Abstract
Vitamin C is an antioxidant that maintains the activity of iron and α-ketoglutarate-dependent dioxygenases. Despite these enzymes being implicated in a wide range of biological pathways, vitamin C is rarely included in common cell culture media. Recent studies show that reprogramming of pluripotent stem cells is enhanced when vitamin C is present, thereby illustrating previous limitations in reprogramming cultures. Here, we summarize understanding of dioxygenase function in reprogramming and epigenetic regulation. The available data suggest a link between dioxygenase function and stem cell differentiation, which is exposed to environmental influence and is relevant for human disease.
Keywords: ascorbate, dioxygenase, histone demethylase, DNA methylation, reprogramming
See the Glossary for abbreviations used in this article.
Glossary.
- 2OG
2-oxoglutarate
- ABH
alkylation repair homologue
- AlkB
alkylated DNA repair protein
- Dio3
deiodinase, iodothyronine type III
- Dlk1
delta-like 1 homologue
- FTO
fat mass and obesity associated
- HDAC2/4
histone deacetylase 2/4
- HIF
hypoxia inducible factor
- INK4/ARF
cyclin-dependent kinase inhibitor 2A
- MEF
mouse embryonic fibroblast
- MLL
myeloid/lymphoid/mixed-lineage leukaemia
- Oct4
octamer-binding transcription factor 4
Introduction
Vitamin C also known as L-ascorbic acid or (5R)-[(1S)-1,2-dihydroxyethyl]-3,4-dihydroxyfuran-2(5H)-one, is an essential nutri-ent for humans and other primates. Most mammals synthesize ascorbate through a common metabolic pathway from glucose. However, primates, bats and guinea pigs have lost activity of the last, vitamin-C-specific enzyme, gulonolactone oxidase, in the pathway due to accumulation of mutations. Therefore, humans have to rely on nutrient sources including fruits and vegetables as a supply of vitamin C [1]. Vitamin C deficiency leads to a condition known as scurvy [2,3], which is characterized by a structural breakdown of the skin and epithelia due to loss of collagen hydroxylase function. In addition, vitamin C is involved in the synthesis of catecholamine, which acts as a neurotransmitter in the sympathic nervous system (Fig 1). Vitamin C is also required for the synthesis of carnitine, linking it to mitochondrial fatty acid transport [4]. Vitamin C has further been proposed to contribute to the regulation of several other pathways, including the hypoxia response, through regulating the activity of HIF prolyl-hydroxylases. One important aspect of vitamin C is that it acts as an electron donor for adjusting the redox state of iron-containing enzymes (Fig 2A,B). Vitamin C is by far the most effective compound for this reaction but is not absolutely essential. At least in some cases, vitamin C can be substituted by other antioxidants. Studies with vitamin-C-deficient mice have shown that glutathione can substitute fully for vitamin C in maintaining the activity of prolyl-hydroxylase function in the hypoxia response pathway [5]. Iron and α-ketoglutarate (2OG)-dependent dioxygenases are only active when iron is present in the Fe(II) state (Fig 2C). These enzymes catalyse oxidative decomposition of 2OG to carbon dioxide and succinate, thereby forming an Fe(IV)-oxo intermediate. The activated oxygen species can then be used to hydroxylate a specific substrate, which also restores the Fe(II) for the next catalytic cycle. In the absence of substrate, the iron atom is left in a higher oxidation state, which prevents further catalysis. Vitamin C acts as an electron donor for reducing iron whereby it is oxidized to dehydroascorbate (Fig 2B). Therefore, vitamin C performs a crucial function in maintaining the catalytic activity of Fe(II) 2OG-dependent dioxygenases.
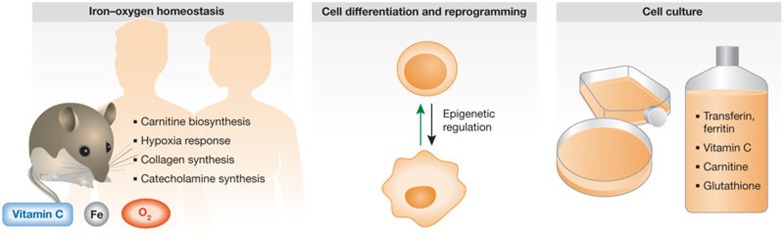
Figure 1.
Overview of processes influenced by vitamin C. Vitamin C regulates crucial enzymatic functions in the organism in cooperation with oxygen and iron metabolism. Dioxygenases are essential for regulating collagen, carnitin and catecholamine synthesis as well as the hypoxia response. In addition, dioxygenases catalyse chromatin, and DNA modifications thereby contribute to epigenetic regulation and cell differentiation. This aspect is also important in reprogramming of cell fates (green arrow). In the organism, iron (Fe) and vitamin C sources are environmental factors that affect the activity of dioxygenases. Similarly, in cell culture systems appropriate supplements can be used to mimic physiological levels of activity.
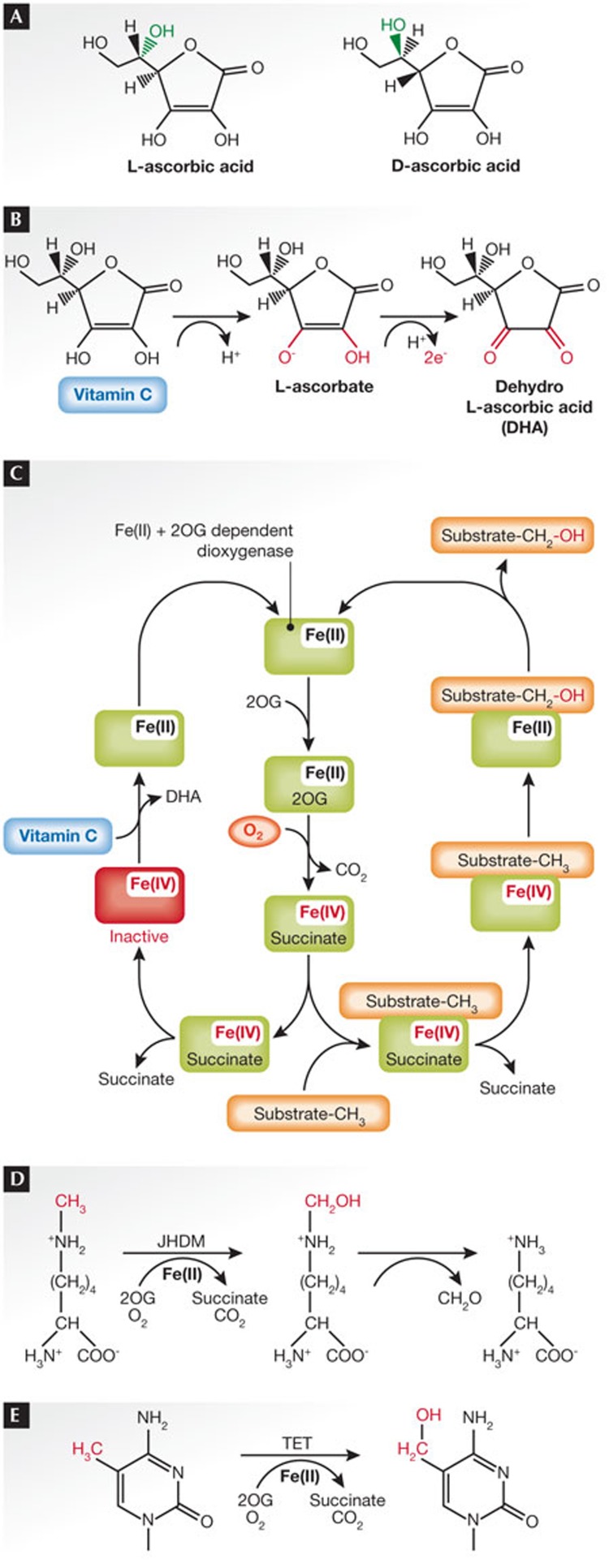
Figure 2.
