Abstract
Ubiquilins (Ubqlns)—a family of ubiquitin-binding proteins—are involved in several protein degradation pathways and have been implicated in various neurodegenerative diseases. Ubqln1 regulates autophagosome maturation during autophagy-mediated degradation. We now show that Ubqln4 mediates the interaction between Ubqln1 and the autophagy machinery by recruiting Ubqln1 to LC3. This targeting of Ubqln1 to autophagosomes requires the Ubqln4 UBL domain and the Ubqln1 UBA domain. This study identifies a new role for Ubqln4, expanding the role for Ubqlns in protein degradation.
Keywords: autophagy, LC3, Ubqln1, Ubqln4
Introduction
Ubiquilins (Ubqlns) are a family of cytosolic proteins that function in protein degradation by carrying cargo to the proteasome, enhancing autophagy-mediated degradation and participating in endoplasmic reticulum-associated protein degradation (ERAD) [[1]. All eukaryotes express Ubqln proteins. While yeast expresses only a single orthologue, Dsk2, there has been an expansion of the family in mammals, and the human genome contains four Ubqln isoforms. Little is known about differences in the function of the four isoforms.
Ubqlns belong to the UBL-UBA family of proteins, as they have both ubiquitin-like (UBL) and a specific type of ubiquitin-binding (ubiquitin-associated, UBA) domain. The amino-terminal UBL domain has homology to ubiquitin and can bind to the proteasome regulatory component s5a [[2, [3, [4]. The carboxy-terminal UBA domain binds both mono- and poly-ubiquitin [[5, [6]. The UBL and UBA domains of all human Ubqln isoforms are highly homologous, while the central regions between the UBL and UBA domains are divergent.
Most studies of mammalian Ubqlns have focused on their roles in protein degradation. Ubqlns have been described as adaptor proteins that deliver substrates to the proteasome [[7, [8]. Their role as a shuttle for specific cargo to the proteasome depends on binding of ubiquitinated proteins through the UBA domain, simultaneous with interaction with s5a through the UBL domain [[3, [4, [5, [6]. This role seems to be the primordial function of Ubqlns, as it is conserved from yeast to man. Unlike its yeast orthologue Dsk2, Ubqln1 has been shown to have a role in autophagy-mediated degradation [[9, [10], suggesting that this extra role has been acquired during expansion of the family in higher eukaryotes. Autophagy is a process that leads to the degradation of proteins and organelles in the cytoplasm through engulfment by a double-membrane organelle, called the autophagosome [[11]. The engulfed components are degraded as the autophagosome fuses with lysosome to form an autolysosome [[12]. Microtubule-associated protein light chain 3 (LC3), an autophagosomal marker, undergoes ubiquitylation-like modifications, which allow it to be inserted into autophagosomal membranes [[13, [14]. Ubqln1 colocalizes and co-immunoprecipitates with LC3, and assists the maturation of autophagosomes to autolysosomes [[9, [10]. Nonetheless, there is no interaction between purified Ubqln1 and LC3 [[10], suggesting that there are extra proteins that mediate the association. The current work shows that Ubqln1 interacts with Ubqln4, Ubqln4 directly associates with LC3, and these associations are necessary and sufficient for Ubqln1 targeting to autophagosomes. Thus, one result of the expansion of the Ubqln family is that the unique isoform Ubqln4 can link the cargo-carrying isoform Ubqln1 to the autophagy machinery, expanding the roles for Ubqlns in protein degradation.
Results and Discussion
Ubqln1 interacts with Ubqln4
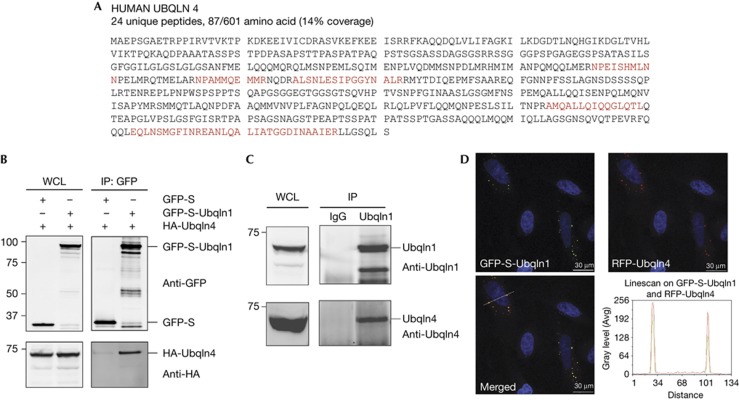
To gain a better understanding of Ubqln1’s function, we characterized proteins associated with Ubqln1 using tandem mass spectrometry. Ubqln4 specifically associated with Ubqln1, as judged by the enrichment of co-immunoprecipitated Ubqln4 peptides when Ubqln1 expression was increased by doxycycline (Fig 1A; supplementary Fig S1C online). Association of Ubqln4 with Ubqln1 was verified by co-immunoprecipitation of haemagglutinin (HA)-Ubqln4 and green fluorescent protein (GFP)-S-Ubqln1 (Fig 1B). To further characterize the Ubqln1–Ubqln4 interaction, antibodies were raised against human Ubqln1 (amino-acid residues 112–132) and human Ubqln4 (amino-acid residues 140–158); their specificities were confirmed using recombinant proteins (supplementary Fig S2 online). Consistent with the above data, endogenous Ubqln1 co-immunoprecipitated with endogenous Ubqln4 (Fig 1C). Thus, Ubqln1 interacts with Ubqln4.
