (Fig 3A). Moreover, overexpression of β-catenin alone and overexpression of both β-catenin and Tcf1E together induced a specific activation of the native Reg region (Fig 3A). This is in contrast to the basal activity conferred by the TERF2 Pr promoter fragment (Fig 3A). Finally, the ability of Reg region to induce firefly expression was inhibited by two independent short interfering RNA (siRNA) sequences, which efficiently target Ctnnb1 mRNA (Fig 3B,C). Importantly, the effect of these siRNA on the ability of the Reg region to drive transcription was rescued by β-catenin overexpression (Fig 3C). The use of both a Wnt-positive reporter containing eight known TCF–LEF sites (TOP) and a Wnt-negative reporter (FOP) validated these assays (Fig 3A,C). The ability of the Reg element to respond to β-catenin was further confirmed by using LiCl treatment, a component that is known to stabilize β-catenin [17] (Supplementary Fig S1 online).
Figure 3.
The TCF–LEF sites of the human TERF2 gene are capable of transactivating a luciferase reporter gene in a β-catenin-dependent manner. (A) The various reporter constructs derived from the pLG-3/Prom plasmid (for the TERF2 Reg sequence) and pLG-3/basic (for the TERF2 Prom sequence). TCF–LEFmut: all the TCF–LEF sites are inactivated by mutation. TOP-Flash and FOP-Flash are positive and negative controls, respectively, for β-catenin/TCF transactivation. Data show average relative luciferase activity in HCT116 cells and represent the mean±s.d. (n=6). Co-transfected plasmids overexpressing activated β-catenin and Tcf1E are identified by black/white scales. (B) Efficiency of inhibition of Ctnnb1 mRNA expression on transfection of two different short interfering RNAs (siRNAs) directed against Ctnn1b mRNA (#1 and #2). SiCtl is a scramble control siRNA. (C) HCT116 cells transfected with various siRNAs as in A. Statistical significance of data was assessed using Bonferroni multiple comparison test. For all panels, error bar represents the mean±s.e.m. for six independent experiments.
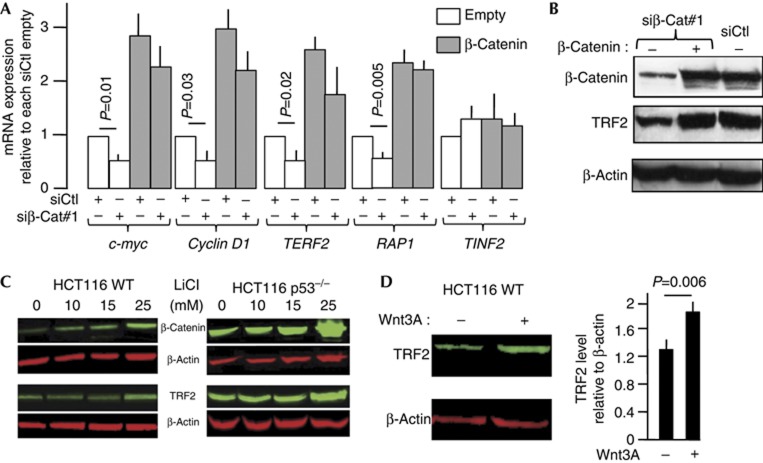
We next investigated whether the Wnt/β-catenin signalling pathway controls expression of the endogenous TERF2 gene. In HCT116 cells, which show constitutive β-catenin-mediated transcriptional activity, the knockdown of Ctnnb1 led to a nearly twofold decrease in the TERF2 mRNA level (Fig 4A). A similar decrease of expression was observed for the known β-catenin targets c-Myc and cyclin D1 (Fig 4A). Conversely, β-catenin overexpression increased c-Myc, cyclin D1 and TERF2 mRNA levels and reversed the downregulation caused by siRNA transfection (Fig 4A). The β-catenin level also determined the expression of the RAP1 gene encoding a TRF2-interacting protein, but did not influence expression of the TINF2 gene, which encodes another shelterin component. The ability of β-catenin to regulate TERF2 and RAP1 expressions appears to be a general phenomenon, as very similar results were obtained using HepG2 cells harbouring a Ctnnb1 mutation (Supplementary Fig S2 online). The fact that no RAP1 overexpression was found associated with mutated Ctnnb1 in the HCCs (Fig 1A) might be due to a threshold effect of microarray data or to cell type-specific differences in RAP1 regulation.