Redox chemistry of vitamin C. (A) Ascorbic acid has chirality. L-ascorbic acid is vitamin C, whereas its enanziomere D-ascorbic acid does not confer the same biological effect. However, chemical antioxidant properties are equivalent in the L- and D-forms. (B) Oxidation of vitamin C to dehydroascorbic acid (DHA) yields two electrons that are used as a reducing agent in redox reactions. For the purpose of this review, these electrons are transferred to an iron atom to reduce its oxidation state. (C) Schematic overview of dioxygenase catalysis indicates different cycles of either substrate hydroxylation or vitamin-C-dependent regeneration. Molecular oxygen is used for the decarboxylation of 2OG, leading to the formation of an activated enzyme with an Fe(IV) intermediate (iron oxidation state is indicated in red). In the presence of substrate, the activated enzymatic complex hydroxylates the methyl group of the substrate and Fe(IV) is reduced to Fe(II) after the release of succinate. In the absence of a substrate, succinate is released without reduction of Fe(IV) leading to an inactive enzyme. Vitamin C can be used to reduce iron to Fe(II), restoring catalytic activity for the next cycle. Several histone- and DNA-modifying enzymes belong to the Fe(II) 2OG-dependent oxidase family. (D) Jumonji C (JmjC)-domain-containing histone demethylases catalyse the removal of methyl groups from lysine residues of histone proteins in a two-step mechanism. The first step, an hydroxylation reaction that converts the methyl group through an oxidative reaction that requires Fe(II) and 2OG, results in an instable intermediate. In a second step, spontaneous elimination of formaldehyde completes the demethylation reaction. (E) TET DNA hydroxylases catalyse the oxidation of methyl groups in the 5′-position of cytosine in DNA. Although the reaction seems to be similar overall to the first step in panel D, the hydroxymethyl group linked through a carbon–carbon bond is stable and is not spontaneously eliminated. 2OG, 2-oxoglutarate; TET, ten-eleven-translocation.
For its general antioxidant properties, vitamin C is a frequent additive in food products and beverages. Its ubiquitous applications ensure a sufficient supply in the human diet. In the light of this, it seems surprising that ascorbate is rarely included in the formulations of cell culture media and supplements (Table 1). The awareness of vitamin C might have been historically dampened by the use of complex media components, such as sera that contain varying levels of vitamin C. Also the fact that common laboratory animals including rodents can synthesize vitamin C in their liver or kidney might have contributed to neglecting it in culture media. However, it needs to be considered that even for mammals that have an active gulonolactone oxidase (Gulo) gene, synthesis of ascorbate might not occur in cultured cells. Yet, attempts to demonstrate a beneficial effect of vitamin C in culture media have generally not been positive. This suggests that either standard cell cultures do not rely on vitamin-C-dependent pathways or that the lack of these pathways does not manifest itself in readily observable phenotypic consequences. Such realization of insufficient amounts of vitamin C comes from studies on reprogramming of induced pluripotent stem (iPS) cells. Addition of vitamin C enhances the generation of mouse and human iPS cells [6]. Furthermore, reprogramming in the presence of vitamin C results in iPS cells with an improved differentiation potential and maintains imprinted DNA methylation patterns on the Dlk1–Dio3 locus [7]. These results are surprising when considering the prevalent view that redox regulation is generally provided by addition of β-mercaptoethanol to culture media. Vitamin-C-dependent processes might have a crucial role in culture systems that involve reprogramming of cell types or change of cell fates. Examining the role of vitamin C in epigenetic regulation might be instructive and provide new ways of thinking.
Table 1. Ascorbate, glutathione and carnitine in common cell culture media and supplements.
| Reference | Medium/supplement | L-ascorbic acid | Glutathione | L-carnitine |
|---|---|---|---|---|
| Sigma Aldrich | DMEM/Ham's F12 | × | × | × |
| M2 | × | × | × | |
| M16 | × | × | × | |
| MEM-α | 50 mg/l* | × | × | |
| CMRL-1066 | 50 mg/l | 10 mg/l | × | |
| Medium 199 | 0.0566 mg/l* | 0.05 mg/l | × | |
| RPMI 1640 | × | 1 mg/l | × | |
| Life Technologies | CMRL | 50 mg/l | 10 mg/l | × |
| Advanced DMEM | 2.5 mg/l** | 1 mg/l | × | |
| RPMI 1640 | × | 1 mg/l | × | |
| GMEM | × | × | × | |
| Ham's F12 | × | × | × | |
| Neurobasal | × | × | × | |
| Leibovitz's L-15 | × | × | × | |
| DMEM | × | × | × | |
| N2 supplement | × | × | × | |
| [7] | KOSR | Yes | N/A | N/A |
| [98] | B27 (NS21) supplement | × | 1 mg/l | 2 mg/l |
CMRL, Connaught Medical Research Laboratories; DMEM, Dulbecco's modified eagle's medium; KOSR, knock-out serum replacement; MEM, minimum essential medium; RPMI, Roswell Park Memorial Institute; ×, not present; N/A, no data available;
*, ascorbic acid sodium salt;
**, ascorbic acid phosphate.
Dioxygenases contribute to epigenetic regulation
Vitamin C has a crucial role for the activity of the family of Fe(II)-2OG-dependent dioxygenases (Fig 2C). The catalytic domain is organized in a double-stranded β-helix (DSBH) fold, which consists of eight β-strands and contains a triad of two histidines and one carboxylate (aspartate or glutamate) that bind to Fe(II) [8]. This typical DSBH fold has been used to identify 2OG-Fe(II) dioxygenases from sequence information, and thereby link this catalytic mechanism with genes that have functions in regulating histone and DNA methylation [9,10].