Figure 1.
Ubqln1 interacts with Ubqln4. (A) Inducible GFP-S-Ubqln1 293 cell line was treated with 1 μg/ml of doxycycline for 24 h. GFP-S-Ubqln1 was immunoprecipitated and sent for mass spectrometry analysis. Red sequences indicate Ubqln4 peptides identified by mass spectrometry. (B) 293 cells were co-transfected with GFP-S or GFP-S-Ubqln1 and HA-Ubqln4. GFP was immunoprecipitated and analysed by western blot using anti-GFP and anti-HA antibodies. Lower molecular weight GFP-tag-containing bands that appear in the GFP-S-Ubqln1 lane might represent degradation products. (C) Endogenous Ubqln1 in 293 cells was immunoprecipitated using a rabbit Ubqln1 antibody or a control rabbit IgG. The immunoprecipitates were analysed by western blotting using anti-Ubqln1 and anti-Ubqln4 antibodies. (D) Confocal images of Hela cells transfected with GFP-S-Ubqln1 and RFP-Ubqln4. Linescan of the white line in the merged image indicates the intensities of the fluorescent molecules in the cross-section. GFP, green fluorescent protein; HA, haemagglutinin; IP, immunoprecipitation; Ubqln, Ubiquilin; WCL, whole-cell lysate.
Ubqln4, also known as UBIN or A1Up, was originally discovered through its binding to ataxin-1, a protein involved in spinocerebellar ataxia type 1 [[15]. In addition to ataxin-1, Ubqln4 regulates the proteasomal degradation of Cx43, a transmembrane protein that forms gap junction channels [[7]. Research on Ubqln4 has mainly focused on the role of Ubqln4 as a shuttle for specific cargo to the proteasome. As Ubqln1 has a role in autophagy and Ubqln4 interacts with Ubqln1, we next tested whether Ubqln4 also has a role in autophagy.
Ubqln4 binds LC3
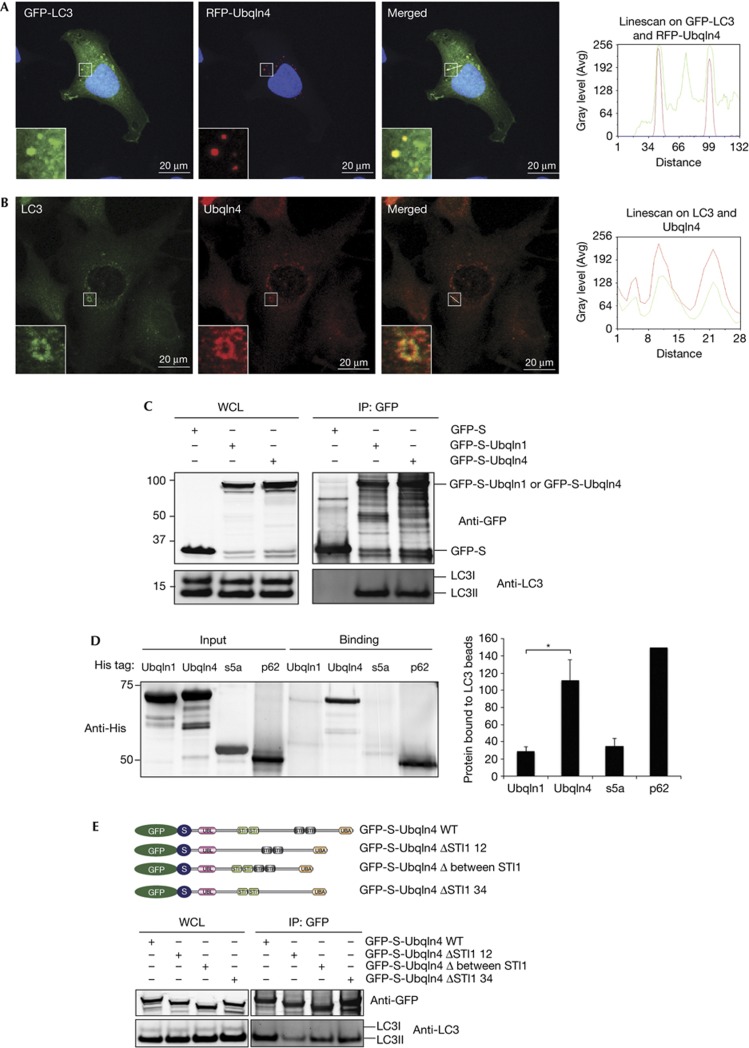
Ubqln1 appears in a punctate distribution, which reflects its association with autophagosomes [[9]. Co-expressed RFP-Ubqln4 colocalized essentially completely with GFP-S-Ubqln1 by immunofluorescence microscopy (Fig 1D), as 89±6% of GFP-Ubqln1 punctate structures also contained RFP-Ubqln4. As previously reported for Ubqln1, the Ubqln4 puncta colocalized with LC3 (Fig 2A,B), suggesting that it, too, colocalized with autophagosomes. 58±7% of RFP-Ubqln4-containing punctate structures also contained GFP-LC3. Like Ubqln1, Ubqln4 co-immunoprecipitated with LC3 (Fig 2C). Both Ubqln1 and Ubqln4 interact predominantly with the LC3II isoform found in autophagosomes (Fig 2C). Ubqlns might favour interaction with LC3II either because of its altered conformation after posttranslational modification or because it is highly concentrated on autophagosomes compared with the more diffuse LC3I. Purified recombinant Ubqln4 and p62 bound to human LC3 on agarose beads in the absence of other proteins, to a much greater extent than Ubqln1 or s5a. p62 was utilized as a positive control, as it binds to LC3 directly and s5a as a negative control, as it has no known role in autophagy (Fig 2D). When either the UBL or the UBA domain of Ubqln4 was