Figure 4.
β-Catenin regulates the expression of TRF2. (A) mRNA expression of β-catenin, two known targets of the canonical Wnt signalling (cyclin D1 and c-Myc), TERF2, RAP1 and TINF2. The P-value was determined using Wilcoxon matched-pairs signed rank test. (B) Western blotting of lysates from HCT116 cells under the indicated conditions. (C) Image and quantitative analysis of β-catenin, TRF2 and β-actin expression in HCT116 and HCT116 p53−/− cells treated with increasing amounts of LiCl. (D) Image and quantitative analysis of TRF2 expression in HCT116 cells treated with the Wnt3A peptide. For all panels, error bar represents the mean±s.e.m. for three independent experiments.
The ability of β-catenin to control endogenous TRF2 expression was confirmed at the protein level, HCT116 cells transfected with an Ctnnb1 siRNA showing a marked decrease of TRF2 protein by western blot (Fig 4B). This effect was rescued by overexpression of β-catenin (Fig 4B). The impact of β-catenin on TRF2 expression in HCT116 cells was further supported by the progressive accumulation of both β-catenin and TRF2 in HCT116 cells treated with increasing amounts of LiCl (Fig 4C). It was recently shown a p53-dependent degradation of TRF2 by Siah1 [18]. Indeed, we detected larger amounts of TRF2 in HCT116 p53–/– cells compared with HCT116 cells, with the amount of TRF2 being further upregulated by LiCl (Fig 4C).
In agreement with the view that TRF2 is a target of the Wnt canonical pathway, addition of purified Wnt3A to the culture medium stimulated the expression of TRF2 in HCT116 cells (Fig 4D), in a variety of hepatoma cell lines, both mutated and non-mutated for Ctnnb1, and in IMR90 cells, a normal fibroblast line (Supplementary Fig S3 online).
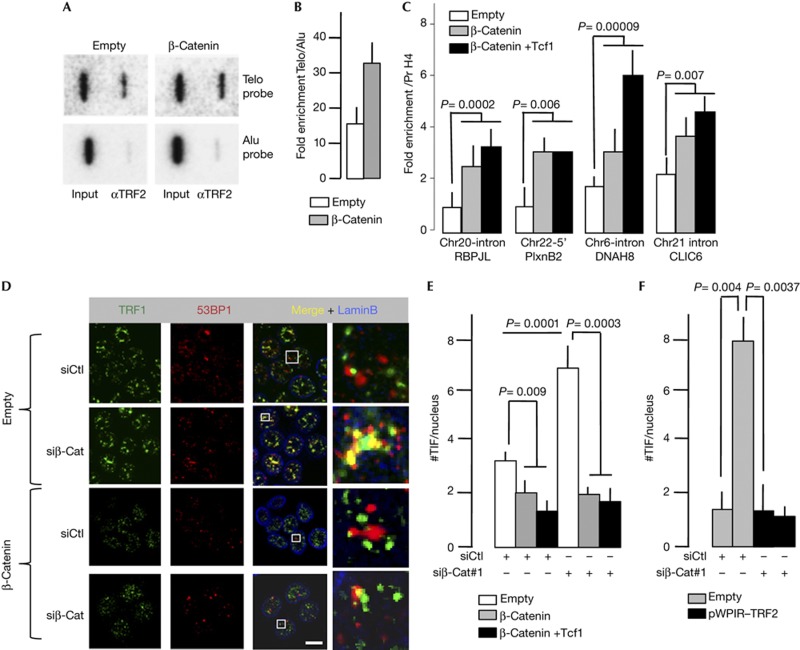
The increase in the level of TRF2 due to β-catenin/Tcf1E overexpression led to an increase in its binding to telomeres, as revealed by slot blotting with a telomeric DNA probe (Fig 5A,B) and several interstitial telomeric sequences [19], as revealed by quantitative PCR (qPCR; Fig 5C). These results indicate that β-catenin regulates telomere protection by modulating the TRF2 level. We therefore tested whether changes in the expression of β-catenin could affect telomere protection by scoring the number of telomere dysfunction-induced foci (TIF), that is, the number of telomere foci (detected by TRF1 antibodies) colocalizing with 53BP1 immuno-signals (Fig 5D). While the overexpression of β-catenin or β-catenin and Tcf1E together reduced both the mean number of TIF per nucleus (Fig 5D,E), Ctnnb1 knockdown caused a marked increase in the TIF number in the control condition (empty). Ruling out an off-target effect, overexpression of β-catenin or β-catenin plus Tcf1E reverse the effect of Ctnnb1 knockdown (Fig 5D,E). The increase of TIF triggered by the reduction of Ctnnb1 expression is accompanied by a decrease in cell viability and an increased percentage of cell-expressing β-galactosidase, a marker of cellular senescence (Supplementary Fig S4 online). These results indicate that β-catenin is required for protection against telomere dysfunction and senescence in HCT116 colon cancer cells.