JmjC-domain-containing histone demethylases. A class of Jumonji C (JmjC)-domain-containing enzymes function in the demethylation of lysine residues of histone proteins through an oxidative reaction that requires iron Fe(II) and 2OG (Table 2; [11,12,13,14,15,16,17,18,19,20,21]). Unlike the flavin cofactor-dependent histone demethylases, which only remove mono- and dimethyl-lysine modifications, some JmjC-domain-containing histone demethylases (JHDMs) can also demethylate trimethylated histone lysines [22,23,24,25,26,27,28,29,30]. The co-factor-bound JmjC domain is thought to produce a highly reactive oxoferryl intermediate that hydroxylates the histone lysine-methyl group, thereby generating an unstable chemical structure. Spontaneous elimination of formaldehyde completes the removal of the methyl group from the histone tail (Fig 2D). The JHDM group consists of many demethylases and can be divided further into several subfamilies based on phylogenetic analysis [31]. Different members perform distinct demethylation reactions with varying specificity for the position of the lysine on the target histone and also for the methylation state. Their hydroxylase activity is mediated by the JmjC domain that contains two histidine residues and one glutamate residue, which chelate the catalytic iron atom and are invariably present in all family members, although not all proteins containing a JmjC domain are catalytically active. The JmjC domain is often present in association with additional domains that confer selective binding based on the methylation status of histones or DNA [32]. Depending on the size of the methyl-binding pocket, different members of the JHDM family are specific for monomethyl-, dimethyl- or trimethyl-modified substrates [20]. In addition to lysine methylation, histone tails can also be mono- and dimethylated on arginine residues. Similarly to lysine methylation, arginine methylation influences chromatin remodelling and gene expression, and has been associated with cell fate transitions and plasticity regulation during development [33]. JHDMs might facilitate multiple functions in the adjustment or change of chromatin modification patterns. In addition, an important role in demethylating non-histone substrates is recognized but less completely understood.
Table 2. Fe(II) 2-oxoglutarate-dependent dioxygenases involved in epigenetic regulation.
| Family | Function | Subfamily | Substrate | Enzyme | Reference | Phenotype in development | Reference | |
|---|---|---|---|---|---|---|---|---|
| JMJC | Histone demethylases | H3K4me1/me2/me3H3K36me2/me3 | No66 | [13] | N/A | |||
| KDM2JHDM1 | H3K36me1/me2 | Kdm2a/Jhdm1aFbxL11 | [11] | N/A | ||||
| Kdm2b/Jhdm1bFbxL10 | [14] | ExencephalyPerinatal death | [99] | |||||
| KDM3JHDM2JMJD1 | H3K9me1/me2 | Kdm3a/Jhdm2aJmjd1a | [12] | Abnormalities in spermatogenesis | [100] | |||
| Kdm3b/Jhdm2bJmjd1b | [15] | N/A | ||||||
| Kdm3c/Jhdm2cJmjd1c | [16] | N/A | ||||||
| KDM4JHDM3JMJD2 | H3K9me3 | H3K36me3 | Kdm4a/Jhdm3aJmjd2a | [22] | N/A | |||
| N/A | Kdm4b/Jhdm3bJmjd2b | [23] | N/A | |||||
| H3K36me3 | Kdm4c/Jhdm3cJmjd2c | [24] | N/A | |||||
| H3K9me2 | Kdm4d/Jmjd2d | [25] | None | [101] | ||||
| KDM5JARID | H3K4me2/me3 | Kdm5a/Jarid1aRbp2 | [26] | Haematopoietic abnormalities (after birth) | [26] | |||
| Kdm5b/Jarid1b | [27] | Embryonic lethal E4.5–E7.5None | [102,103] | |||||
| Kdm5c/Jarid1cSmcx | [28] | Posterior patterning and cardiac defects at E9.5 (male) | MGI | |||||
| Kdm5d/Jarid1dSmcy | [28] | N/A | ||||||
| KDM6 | H3K27 me3/me2 | Kdm6a/Utx | [29] | Cardiac defect, growth retardation,cranioschisis and female in utero deathHaematopoietic defects (after birth)Male: embryonic lethal with variable penetranceFemale: embryonic lethal before E11.5 | [104,105] | |||
| Kdm6b/Jhdm3dJmjd3 | [30] | Perinatal death | [106] | |||||
| PHD | H3K9me2 | Phf2 | [17] | Perinatal death with variable penetranceGrowth retardation | [107] | |||
| H4K20me1H3K9me1/me2 | Phf8 | [18][19] | Cleft lip/palateMental retardation | [108] | ||||
| H4K20me1H3K9me1/me2H3K27me2 | Kdm7a/Jhdm1dKiaa1718 | [20] | N/A | |||||
| KDM8 | H3K36me2 | Kdm8/Jmjd5 | [21] | Embryonic lethal around E11 | [109] | |||
| TET | DNA demethylases | 5mC5hmC5fC | Tet1* | [34] | Slightly smaller body size at birth | [42] | ||
| Tet2* | [34] | Haematopoietic abnormalities (after birth) | [50] | |||||
| Tet3 | [34] | Perinatal lethality | [46] | |||||
| ABH | H2A demethylase | Methylated H2A | Alkbh1 | [60] | Placental defects | [59] | ||
| DNA demethylases | 1meA, 3meCdsDNA | ABH2 | [53], [58] | None | [54] | |||
| 1meA, 3meCssDNA/RNA | ABH3 | [53], [58] | None | [54] | ||||
| 3meT ssDNA | FTO | [52] | Reduced size, early death | [63] |
*Tet1 and Tet2 double-knockout [43]. ABH, alkylation repair homologue; Alkbh1, alkylation repair homologue 1; ds: double-stranded; E, embryonic day; FTO, fat mass and obesity-associated; JARID, jumonji/AT-rich interaction domain; JHDM, Jumonji family of histone demethylase; JMJC, jumonji C; KDM, lysine demethylase; N/A, no data available; PHD, plant homeodomain finger motif; ss, single-stranded; TET, ten-eleven-translocation.