deleted, co-immunoprecipitation with LC3 was unaffected, suggesting that the LC3-binding site in Ubqln4 resides in the central region of the protein (supplementary Fig S3A online). The LC3-interacting region (LIR) of p62 is composed of a highly conserved acidic cluster (DDD) and two hydrophobic amino-acid motifs W/Y–X–X–L/I [[16]. In the central region of Ubqln4, YNAL and YTDI sequences consistent with this motif can be found, but they lack the acidic cluster that is necessary for LC3 binding. Mutation of Y in each of these sequences to A did not affect LC3 binding, suggesting that these tetrapeptides are not within true LC3-binding motifs (supplementary Fig S3B online). To better localize the LC3-binding domain within Ubqln4, deletions in the central region of Ubqln4 were made. The only recognized motifs within the central region of Ubqln4 are four repeats of the STI1 motif, which is involved in the interaction of a stress-inducible protein with heat shock chaperones [[17]. The four STI1 motifs are organized into two groups with a long intervening sequence (Fig 2E). When either the intervening region between the STI1 motifs or the two STI1 domains near the C-terminus was deleted, co-immunoprecipitation with LC3 was unaffected (Fig 2E). However, deletion of the N-terminal STI1 repeats diminished the interaction of Ubqln4 and LC3 (Fig 2E). This suggests that these two STI1 repeats are needed for Ubqln4 binding to LC3, but the more C-terminal motifs are not.
Figure 2.
Ubqln4 interacts with LC3. Confocal images of (A) Hela cells transfected with GFP-LC3 and RFP-Ubqln4 and starved for 1 h or (B) Hela cells starved for 1 h and stained for endogenous LC3 and Ubqln4. The squares show colocalization. The insets show the enlargement of the indicated areas. Linescan of the white line in the merged image indicates the intensities of the fluorescent molecules in the cross-section. (C) 293 cells were transfected with GFP-S, GFP-S-Ubqln1 or GFP-S-Ubqln4 and treated with 50 μM chloroquine for 14 h. GFP was immunoprecipitated and analysed by western blot using anti-GFP and anti-LC3 antibodies. (D) Recombinant His-Ubqln1, His-Ubqln4, His-S5a or His-p62 was mixed with LC3 covalently coupled to agarose beads. The beads were washed, eluted and detected by western blot using anti-His antibody. The extent of Ubqln1, Ubqln4, s5a or p62 co-immunoprecipitation with LC3 was measured. Bars for Ubqln1, Ubqln4 and s5a represents the average and s.e.m. of four independent experiments. *P-value=0.0094. (E) 293 cells were transfected with GFP-S-Ubqln4 WT, GFP-S-Ubqln4 ΔSTI1 12, GFP-S-Ubqln4 Δ between STI1 or GFP-S-Ubqln4 ΔSTI1 34 and treated with 50 μM chloroquine for 14 h. GFP was immunoprecipitated and analysed by western blot using anti-GFP and anti-LC3 antibodies. GFP, green fluorescent protein; IP, immunoprecipitation; LC3, microtubule-associated protein light chain 3; Ubqln, Ubiquilin; WCL, whole-cell lysate; WT, wild-type.