Figure 5.
Telomere changes caused by modulation in β-catenin levels. Images (A) and quantification (B) of chromatin immunoprecipitation (ChIP) analysis of HCT116 cells overexpressing β-catenin and Tcf1E (n=3), performed using TRF2 antibodies and revealed by slot blotting with a telomeric DNA probe. Error bar represents the mean±s.e.m. (C) The ChIP-immunoprecipitated DNA from A and B was subjected to PCR with primers specific for different interstitial telomeric sequences (ITSs). The name of the ITS-proximal gene is shown. P-value was determined using Mann–Whitney test. Error bar represents the mean±s.e.m. for six independent experiments. (D) Representative images from immunofluorescence experiments used to analyse TIF. (E) Number of TIF in HCT116 cells on Ctnnb1 knockdown. At least 40 nuclei per condition were analysed. P-value was determined using Student’s t-test. (F) Number of TIF in HCT116 cells combining Ctnnb1 knockdown and TRF2 overexpression. At least 40 nuclei per condition were analysed. P-value was determined using Student’s t-test. For panels E and F, error bar represents the mean±s.e.m. for three independent experiments. H4 Pr, histone H4 promoter.
On the basis of our results showing that TRF2 expression is regulated by β-catenin (Figs 2), we tested whether the mechanism by which β-catenin protects telomeres involves TRF2. The overexpression of TRF2 in β-catenin-compromised HCT116 cells triggers the appearance of significantly less TIFs than in cells transduced with the empty vector (Fig 5F). This shows that an overdosage of TRF2 rescues the telomere dysfunction triggered by Ctnnb1 knockdown. We conclude that β-catenin maintains a level of TRF2 critical for telomere protection in HCT116 cells.
Overall, our findings show that in normal and cancer cells, both from mouse and human, the activation of Wnt/β-catenin signalling leads to an increased TRF2 level as well as an enhanced telomere protection. Previously, TERT was shown to enhance the transcriptional output of the Wnt pathway [20, 21]. Therefore, it would be interesting to determine whether TERT cooperates with β-catenin to induce TRF2 overexpression during tumour progression. Along this line, three groups recently showed that the expression of the protein component of telomerase TERT is also regulated by the Wnt/β-catenin signalling pathway [22,23,24]. Altogether, these findings indicate that an activation of the Wnt signalling pathway can reinforce telomere stability by coupling and enhancing the two main telomere maintenance pathways that are the telomerase activity and the shelterin capping complex. This Wnt-mediated telomere protective effect is expected to have an important role during development, adult stem cell function and oncogenesis.
In conclusion, TRF2 should not merely be viewed as a ‘housekeeping’ protein required for telomere protection, but as a regulated factor that couples the functional state of telomeres to the long-range organization of chromosomes and gene regulation networks.
METHODS
Patients, tumour samples and RNA extraction procedure. The tumour specimens were collected from four different surgical centres (Paris, Milan, Rennes and Shanghaï). Adjacent non-neoplastic liver tissues were obtained at the time of surgery. After resection, tumour specimens were immediately frozen in liquid nitrogen and continuously stored at −80 °C until processed for array profiling. Tissue specimens were obtained retrospectively under strict anonymity from historical collections established in surgery or pathology units of the participating hospitals. This study, respectful of the declaration of Helsinki, was approved by Institutional Human Research review boards. Only samples with a ribosomal RNA ratio 28S/18S >1.8 were included in the analysis.