TET proteins mediate oxidation of 5mC in DNA. Homology searches in mammalian genomes, for genes related to the hydroxylases involved in base J synthesis in trypanosomes, have led to the identification of the ten-eleven-translocation genes 1–3 (TET1–3), as members of a family of DNA hydroxylases that convert 5-methyl-cytosine (5mC) into 5-hydroxymethyl-cytosine (5hmC) (Fig 2E; [9,34]). Further conversion through iterative oxidation of 5mC results in the formation of 5-formylcytosine and 5-carboxylcytosine [9,34,35,36]. All TET proteins have a catalytic domain that contains a DSBH fold and binds to Fe(II) and 2OG. In addition, TET1 and TET3 but not TET2 have a zinc finger domain that might facilitate binding to or interaction with other proteins and DNA. 5hmC was found to be enriched at transcription start sites of bivalent gene promoters and exons in embryonic stem cells, indicating that TET enzymes contribute to the regulation of Polycomb group complex target genes [37,38]. This finding further suggests a potential role in the regulation of cellular identity [39,40,41]. Tet1 and Tet2 are expressed in mouse embryonic stem cells, within which Tet3 expression is barely detected. The converse pattern is observed in fibroblasts, in which high expression of Tet3, intermediate expression of Tet2 and almost no expression of Tet1 is observed. It has further been shown that Tet genes are regulated by transcription factors associated with pluripotency. Depletion of Oct4 by RNA interference leads to the downregulation of Tet1 and upregulation of Tet3 with a concomitant decrease in the levels of 5hmC [39]. This indicates a crucial role for Tet1 in mouse embryonic stem cells. However, a targeted disruption of Tet1 in mice is compatible with normal development [42], and a fraction of Tet1- and Tet2-double-mutants, despite presenting multiple developmental defects, can survive to adulthood [43]. These data suggest that Tet3 might also have overlapping functions with Tet1 and Tet2.
Mutations in TET1 and TET2 are also implicated in the development of several types of cancer and are associated with a decrease in 5hmC. TET1 is an MLL partner in lymphoid and myeloid acute leukaemia, and mutations in Tet2 are associated with myeloid leukaemias or myeloproliferative disorders, highlighting potential roles for TET enzymes in the regulation of cell identity [44]. Importantly, TET enzymes have been proposed as a crucial component of a DNA demethylation pathway [35]. Iterative oxidation of 5mC by Tet dioxygeneases results in the formation of 5-carboxylcytosine (5caC), which is recognized and excised by thymine DNA glycosylase. Tet1 has been implicated in DNA demethylation associated with genome-wide reprogramming during germline development in mice [45]. Tet3 has been shown to mediate hydroxylation of 5mC on the sperm-derived chromosomes in zygotes [46], whereas the maternally inherited chromosomes of the oocyte are protected from Tet3 activity by Stella binding [47]. This results in a differential modification of maternal chromosomes with 5mC and paternal chromosomes with 5hmC in preimplantation embryos. Interference with this marking by depletion of Tet3 in oocytes results in embryonic lethality after implantation that manifests itself with variable penetrance [46]. These findings indicate that Tet enzymes are associated with transitions in cell identity and suggest a role for Tet proteins in the reprogramming of epigenetic patterns. However, none of the Tet genes are essential for development in mice [42,46,48,49,50], indicating that a potential redundancy or overlapping of functions with other epigenetic pathways might exist.
AlkB family proteins in DNA repair. Computational searches for members of the Fe(II)-2OG-dependent hydroxylase family have revealed homology in the bacterial AlkB DNA repair protein [10]. There are at least nine members of the AlkB protein family in mammals: ABH1–ABH8 and FTO [51,52]. AlkB and its mammalian homologues ABH2 and ABH3 mediate DNA repair by oxidation of 3-methylcytosine and 1-methyladenine [53]. Studies in mice have shown that ABH2 is the crucial enzyme for repairing 3-methylcytosine- and 1-methyladenine-modified DNA [54]. FTO has further been associated with repair of 3-methyl-thymidine in DNA [52]. AlkB enzymes use metal catalysis to oxidize stable methyl adducts that are attached to nitrogens of the heterocyclic bases [53,55,56]. The oxidized methyl groups are released as formaldehyde, and unmodified bases are then regenerated. The mechanism holds similarities to the catalytic mechanism of JHMDs. However, AlkB enzymes have additional domains binding to nucleic acids and distorting DNA structure to flip out bases, making them accessible to chemical reactions [57]. In addition to having an important role in DNA repair, AlkB proteins have also been implicated in repairing RNA in response to exposure to alkylating agents [58]. In vitro, ABH2 presents a strong preference for double-stranded DNA contrary to ABH3 that demethylates RNA preferentially [54,58]. Similarly to the JmjC domain protein family, not all AlkB homologues have catalytic activity. Although ABH1 seems to lack measurable DNA/RNA repair activity, it is developmentally regulated and essential for placental development in mice [59]. Mutation of Abh1 interferes with trophoblast differentiation in mice and results in an increased number of trophoblast stem cells. It has been suggested that Abh1 modulates histone deacetylation by mutually antagonistic binding with HDAC4 to the essential trophoblast factor Mrj [59] and histone H2A demethylation [60].
FTO is a particularly interesting member of the AlkB family. It has been shown to mediate repair of 3-methyl thymidine in DNA [52]. In addition, mutations in FTO have been correlated with obesity and metabolic disorders in humans. Notably, engineered mutations in FTO have been shown to affect body fat mass in mice [61]. Deletion of FTO leads to postnatal growth retardation [62,63]. Importantly, growth retardation is also observed when FTO is specifically mutated in the brain suggesting a central function in the physiological regulation of energy metabolism and growth. Developmental regulation and phenotypic consequences of mutation of AlkB proteins suggest that some members of this protein family could potentially also have roles in epigenetic regulation, thus, providing functions beyond DNA repair.
Histone demethylases in iPS cell reprogramming
The ability of vitamin C to enhance iPS cell generation has prompted investigations into the mechanism of its action. Notably, the reprogramming process leads to an increased level of reactive oxygen species, suggesting that antioxidant properties of vitamin C might contribute to its effect. However, enhanced reprogramming is not observed with other antioxidants including vitamin B1, reduced gluthatione, sodium selenite, amino-acetylcysteine, resveratrol, α-lipoic acid, vitamin E and L-carnitine [6]. Therefore, the activity of vitamin-C-dependent enzymes is probably more important. Vitamin C promotes cell proliferation and fibroblasts show a striking increase in their lifespan when cultured in its presence. Part of this effect is probably due to counteracting senescence by preventing the activation of the INK4/ARF locus [6], which has been previously described as a road block for iPS cell generation [64]. Inactivating mutations of tumour-suppressor genes or transformation have also enhanced iPS cell formation potentially by increasing cell proliferation [64,65,66].