Ubqln1 punctate distribution is dependent on Ubqln4
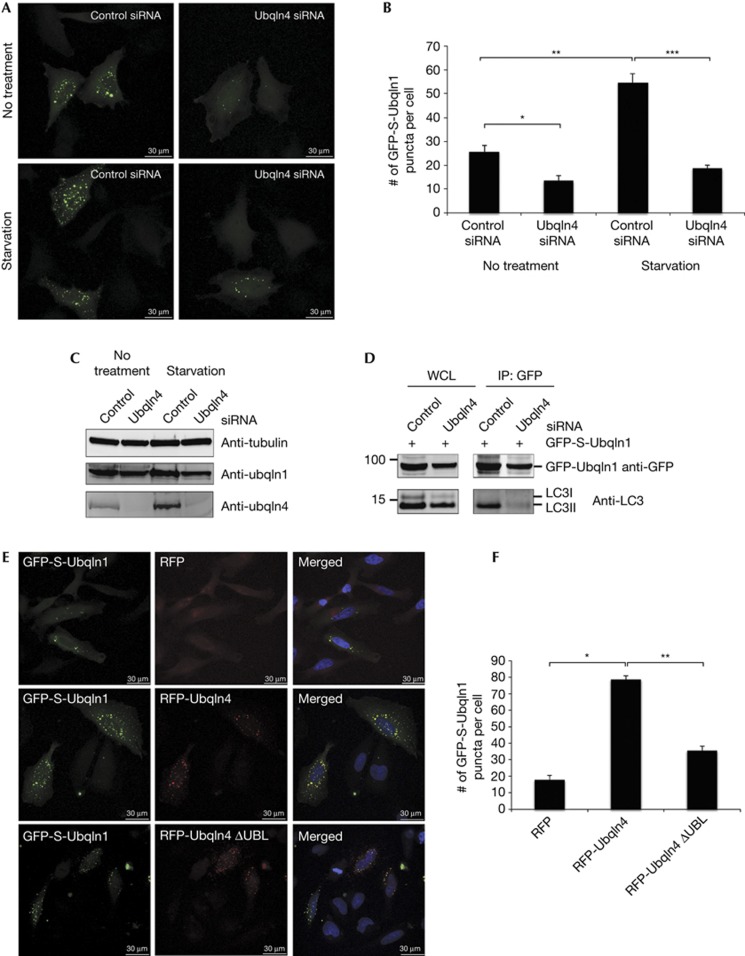
These data suggested the possibility that Ubqln4 is the missing component that mediates interaction of Ubqln1 with LC3. To determine whether Ubqln4 facilitates the interaction of Ubqln1 with LC3 and thus is involved in Ubqln1’s punctate distribution in cells, we used a pool of four short interfering RNA duplexes to silence the expression of Ubqln4; this depleted the cellular level to ∼90%, without any off-target effect on the expression of Ubqln1 (Fig 3C). In Hela cells transfected with control siRNA, about 30 Ubqln1 puncta per cell were observed (Fig 3A,B). The number of Ubqln1 puncta detected decreased by half in Ubqln4-depleted cells, suggesting that Ubqln4 has a role in the cellular distribution of Ubqln1 (Fig 3A,B). A similar requirement for Ubqln4 in generation of Ubqln1 puncta was observed in 293 cells (supplementary Fig S4 online). Consistent with a role for Ubqln4 in Ubqln1 association with autophagosomes, expression of RFP-Ubqln4 in Hela cells increased the number of Ubqln1 puncta to an average of 80 puncta per cell (Fig 3E,F). Starvation, a known initiator of autophagy, increased expression of Ubqln4 by about two-fold (1.9±0.05, n=4), while Ubqln1’s expression did not change with cell starvation (0.9±0.2-fold, n=4) (supplementary Fig S5 online). Thus, if Ubqln4 carries Ubqln1 to autophagosomes, the effect of its depletion by siRNA should be even greater in starved cells. During starvation, cells transfected with control siRNA doubled the number of Ubqln1 puncta to about 60/cell (Fig 3A,B). This increase was abrogated almost completely in Ubqln4-depleted cells. To verify this observation biochemically, we immunoprecipitated GFP-S-Ubqln1 and looked for its interaction with LC3 in the presence or absence of Ubqln4. In the absence of Ubqln4, we observed a decrease in LC3 association to Ubqln1 (Fig 3D). Thus, Ubqln4 mediates association of Ubqln1 with autophagosomes. Whether Ubqln1 and Ubqln4 first interact in the cytoplasm and then Ubqln4 carries the complex to the autophagosome, or Ubqln4 already resides on the autophagosome and then recruits Ubqln1 to its location still remains to be resolved.
Figure 3.