Gene expression arrays. Microarray analysis was carried out using Affymetrix U133A GeneChips according to the manufacturer’s instructions. Biotin-labelled cRNA, produced by in vitro transcription, was fragmented and hybridized to the Affymetrix U133A GeneChip arrays (www.affymetrix.com_products_arrays_specific_Hu133A.affx) at 45 °C for 16 h and then washed and stained using the GeneChip Fluidics. The arrays were scanned by a GeneArray Scanner and patterns of hybridization detected as light emitted from the fluorescent reporter groups incorporated into the target and hybridized to oligonucleotide probes. All analyses were performed in a MIAME (minimal information about a microarray experiment)-compliant fashion, as defined in the guidelines established by MGED (www.mged.org).
Cell treatment and protein analysis. For knockdown studies, the cells were transfected with two validated human β-catenin (Ctnnb1) siRNAs (named siβ-Cat#1 and siβ-Cat#2) or negative control siRNA (named siCtl; Ambion). We used as expression vectors pCI-NEO-β-CATENINXL for the S33Y mutant β-catenin construct and Evr2-Tcf1E for Tcf1E. Telomere dysfunction-induced foci assay by immunofluorescence was performed as previously published [25]. Primary antibodies against TRF2 (Imgenex), β-catenin (BD Transduction Laboratories) and β-actin (Abcam) were used. For ECL detection, horseradish peroxidase-conjugated secondary antibodies were used; for fluorescence western blotting, we used DyLight-conjugated secondary antibodies (Thermo Scientific) and an Odyssey Infrared Imaging system (LI-COR).
Generation of conditional knockout mice, skin sample collection and analysis. Ctnnb1fl/+ strain was described previously [26]. For acute deletion of β-catenin, K14CreERT2 mice were intercrossed with Ctnnb1fl/fl alleles. Entire litters of newborn pups derived from Ctnnb1fl/+ or Ctnnb1fl/fl females crossed with K14CreERT2;Ctnnb1fl/+ or K14CreERT2;Ctnnb1fl/fl males were topically treated with 40 μl of saturated (200 mg/ml) Tamoxifen solution (Sigma) dissolved in 9:1 dimethylsulphoxide/ethanol at postnatal days 0.5 and 1.5. All mice were treated in accordance with AAALAC approved guidelines at Stanford University. Dorsal skin samples were obtained from mice post mortem.
Immunohistochemistry. Two-μm paraffin sections were used for immunohistological investigations. An indirect immunofluorescence double-labelling technique was used to mark TRF2 (antibodies orf2 [8]) and β-catenin (antibodies cat.# 610154, BD Transduction Laboratories, Le-Pont-de-Claix, France) expressing cells. The reaction products were visualized by incubation (1.5 h at room temperature) with Cy3- and Cy2-conjugated secondary antibodies. Counterstaining of the nuclei was done with 4′-6-diamidino-2-phenylindole.
Luciferase transcription reporter assay. The TERF2 gene fragments cloned in the luciferase plasmids have the following coordinate (human genome hg19 release): Pr=69,420,372–69,419,875 and Reg=69,419,615–69,417,951
To normalize transfection efficiency in reporter assays, the HCT116 cells were co-transfected with 0.1–0.2 μg of a plasmid carrying the internal control reporter Renilla reniformis luciferase driven by a TK promoter (pRL-TK; Promega). Then 72 h after transfection, a luciferase assay was performed using a Dual Luciferase Assay System kit (Promega) in accordance with the manufacturer’s protocols. Statistical significance was determined using Student’s t-test. P<0.05 was considered as significant.
Chromatin immunoprecipitation assay. Chromatin immunoprecipitation was performed as previously published [19]. We used the following antibodies: TRF2m (Imgenex 124A; mouse monoclonal) and β-catenin (BD Transduction Laboratories; mouse monoclonal). After DNA precipitation, purified samples for β-catenin ChIP were used for qPCR. TRF2 ChIP was analysed by slot blotting with telomere versus Alu probes for telomeric localization and by qPCR for interstitial telomeric sequence enrichment [19]. The primers used for qPCR are given in Supplementary Table S2 online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported by grants from the Association de la Recherche contre le Cancer (ARC), L’Institut National du Cancer (TELOFUN program), Agence Nationale de la Recherche (ANR) (TELOREP and INNATELO programs), the Ligue Nationale Contre le Cancer (the teams of E.G. and A.D. are ‘équipes labellisées’) and the European Community (TELOMARKER Health-F2-2007-200950).