To investigate whether vitamin C also facilitates epigenetic changes, Wang et al have analysed chromatin modifications during reprogramming in the presence or absence of vitamin C [67]. Their study showed that vitamin C induces a marked reduction in histone H3 Lys 36 dimethylation and trimethylation. Further analysis identified two Fe(II) 2OG-dependent histone demethylases, Jhdm1a and Jhdm1b, as key regulators of Lys 36 demethylation during reprogramming [67]. Combined depletion of Jhdm1a and Jhdm1b by RNA interference impairs reprogramming. Conversely, expression of Jhdm1b and Oct4 is sufficient for reprogramming of fibroblasts to iPS cells [67].
Notably, reduced levels of histone H3 Lys 36 dimethylation and increased Lys 27 trimethylation at the INK4/ARF locus are observed in cells reprogrammed in the presence of vitamin C, in agreement with reports showing that Jhdm1b regulates cell proliferation and senescence by repressing the INK4/ARF locus through histone H3 Lys 36 demethylation [14,64,68]. The INK4/ARF locus is an important tumour-suppressor locus that encodes three functionally related proteins: p19ARF, p15INK4b and p16INK4a. Aberrant mitogenic signals induce expression of the INK4/ARF locus and thereby activate the anti-proliferative retinoblastoma protein and p53 pathways, which mediate cell cycle exit and senescence [69]. INK4/ARF is basally expressed in normal somatic cells and silenced completely in embryonic stem and iPS cells, in which it is also marked by bivalent chromatin [64]. Increased INK4/ARF expression has further been associated with ageing [70]. Somatic cell reprogramming is a proliferation-dependent process and the activity of the INK4/ARF locus has a rate-limiting effect, which is illustrated by the low efficiency of reprogramming of cells from aged tissues. Notably, depletion of mouse Arf has been shown to increase the yield of iPS cells, albeit the same effect is not observed during reprogramming of human cells [64].
Reprogramming by using Oct4 and JHDM1B expression in the presence of vitamin C also strongly activates the microRNA (miR) 302/367 cluster [67]. JHDM1B decreases the methylation levels of histone H3 Lys 36 surrounding the Oct4 binding sites of the miR302/367 gene and facilitates gene expression [67]. The miR302/367 cluster regulates pluripotency through inhibition of genes essential for embryonic development and differentiation [71] and is a direct target of Oct4 and Sox2 in embryonic stem cells [72,73]. Consistent with a crucial role in pluripotent cells miR302/367 expression is lost rapidly on differentiation [74]. Notably, expression of the entire miR302/367 cluster is sufficient for reprogramming of fibroblasts, whereby miR367 is essential for activation of Oct4 [75]. This suggests that miR302/367 might be a crucial target of Jhdm1a and Jhdm1b in iPS cell generation.
However, vitamin C has additional functions. When pre-iPS cells, which are trapped at an intermediate state of reprogramming, are treated with vitamin C, conversion to fully reprogrammed iPS cells can be induced [6]. Examination of epigenetic factors that restrict pluripotency has led to the identification of repressive histone H3 Lys 9 methylation marks on transcription factor genes that are required for pluripotent stem cells [76]. Chen et al show that, in the presence of vitamin C, histone H3 Lys 9 dimethylation decreases, which correlates with the upregulation of gene expression. This observation suggests that the histone-methylation-mediated epigenetic barrier is sensitive to the presence of vitamin C. Furthermore, inhibition of the Lys 9-specific methyltransferases Suv39h1, Suv39h2, G9a and Setdb1 synergizes with vitamin C in this system. Combined treatment substantially improves the kinetics and efficiency of the reprogramming process. When Setdb1 activity is depleted full reprogramming of pre-iPS cells can also be observed in the absence of vitamin C, suggesting that Setdb1 is crucial for the epigenetic restriction of pre-iPS cells [76]. A targeted RNA-interference-based screen identified the histone demethylase Kdm3b (Jhdm2b) as the primary target of vitamin C. Furthermore, Kdm3a, Kdm4b and Kdm4c also contribute to reprogramming. These results suggest demethylation of histone H3 Lys 9 to be an important step in reprogramming of pre-iPS cells to iPS cells, which is facilitated by enzymes that depend on vitamin C for maintaining activity. The JmjC-domain-containing H3K27 demethylase Utx has also been identified as a crucial regulator for induction of pluripotency during somatic cell reprogramming [77]. Interestingly, different histone demethylases are required for the reprogramming of fibroblast or pre-iPS cells. Contrary to what is observed in the INK4/ARF locus during the reprogramming of MEFs, upregulation of Jhdm1b (Kdm2b) does not seem to be associated with a reduction in Lys 36 methylation when pre-iPS cells are reprogrammed to iPS cells [76]. Differences in genetic requirements for these reprogramming systems potentially reflect the differentiation state of the starting cell population and the relative time-point in the reprogramming process. Taken together, these results show that regulation of Fe(II) 2OG-dependent histone demethylases is an important factor for reprogramming. Culture conditions without vitamin C apparently limit the activity of these enzymes and thereby interfere with the adjustment of epigenetic patterns.
Tet2 facilitates haematopoietic differentiation
An important insight into the role of Fe(II) 2OG-dependent dioxygenases in cell fate transitions is provided by studies of Tet2 function in haematopoietic differentiation. Mutations of Tet2 are frequently associated with myeloid malignancies. Similarly, disruption of the Tet2 gene in mice results in an expansion of haematopoietic progenitors and stem cells, and a defect of differentiation of the blood lineages [48,49,50]. This phenotype is associated with decreased 5hmC suggesting that Tet2-mediated DNA hydroxymethylation is crucial for changing cell identity. Independent support for this view comes from studies of mutations in the isocitrate dehydrogenase genes. IDH1 and IDH2 mutations are found in a range of tumour types. These mutations act in a dominant manner and cause a change in substrate specificity that leads to the conversion of 2OG to 2-hydroxyglutarate (2HG; [78,79]). 2HG is not normally present in the cell but acts as an onco-metabolite. It is thought that 2HG inhibits Fe(II) 2OG-dependent dioxygenases by binding in the position that would normally confer 2OG binding [80,81]. An oncogenic mutation in Idh1 has been associated with impaired haematopoietic differentiation in mice [82]. This phenotype is similar to and can be explained by inhibition of Tet2 function through 2HG [80]. 2HG has also been associated with oncogenic transformation in other tissues. Notably, IDH mutations and 2HG are associated with inhibition of histone demethylation in glioma [83,84]. This observation is consistent with independent work showing that 2HG can inhibit a broad spectrum of α-ketoglutarate-dependent enzymes [85]. Mutations in IDH1 and IDH2 might thereby reduce the activity of crucial epigenetic enzymes, thereby inducing errors in cell differentiation that can potentially result in tumour formation [83]. Notably, production of 2HG by oncogenic IDH mutations correlates with epithelial-to-mesenchymal transition in cultured cells [86]. This is an interesting observation that raises the question about the usefulness of 2HG for manipulating the phenotypic state of cells in culture and its potential applications in strategies for directing differentiation (Sidebar A). It is probable that the group of enzymes that are inhibited by 2HG overlap to a large extent with enzymes that are dependent on vitamin C (Table 2; [81]). Although both agents act on various levels, 2HG might enforce a situation that resembles a marked and prolonged deficiency of vitamin C, suggesting mutually antagonistic effects of 2HG and vitamin C on the activity of epigenetic regulators and cellular plasticity.