Ubqln1 punctate distribution is dependent on Ubqln4. (A) Confocal images of Hela cells transfected with GFP-S-Ubqln1 and also transfected with either control or Ubqln4 siRNA. The cells were either untreated or were transferred to starvation medium for 1 h. (B) Quantification of the number of GFP-S-Ubqln1 WT puncta per cell in (A). Each bar represents the average and s.e.m. of 30 cells per condition in three independent experiments. *P-value=0.011, **P-value=0.0005 and ***P-value=0.0001. (C) Depletion of Ubqln4 with siRNA in (A) was verified by western blot using anti-Ubqln1, anti-Ubqln4 and anti-tubulin antibodies. (D) Hela cells transfected with GFP-S-Ubqln1 and also transfected with either control or Ubqln4 siRNA were starved for 1 h. GFP was immunoprecipitated and analysed by western blot using anti-GFP and anti-LC3 antibodies. (E) Confocal images of GFP-S-Ubqln1 WT in Hela cells transfected with RFP, RFP-Ubqln4 or RFP-Ubqln4 ΔUBL. (F) Quantification of the number of GFP-S-Ubqln1 WT puncta per cell in (E). Each bar represents the average and s.e.m. of 30 cells per condition in three independent experiments. *P-value=0.0001 and **P-value=0.0003. GFP, green fluorescent protein; IP, immunoprecipitation; LC3, microtubule-associated protein light chain 3; UBL, ubiquitin-like; Ubqln, Ubiquilin; WCL, whole-cell lysate; WT, wild-type.
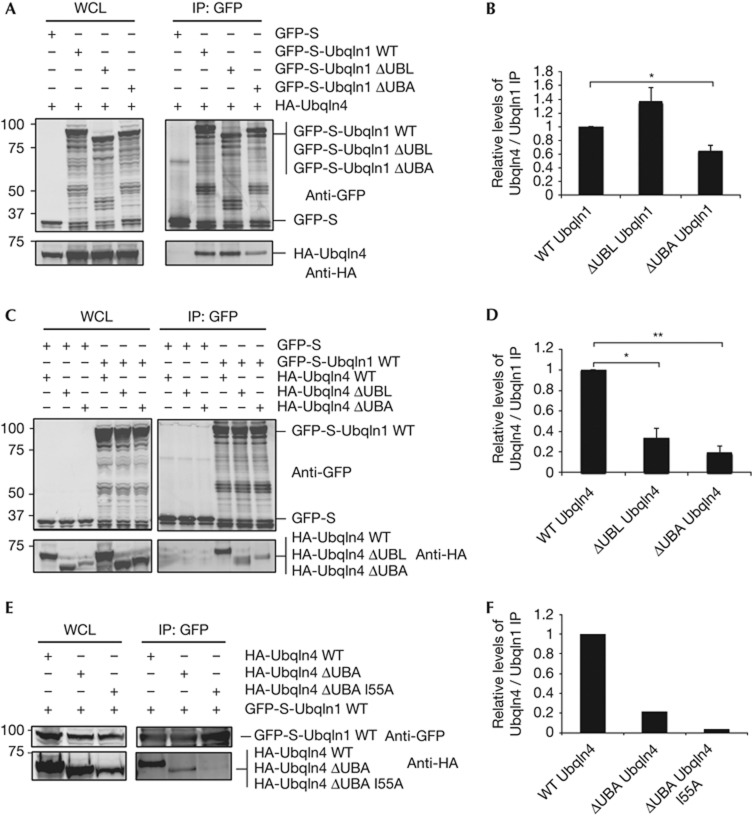
To further understand how Ubqln4 mediates Ubqln1 interaction with autophagosomes, the roles for the UBL and UBA domains of each isoform were determined. Deletion of the UBL domain in Ubqln1 had no effect on its interaction with Ubqln4; by contrast, deletion of the UBA domain decreased its association with Ubqln4 (Fig 4A,B). The requirement for the Ubqln1 UBA domain suggested that interaction might be mediated through interaction with the UBL domain of Ubqln4. Deletion of the Ubqln4 UBL domain decreased association with Ubqln1 to a similar extent as deletion of the Ubqln1 UBA domain (Fig 4C,D). All of the UBA domains that have been characterized so far contact a face on ubiquitin that includes I44 [[18]. The equivalent of I44 in Ubqln4’s UBL is I55. Mutation of I55 abolished Ubqln1–Ubqln4 binding, which suggests that the UBA of Ubqln1 binds the UBL domain of Ubqln4 in a manner very similar to UBA binding to ubiquitin (Fig 4E). When the equivalent I79 in Ubqln1 was mutated to alanine, the interaction of Ubqln1 and Ubqln4 was not disrupted (supplementary Fig S6B online). Deletion of the UBA domain of Ubqln4 also decreased interaction with Ubqln1 (Fig 4C,D), potentially because it mediates Ubqln4 homodimerization (supplementary Fig S6A online), as has been shown for several UBA domain-containing proteins, including Dsk2 [[19, [20, [21, [22]. If Ubqln4 mediates Ubqln1 interaction with autophagosomes, then ΔUBL Ubqln4, which has decreased ability to interact with Ubqln1, should also have a decreased ability to direct Ubqln1 into punctate structures. Compared with wild-type Ubqln4, expression of ΔUBL Ubqln4 only induced about half the number of Ubqln1 puncta (Fig 3E,F), about the same extent of reduction as the reduction in Ubqln1–Ubqln4 interaction (Fig 4C,D). Thus, the interaction between Ubqln1 and Ubqln4, dependent on the UBA domain of Ubqln1 and the UBL domain of Ubqln4, is required for appropriate localization of Ubqln1 to autophagosomes. This is the first evidence that interactions among different Ubqln isoforms has functional significance and that one Ubqln’s UBL domain can bind a different Ubqln’s UBA.