Author contributions: I.D. designed and performed most of the experiments; N.W. designed, performed and analysed the mouse skin microscopy; M.S. and S.A. conceived the mouse skin experiments; J.C. performed the mouse skin experiments; M.S. performed the APC mouse experiments; M.P. conceived and analysed APC mouse experiments; S.B., J.Y. and A.Dj contributed to TIF analysis; C.S.-B., T.S., J.Y. and F.M. contributed to ChIP experiments; V.M.R. designed molecular and cellular experiments; P.P. and A.D. conceived, performed and analysed the microarray experiments; and E.G. conceived, coordinated and analysed most of the experiments, as well as wrote the manuscript with the input from the co-authors. M.Si and M.P. conducted the experiments with AMP mice. M.P. contributed to the design of the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Blackburn EH, Greider CW, Szostak JW (2006) Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 12: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Cech TR (2004) Beginning to understand the end of the chromosome. Cell 116: 273–279 [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Pisano S, Poulet A, Le Du MH, Gilson E (2010) Structural identity of telomeric complexes. FEBS Lett 584: 3785–3799 [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Blackburn EH (2000) Telomere states and cell fates. Nature 408: 53–56 [DOI] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat Genet 17: 236–239 [DOI] [PubMed] [Google Scholar]
- Diehl MC, Idowu MO, Kimmelshue KN, York TP, Jackson-Cook CK, Turner KC, Holt SE, Elmore LW (2011) Elevated TRF2 in advanced breast cancers with short telomeres. Breast Cancer Res Treat 127: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Wang L, Chen X, Sun P, Wu Y (2009b) Upregulation and CpG Island hypomethylation of the TRF2 gene in human gastric cancer. Dig Dis Sci 55: 997–1003 [DOI] [PubMed] [Google Scholar]
- Munoz P, Blanco R, Flores JM, Blasco MA (2005) XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet 37: 1063–1071 [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Kawai T, Kumaki F, Hiroi S, Mukai M, Ikeda E, Koering CE, Gilson E (2003) Expression of mRNAs for telomeric repeat binding factor (TRF)-1 and TRF2 in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Clin Cancer Res 9: 1105–1111 [PubMed] [Google Scholar]
- Oh BK, Kim YJ, Park C, Park YN (2005) Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am J Pathol 166: 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Wu Y, Gilson E (2010b) Dynamics of telomeric chromatin at the crossroads of aging and cancer. Essays Biochem 48: 147–164 [DOI] [PubMed] [Google Scholar]
- Dong W, Shen R, Wang Q, Gao Y, Qi X, Jiang H, Yao J, Lin X, Wu Y, Wang L (2009a) Sp1 upregulates expression of TRF2 and TRF2 inhibition reduces tumorigenesis in human colorectal carcinoma cells. Cancer Biol Ther 8: 2166–2174 [DOI] [PubMed] [Google Scholar]
- Gavert N, Ben-Ze'ev A (2007) β-Catenin signaling in biological control and cancer. J Cell Biochem 102: 820–828 [DOI] [PubMed] [Google Scholar]
- Stambolic V, Ruel L, Woodgett JR (1996) Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr Biol 6: 1664–1668 [DOI] [PubMed] [Google Scholar]
- Fujita K, Horikawa I, Mondal AM, Jenkins LM, Appella E, Vojtesek B, Bourdon JC, Lane DP, Harris CC (2010) Positive feedback between p53 and TRF2 during telomere-damage signalling and cellular senescence. Nat Cell Biol 12: 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet T et al. (2011) The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res 21: 1028–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JI et al. (2009) Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkreli M et al. (2012) Reversible cell-cycle entry in adult kidney podocytes through regulated control of telomerase and Wnt signaling. Nat Med 18: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Toh L, Lau P, Wang X (2012) Human telomerase reverse transcriptase (hTERT) is a novel target of the Wnt/β-catenin pathway in human cancer. J Biol Chem 287: 32494–32511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitner S, Reiche JA, Schaffauer AJ, Hiendlmeyer E, Herbst H, Brabletz T, Kirchner T, Jung A (2012) Human telomerase reverse transcriptase (hTERT) is a target gene of β-catenin in human colorectal tumors. Cell Cycle 11: 3331–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, Hein K, Vogt R, Kemler R (2012) Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336: 1549–1554 [DOI] [PubMed] [Google Scholar]
- Ye J et al. (2010a) TRF2 and apollo cooperate with topoisomerase 2α to protect human telomeres from replicative damage. Cell 142: 230–242 [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R (2001) Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128: 1253–1264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.