Sidebar A | In need of answers.
Several enzymes have been identified that contribute to changing chromatin and DNA modifications during cell fate transition. For these enzymes, specific, biologically relevant functions have been demonstrated. However, the full range of regulators is unknown. The genome contains many dioxygenases that can be seen as putative epigenetic regulators. It is important to analyse further their biological function in future studies. In some cases, functional redundancies might obscure the real impact, as is probable in the case of the TET enzymes.
Iron and oxygen metabolism is regulated by a complex circuitry in the organism. If a dynamic equilibrium is disrupted, several diseases can emerge. Future work needs to explore further the molecular links between this metabolic equilibrium and epigenetic regulatory mechanisms. This requires integration of a wide range of regulatory networks including hypoxia response, collagen biosynthesis, epithelial polarity and epigenetic memory. Recent results suggest that these apparently unrelated biological processes are linked in cell culture systems and the organism. What needs to be explored is how this link is constructed and whether it is amenable for clinical intervention. Future research into the wider aspects of iron oxygen metabolism could provide a better understanding of human diseases including cancer, metabolic problems and diabetes.
The fact that an important network of epigenetic regulators is exposed to environmental influences is surprising and interesting. It is also not easy to explain why evolution has selected against the endogenous synthesis of vitamin C in primates. Loss of the Gulo gene could be accidental through genetic drift, but this seems unlikely. It will be interesting to explore physiological roles of environmental regulation of Fe(II) 2OG-dependent dioxygenases. Fe and vitamin C uptake are considered the main regulatory factors and could be related to seasonal adaptations of energy metabolism. In addition, oxygen levels are expected to be of some significance in certain situations, particularly as oxygen pressure is reduced through the placental system in the developing embryo and in populations living in high altitude regions. It remains to be seen whether these conditions evoke alternative or additional mechanisms for the epigenetic regulation of stem cells during fate transitions.
Vitamin C improves iPS cell quality
Vitamin C has been shown to induce widespread DNA demethylation in human embryonic stem cells. This effect is most clearly observed on genes that are marked by bivalent chromatin modifications and genes that become demethylated during reprogramming [87]. Reprogramming in the absence of vitamin C induces hypermethylation of the imprinted Dlk1–Dio3 locus, which is negatively associated with the quality of mouse iPS cells [7]. The resulting iPS cells are compromised in their ability to form embryonic stem cell-derived mice in tetraploid complementation experiments [88]. Aberrant methylation can be reduced when the relative expression of reprogramming factors is optimized [88]. This suggests that vitamin C could protect from changes induced by suboptimal expression ratios or culture stress. Although vitamin C can prevent methylation defects, it is not sufficient to reverse hypermethylation of the Dlk1–Dio3 locus once established [7].
The Dlk1–Dio3 gene cluster consists of several paternally or maternally imprinted genes. Several pathways contribute to its inactivation in iPS cells. A loss of activating histone H3 acetylation and Lys 4 dimethylation and trimethylation marks, as well as a gain of DNA methylation on the maternal allele, have been observed [89]. DNA hypermethylation has further been shown to require the DNA methyltransferase Dnmt3a. Addition of valproic acid, which is a chemical inhibitor of histone deacetylation can increase histone H3 acetylation and Lys 4 methylation on the silent Dlk1–Dio3 locus but cannot fully rescue the developmental phenotype. By contrast, vitamin C prevents the loss of histone H3 acetylation and Lys 4 methylation resulting in the maintenance of correct Dlk1–Dio3 imprinting [7]. These results emphasize the role of Fe(II) 2OG-dependent dioxygenase function in the regulation of DNA methylation patterns. It is tempting to speculate that Tet1 and Tet2 activities are involved in maintaining DNA methylation patterns in iPS cells. Consistent with this idea, aberrant imprinting marks in the Dlk1 locus have been reported in the progeny of mice carrying mutations in both Tet1 and Tet2 [43]. Recent studies have also provided evidence that Tet1 and Tet2 contribute to reprogramming efficiency [90,91]. However, a complete understanding of the role of Tet enzymes during somatic cell reprogramming requires further studies (Sidebar A).
Concluding remarks and future outlook
Somatic cell reprogramming requires the repression of differentiation genes and the activation of the genes that regulate pluripotency. On the transduction of cells with reprogramming factors, repression of the somatic markers is efficiently achieved but induction of the pluripotency gene network occurs with low efficiency. At this point the epigenetic landscape of the cells is crucial for the success or the failure of the process [92]. Valproic acid is a widely used HDAC2 inhibitor that increases the acetylation state of chromatin and the efficiency of reprogramming [7,75]. This illustrates that compounds promoting changes of the chromatin configuration can potentially enhance reprogramming. Vitamin C contributes to overcoming at least some of the epigenetic constraints towards pluripotency by regulating the INK4/ARF, miR302/367 and Dlk1–Dio3 loci. Furthermore, vitamin C is implicated in a long list of metabolic functions and it could also cause indirect effects through other pathways [4,5,85,93,94].
Recent studies have implicated Fe(II)-dependent oxidative modification activities in normal tissue homeostasis and experimentally induced reprogramming. Histone- and DNA-modifying enzymes have been identified that contribute crucially to epigenetic regulation in a cell-type- and differentiation-stage-specific manner. Loss of these activities is associated with epigenetic defects and compromised cell differentiation or developmental potential. One of the most important aspects of Fe(II) 2OG-dependent dioxygenases is their susceptibility to environmental factors. These enzymes require vitamin C for maintaining catalytic activity and are inhibited by onco-metabolites [78,79,95]. Insufficient activity of this class of enzymes can lead to irreversible aberrations similar to the defects in DNA methylation patterns observed in some iPS cells. Dietary sources of vitamin C and iron, as well as exposure to onco-metabolites, could have long-term effects on human health. Understanding the molecular links will undoubtedly facilitate future association studies and clarify the wide range of effects attributed to vitamin C (Sidebar A).