Figure 4.
Ubqln1 interacts with Ubqln4 through its UBA domain. (A) 293 cells were co-transfected with GFP-S-Ubqln1, GFP-S-Ubqln1 ΔUBL (deletion of UBL domain) or GFP-S-Ubqln1 ΔUBA (deletion of UBA domain) and HA-Ubqln4. GFP was immunoprecipitated and analysed by western blot using anti-GFP and anti-HA antibodies. (B) The extent of Ubqln4 co-immunoprecipitation with Ubqln1 was measured and the ratio of Ubqln4 to Ubqln1 was calculated for WT Ubqln1, Ubqln1ΔUBL and Ubqln1ΔUBA. The numbers represent the ratio of Ubqln4 to Ubqln1 relative to WT. Each bar represents the average and s.e.m. of four independent experiments. *P-value=0.0041. (C) 293 cells were co-transfected with GFP-S or GFP-S-Ubqln1 and HA-Ubqln4, HA-Ubqln4 ΔUBL or HA-Ubqln4 ΔUBA. GFP was immunoprecipitated and analysed by western blot using anti-GFP and anti-HA antibodies. (D) The extent of Ubqln4 co-immunoprecipitation with Ubqln1 was measured and the ratio of Ubqln4 to Ubqln1 was calculated for WT Ubqln4, Ubqln4ΔUBL and Ubqln4ΔUBA. The numbers represent the ratio of Ubqln4 to Ubqln1 relative to WT. Each bar represents the average and s.e.m. of four independent experiments. *P-value=0.0003 and **P-value=0.0001. (E) 293 cells were co-transfected with GFP-S-Ubqln1 and HA-Ubqln4, HA-Ubqln4 ΔUBA or HA-Ubqln4 ΔUBA I55A (I55 in the UBL domain of Ubqln4 was mutated to A). GFP was immunoprecipitated and analysed by western blot using anti-GFP and anti-HA antibodies. (F) The extent of Ubqln4 co-immunoprecipitation with Ubqln1 was measured and the ratio of Ubqln4 to Ubqln1 was calculated for WT Ubqln4, Ubqln4ΔUBA and Ubqln4ΔUBA I55A. The numbers represent the ratio of Ubqln4 to Ubqln1 relative to WT. Each bar represents the average of two independent experiments. GFP, green fluorescent protein; HA, haemagglutinin; IP, immunoprecipitation; UBA, ubiquitin-associated; UBL, ubiquitin-like; Ubqln, Ubiquilin; WCL, whole-cell lysate; WT, wild-type.
Consistent with this model, Ubqln1 lacking its UBA domain (Ubqln1ΔUBA) does not localize to autophagosomes [[9], presumably because the UBA domain of Ubqln1 is necessary for its interaction with Ubqln4. To determine whether this requirement can be bypassed, we introduced p62’s LIR [[16] into the central region of Ubqln1ΔUBA (Ubqln1ΔUBA/LIR). Ubqln1ΔUBA/LIR appeared in a punctate distribution (supplementary Fig S7A online), which colocalized with Ubqln1 (supplementary Fig S7B online). LC3 could be co-immunoprecipitated with Ubqln1 and Ubqln1ΔUBA/LIR, but not with Ubqln1ΔUBA (supplementary Fig S7D online). While autophagosome association of Ubqln1 is dependent on Ubqln4, depletion of Ubqln4 with siRNA had no effect on localization of Ubqln1ΔUBA/LIR (supplementary Fig S7C online).