In the light of the present literature, the effect of vitamin C in reprogramming might be best explained by a combined enhancement of JHDM and TET activities, which facilitate changing histone methylation and DNA methylation patterns, respectively. Altering epigenetic patterns might be less crucial for traditional cell culture systems that have focused on maintaining cells in a defined state. This could explain the absence of vitamin C from many classical media formulations. Yet, established cell lines are known to frequently acquire methylation patterns that are not typical of their tissue of origin [96]. Lack of crucial enzyme activities could contribute to a gain of aberrant CpG island methylation. In addition to published literature on embryonic stem cells and MEFs [87,97], it will be interesting to investigate the effects of vitamin C in other culture systems. Similarly, culture under low oxygen pressure could contribute to preserving small amounts of vitamin C in media components such as serum, which would be rapidly oxidized in atmospheric conditions.
Notably, vitamin C and 2HG have opposing effects not only on the activity of Fe(II) 2OG dioxygenases but also on cell fate changes. Whereas vitamin C facilitates reprogramming, 2HG seems to impair differentiation in the haematopoietic system and potentially also in other tissues (Fig 3). Thus, 2HG might be useful as an inhibitor of reprogramming and for stabilizing stem cell fate in culture. Future work will show if 2HG and vitamin C could be potentially useful for regulating stem cell differentiation for applications in regenerative medicine. Furthermore, the development of compounds based on 2OG for potential clinical applications will provide additional tools for the manipulation of cell fates in culture and organisms.
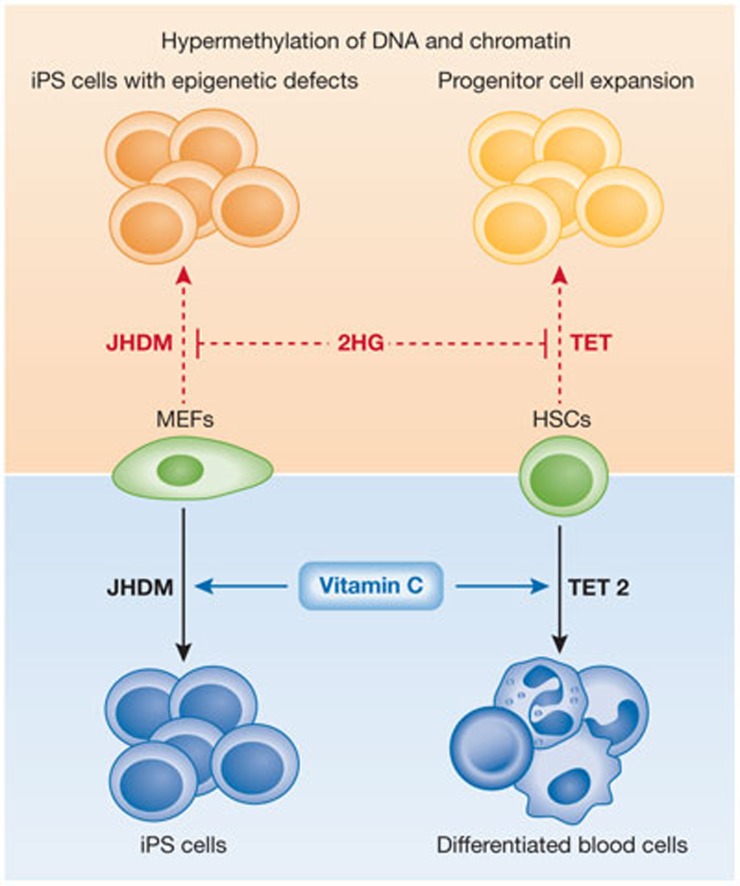
Figure 3.
Summary of ten-eleven-translocation and Jumonji family of histone demethylase function in reprogramming and differentiation. JHDM activity has been shown to be crucial for reprogramming of fibroblasts to iPS cells through heterologous expression of reprogramming factors. Conversely, TET2 has been implicated in facilitating differentiation of haematopoietic stem cells and progenitors. Both activities are dependent on vitamin C and are inhibited by the onco-metabolite 2HG. Loss of these activities, either due to inhibition of 2HG or unphysiologically low levels of vitamin C and iron results in aberrant epigenetic patterns and can block a change in cell identity. 2HG, 2-hydroxyglutarate; iPS, induced pluripotent stem; MEF, mouse embryonic fibroblast; JHDM, Jumonji family of histone demethylase; TET, ten-eleven-translocation.
Finally, the puzzling question remains why primates have lost the ability to synthesize vitamin C (Sidebar A). Could environmental influence be selected for when vitamin-C-dependent enzymes perform central functions in cell differentiation? Seasonal changes in vitamin C supply would be predicted to translate into alternating stem cell activity. Low levels of vitamin C might help to stabilize and preserve stem cells in episodes of low metabolic activity similar to stem cell hibernation. If it turns out that evolution has provided for environmental access to stem cell physiology through Fe(II) and 2OG-dependent dioxygenase activity, this could be highly relevant for regenerative medicine. Future work will undoubtedly uncover further molecular details of this important interplay between environment and tissue homeostasis, and provide a clearer picture for potential clinical applications.
Asun Monfort & Anton Wutz
Acknowledgments
This work was supported by a Wellcome Trust Senior Research Fellowship awarded to A.W. (grant reference 087530/Z/08/A).
Footnotes
The authors declare that they have no conflict of interest.