Ubqln4 knockdown inhibits autophagosome acidification
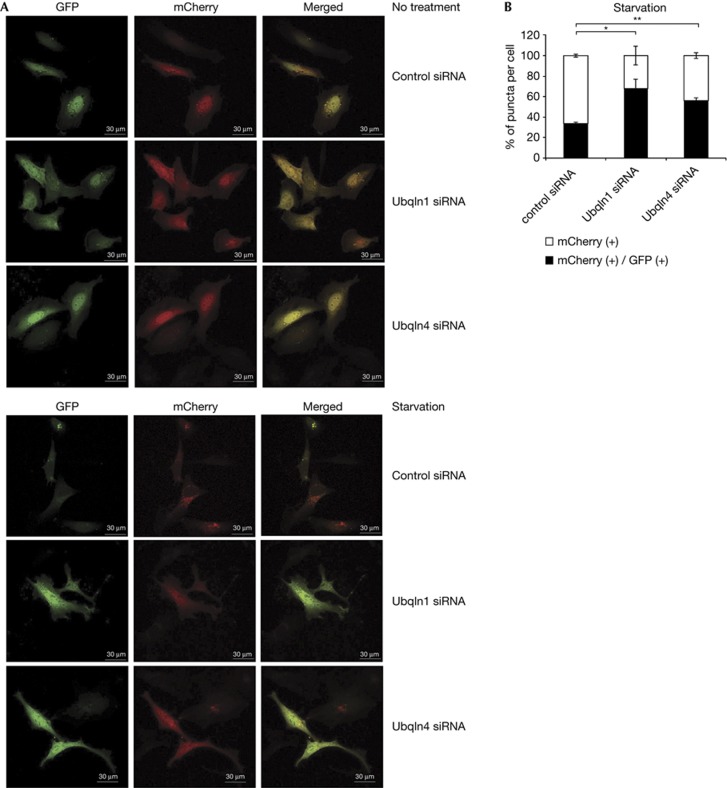
Ubqln1 depletion inhibits acidification of autophagosomes [[9]. This was shown using a LC3 construct fused to both GFP (acid-sensitive) and mCherry (acid stable) fluorophores. Initial LC3 puncta after initiation of autophagy show both GFP and mCherry fluorescence; as fusion occurs, the number of mCherry single-positive LC3 puncta increases because autophagosomal acidification inhibits GFP fluorescence. Depletion of Ubqln1 decreased the rate and extent of loss of LC3 GFP fluorescence, suggesting a role for Ubqln1 in autophagosome–lysosome fusion [[9]. To test whether depletion of Ubqln4 also delays autophagosome–lysosome fusion, Ubqln4 was depleted from cells expressing the dual fluorescent LC3. During normal basal conditions, only 1–3 double-positive puncta were observed, whether cells were transfected with control siRNA, Ubqln1 siRNA or Ubqln4 siRNA. In control cells after 8 h of starvation, the number of LC3 puncta increased, and 67% were mCherry single-positive, suggesting autophagosome acidification (Fig 5A,B). As previously shown, Ubqln1 siRNA significantly decreased the number of mCherry single-positive LC3 puncta (Fig 5A,B). Ubqln4 siRNA decreased the number of mCherry single-positive puncta to the same extent as depletion of Ubqln1 (Fig 5A,B). These functional data support the hypothesis that Ubqln4 mediates association of Ubqln1 with autophagosomes and regulates Ubqln1’s role in autophagosome–lysosome fusion. The current data cannot rule out that Ubqln4 has a role in autophagosome–lysosome fusion in addition to its recruitment of Ubqln1.
Figure 5.
Depletion of Ubqln4 inhibits autophagosome–lysosome fusion. (A) Confocal images of Hela cells transfected with mCherry-GFP-LC3 and also transfected with control, Ubqln1 or Ubqln4 siRNA. The cells were either untreated or were transferred to starvation medium for 8 h. (B) Quantification of the number of mCherry/GFP double-positive and mCherry single-positive puncta per cell in (A). Each bar represents the average and s.e.m. of 30 cells per condition in three independent experiments. The number of mCherry single-positive puncta of Ubqln1 and Ubqln4 siRNA was compared with that of control siRNA. *P-value=0.0475 and **P-value=0.0373. GFP, green fluorescent protein; LC3, microtubule-associated protein light chain 3; Ubqln, Ubiquilin.
As Ubqlns function both in proteasomal and autophagosomal degradation, one important question is how Ubqln targeting is regulated, that is, how do Ubqlns choose to direct their cargo to proteasomes or autophagosomes? We propose that the interaction between Ubqln1 and Ubqln4 might be the signal that redirects Ubqln1 and its ubiquitinated cargo from the proteasome to the autophagosome. Consistent with this model, the cellular level of Ubqln4 doubles during starvation, which is a potent signal for autophagy. This increase leads to a two-fold increase in the interaction between Ubqln1 and Ubqln4, with its potential to redirect Ubqln1 traffic to autophagosomes. As Ubqln4 also interacts with Ubqln2 (unpublished data), it is possible that Ubqln4 might also act as an adapter between Ubqln2 and its effectors. Further studies will be needed to determine the mechanism by which the concentration of Ubqln4 protein increases during starvation and to determine whether Ubqln1–Ubqln4 interaction is the signal that redirects Ubqln1 from the proteasome pathway towards autophagosomes. One further implication of our work is that while the most prominent role for Ubqln1 and Ubqln2 might be binding to ubiquitinated proteins, this might be less important for Ubqln4, which acts instead as an adapter for the cargo-bearing Ubqlns to new degradation pathways.
In summary, this work has demonstrated a new interaction between two members of the Ubqln family that is necessary for completion of autophagy. Interactions among members of this intriguing family likely are important in regulation of several mechanisms of protein degradation in cells.
Methods
See supplementary materials and methods for full details.
Cell culture. 293 and Hela cells were grown in Dulbecco modified Eagle medium+10% fetal bovine serum, 10 mM HEPES, 100 μM MEM non-essential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin and 0.29 mg/ml L-glutamine. Inducible GFP-S-Ubqln1 293 cell line was created and maintained as described previously [[23].
siRNA transfection. Cells were transfected with 50 nM standard siGenome siRNA oligos (Dharmacon) using Oligofectamine (Life Technologies).