References
- Drouin G, Godin JR, Pagé B (2011) The genetics of vitamin C loss in vertebrates. Curr Genomics 12: 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkofsky B (1991) Ascorbate requirement for hydroxylation and secretion of procollagen: relationship to inhibition of collagen synthesis in scurvy. Am J Clin Nutr 54: 1135S–1140S [DOI] [PubMed] [Google Scholar]
- Baron JH (2009) Sailors' scurvy before and after James Lind—a reassessment. Nutr Rev 67: 315–332 [DOI] [PubMed] [Google Scholar]
- Nelson PJ, Pruitt RE, Henderson LL, Jenness R, Henderson LM (1981) Effect of ascorbic acid deficiency on the in vivo synthesis of carnitine. Biochim Biophys Acta 672: 123–127 [DOI] [PubMed] [Google Scholar]
- Nytko KJ, Maeda N, Schläfli P, Spielmann P, Wenger RH, Stiehl DP (2011) Vitamin C is dispensable for oxygen sensing in vivo. Blood 117: 5485–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA et al. (2010) Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6: 71–79 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M et al. (2012) Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet 44: 398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ (2006) Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem 100: 644–669 [DOI] [PubMed] [Google Scholar]
- Tahiliani M et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (2001) The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol 2: RESEARCH0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125: 483–495 [DOI] [PubMed] [Google Scholar]
- Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B (2010) Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J 29: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y, Zhang Y (2008) The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 15: 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY et al. (2012) KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol Cell Biol 32: 2917–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM et al. (2010) Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance. Nucleic Acids Res 38: 6389–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li J, Song T, Lu M, Kan PY, Lee MG, Sha B, Shi X (2010) Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J Biol Chem 285: 9322–9326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L et al. (2010) Structural insights into a novel histone demethylase PHF8. Cell Res 20: 166–173 [DOI] [PubMed] [Google Scholar]
- Qi HH et al. (2010) Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466: 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR et al. (2010) Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 17: 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia DA et al. (2010) KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci USA 107: 9671–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y (2006) The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442: 312–316 [DOI] [PubMed] [Google Scholar]
- Fodor BD et al. (2006) Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev 20: 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Majolica A, Rappsilber J, Antal T, Hansen KH, Helin K (2006) The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442: 307–311 [DOI] [PubMed] [Google Scholar]
- Whetstine JR et al. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilland DG, Zhang Y, Kaelin WG Jr (2007) The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128: 889–900 [DOI] [PubMed] [Google Scholar]
- Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL (2007) Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol 14: 240–242 [DOI] [PubMed] [Google Scholar]
- Iwase S et al. (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Agger K et al. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734 [DOI] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA 104: 18439–18444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7: 715–727 [DOI] [PubMed] [Google Scholar]
- Upadhyay AK, Horton JR, Zhang X, Cheng X (2011) Coordinated methyl-lysine erasure: structural and functional linkage of a Jumonji demethylase domain and a reader domain. Curr Opin Struct Biol 21: 750–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M (2007) Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445: 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF et al. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA et al. (2011) Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473: 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y et al. (2011) Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42: 451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP et al. (2011) Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K (2011) TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y (2011) Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25: 679–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM et al. (2011) Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 9: 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM et al. (2013) Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24: 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F et al. (2009) Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301 [DOI] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, Surani MA (2012) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339: 448–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP et al. (2011) The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610 [DOI] [PubMed] [Google Scholar]
- Nakamura T et al. (2012) PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486: 415–419 [DOI] [PubMed] [Google Scholar]
- Ko M et al. (2011) Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci USA 108: 14566–14571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z et al. (2011) Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118: 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K et al. (2011) Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM (2003) Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics 4: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T et al. (2007) The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318: 1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan T et al. (2002) Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci USA 99: 16660–16665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringvoll J et al. (2006) Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J 25: 2189–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PØ, Johansen RF, Seeberg E (2002) AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419: 178–182 [DOI] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419: 174–178 [DOI] [PubMed] [Google Scholar]
- Sundheim O et al. (2008) AlkB demethylases flip out in different ways. DNA Repair (Amst) 7: 1916–1923 [DOI] [PubMed] [Google Scholar]
- Aas PA et al. (2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 [DOI] [PubMed] [Google Scholar]
- Pan Z, Sikander S, Witherspoon M, Dizon D, Nguyen T, Benirsche K, Wiley C, Vrana P, Lipkin SM (2008) Impaired placental trophoblast lineage differentiation in Alkbh1−/− mice. Dev Dyn 237: 316–327 [DOI] [PubMed] [Google Scholar]
- Ougland R et al. (2012) ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells 30: 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C et al. (2009) A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet 5: e1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P (2006) The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS ONE 5: e14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Kock L, Emmerling C, Vierkotten J, Peters T, Brüning JC, Rüther U (2009) Inactivation of the Fto gene protects from obesity. Nature 458: 894–898 [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K (2009) Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisúa Belmonte JC (2009) Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T et al. (2011) The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9: 575–587 [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN (2009) Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci USA 106: 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE (2006) The regulation of INK4/ARF in cancer and aging. Cell 127: 265–275 [DOI] [PubMed] [Google Scholar]
- Agherbi H, Gaussman-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M (2009) Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS ONE 4: e5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA (2003) Embryonic stem cell-specific MicroRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK (2008) Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol 28: 6426–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A et al. (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR et al. (2004) Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270: 488–498 [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F et al. (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J et al. (2012) H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet 45: 34–42 [DOI] [PubMed] [Google Scholar]
- Mansour AA et al. (2012) The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 488: 409–413 [DOI] [PubMed] [Google Scholar]
- Ward PS et al. (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L et al. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME et al. (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W et al. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M et al. (2012) IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488: 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C et al. (2012) IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S et al. (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R et al. (2011) The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassian AR et al. (2012) Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/microRNA (miR)-200-dependent epithelial-mesenchymal transition (EMT). J Biol Chem 287: 42180–42194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung TL, Brena RM, Kolle G, Grimmond SM, Berman BP, Laird PW, Pera MF, Wolvetang EJ (2010) Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells 28: 1848–1855 [DOI] [PubMed] [Google Scholar]
- Carey BW et al. (2011) Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 9: 588–598 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedliner K (2010) Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465: 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege CA et al. (2012) Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488: 652–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y et al. (2013) NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature [Epub ahead of print] doi:; DOI: 10.1038/nature11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS et al. (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Cullen JJ, Buettner GR (2012) Ascorbic acid: chemistry, biology and the treatment of cancer. Biochim Biophys Acta 1826: 443–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles HJ, Raval RR, Harris AL, Ratcliffe PJ (2003) Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res 63: 1764–1768 [PubMed] [Google Scholar]
- Yang M, Soga T, Pollard PJ, Adam J (2012) The emerging role of fumarate as an oncometabolite. Front Oncol 2: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Boyes J, Bird A (1990) High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 62: 503–514 [DOI] [PubMed] [Google Scholar]
- Kuo SM, Burl LR, Hu Z (2012) Cellular phenotype-dependent and -independent effects of vitamin C on the renewal and gene expression of mouse embryonic fibroblasts. PLoS ONE 7: e32957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y et al. (2008) NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods 171: 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Tokunaga A, Sakamoto R, Yoshida N (2011) Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol Cell Neurosci 46: 614–624 [DOI] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y (2007) Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450: 119–123 [DOI] [PubMed] [Google Scholar]
- Iwamori N, Zhao M, Meistrich ML, Matzuk MM (2011) The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol Reprod 84: 1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole S et al. (2011) PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol 38: 1267–1277 [DOI] [PubMed] [Google Scholar]
- Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarratequi I, Helin K (2011) Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J 30: 4586–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme S et al. (2013) The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood [Epub ahead of print] doi:; DOI: 10.1182/blood-2012-08-452003 [DOI] [PubMed] [Google Scholar]
- Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, Ge K (2012) UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci USA 109: 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T et al. (2010) The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 11: 936–944 [DOI] [PubMed] [Google Scholar]
- Okuno Y et al. (2012) Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes [Epub ahead of print] doi:; DOI: 10.2337/db12-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ, Ratcliffe PJ, Schofield CJ (2010) PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet 19: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura A, Minehata K, Terashima M, Kondogh G, Hara T, Suzuki T (2012) Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development 139: 749–759 [DOI] [PubMed] [Google Scholar]