Mass spectrometry. Reduced and S-alkylated protein complexes were fractionated by SDS–PAGE and the entire lane cut into bands for trypsin digestion in situ. The resulting peptides were analysed by capillary LC-tandem mass spectrometry in a data-dependent experiment [[24]. Tandem mass spectra were searched against a database of human proteins using the Mascot programme and filtered to a false discovery rate of <1%.
Immunoprecipitation. Immunoprecipitation with anti-GFP antibody beads was described previously [[23]. For immunoprecipitation of endogenous proteins, lysates were first mixed with rabbit IgG or rabbit anti-Ubqln antibody for 2 h at 4 °C, before addition of protein A agarose for 2 h at 4 °C.
Interaction of recombinant proteins. His-tagged proteins were purified using nickel-nitrilotriacetic acid resin and stored in 25 mM Tris pH 7.5, 300 mM NaCl and 10% glycerol. Equal aliquots of human LC3 covalently coupled to agarose beads (Boston Biochem) were incubated with recombinant His-ubqln1, His-ubqln4, His-s5a (Boston Biochem) or His-p62 (Biotang Inc.) at 4 °C for 4 h. Beads were washed 5 × with NP40 buffer and eluted with 1 × NuPAGE sample buffer.
Western blotting. Proteins transferred from SDS–PAGE gels to polyvinylidene difluoride membranes were visualized with the Odyssey system (LI-COR Biotechnology).
Immunofluoresence. Images were obtained with a Leica SPE laser scanning confocal microscope with 63 × /1.30 CS oil objective. Leica LAS AF software (Leica, Germany) and Metamorph were utilized for analysis.
Statistical analysis. Statistical significance was determined by unpaired, two-tailed t-tests.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We express our gratitude to Donald Kirkpatrick for comments on the manuscript. We thank Nicholas Lewin-koh for helpful input in statistical analysis.
Author Contributions: D.L. designed and performed the experiments, interpreted the data and wrote the manuscript. D.A. performed the mass spectrometry experiment. E.J.B. interpreted the data and edited the manuscript.
Footnotes
The authors are all employees of Genentech Inc.
References
- Lee DY, Brown EJ (2012) Ubiquilins in the crosstalk among proteolytic pathways. Biol Chem 393: 441–447 [DOI] [PubMed] [Google Scholar]
- Ko HS, Uehara T, Tsuruma K, Nomura Y (2004) Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett 566: 110–114 [DOI] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, Glickman MH, Fushman D (2009) Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell 36: 1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Raasi S, Fushman D (2008) Affinity makes the difference: nonselective interaction of the UBA domain of ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol 377: 162–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM (2005) Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol 12: 708–714 [DOI] [PubMed] [Google Scholar]
- Li X, Su V, Kurata WE, Jin C, Lau AF (2008) A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. J Biol Chem 283: 5748–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM (2000) The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell 6: 409–419 [DOI] [PubMed] [Google Scholar]
- N'Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ (2009) PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep 10: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, Fang S, Cuervo AM, Nixon RA, Monteiro MJ (2010) Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet 19: 3219–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T (2004) Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun 313: 453–458 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E (2004) LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 36: 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson JD, Riley B, Burright EN, Duvick LA, Zoghbi HY, Orr HT (2000) Identification and characterization of an ataxin-1-interacting protein: A1Up, a ubiquitin-like nuclear protein. Hum Mol Genet 9: 2305–2312 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Lassle M, Blatch GL, Kundra V, Takatori T, Zetter BR (1997) Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J Biol Chem 272: 1876–1884 [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Isogai S, Morimoto D, Arita K, Unzai S, Tenno T, Hasegawa J, Sou YS, Komatsu M, Tanaka K, Shirakawa M et al. (2011) Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J Biol Chem 286: 31864–31874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Peschard P, Zimmerman B, Lin T, Moldoveanu T, Mansur-Azzam N, Gehring K, Park M (2007) Structural basis for UBA-mediated dimerization of c-Cbl ubiquitin ligase. J Biol Chem 282: 27547–27555 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Funakoshi M, Endicott JA, Kobayashi H (2005) Budding yeast Dsk2 protein forms a homodimer via its C-terminal UBA domain. Biochem Biophys Res Commun 336: 530–535 [DOI] [PubMed] [Google Scholar]
- Tse MK, Hui SK, Yang Y, Yin ST, Hu HY, Zou B, Wong BC, Sze KH (2011) Structural analysis of the UBA domain of X-linked inhibitor of apoptosis protein reveals different surfaces for ubiquitin-binding and self-association. PloS One 6: e28511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JZ, Miller JJ, Jackson PK (2009) High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 9: 2888–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana NE, McCutcheon K, Pham VC, Harden K, Nguyen A, Young J, Adams C, Schroeder K, Arnott D, Bafna V et al. (2011) Resurrection of a clinical antibody: template proteogenomic de novo proteomic sequencing and reverse engineering of an anti-lymphotoxin-alpha antibody. Proteomics 11: 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.