Abstract
Trimethylation of histone 3 lysine 9 (H3K9me3) is a marker of repressed transcription. Cells transfected with mutant isocitrate dehydrogenase (IDH) show increased methylation of histone lysine residues, including H3K9me3, due to inhibition of histone demethylases by 2-hydroxyglutarate. Here, we evaluated H3K9me3 and its association with IDH mutations in 284 gliomas. H3K9me3 was significantly associated with IDH mutations in oligodendrogliomas. Moreover, 72% of World Health Organization grade II and 65% of grade III oligodendrogliomas showed combined H3K9me3 positivity and 1p19q co-deletion. In astrocytic tumors, H3K9me3 positivity was found in all grades of tumors; it showed a significant relationship with IDH mutational status in grade II astrocytomas but not in grade III astrocytomas or glioblastomas. Finally, H3K9me3-positive grade II oligodendrogliomas but not other tumor subtypes showed improved overall survival compared to H3K9me3-negative cases. These results suggest that repressive trimethylation of H3K9 in gliomas may occur in a context-dependent manner and is associated with IDH mutations in oligodendrogliomas, but may be differently regulated in high-grade astrocytic tumors. Further, H3K9me3 may define a subset of grade II oligodendrogliomas with better overall survival. Our results suggest variable roles for IDH mutations in the pathogenesis of oligodendrogliomas vs. astrocytic tumors.
Keywords: Astrocytoma, Glioma, H3K9me3, Histone methylation, Isocitrate dehydrogenase (IDH), Oligodendroglioma
INTRODUCTION
Epigenetic mechanisms such as DNA methylation and histone methylation play a significant role in the pathogenesis and progression of tumors. DNA methylation has been extensively studied in glioblastomas (1–3). In contrast, histone modifications in glioma pathogenesis have remained poorly understood. Recent studies have emerged implicating histone modifications in adult gliomas (4–6) and histone gene mutations in pediatric glioblastomas (7, 8). Histones are proteins that form octamers consisting of H2A, H2B, H3 and H4 around which DNA is wrapped. Histones have amino acid tails that are subject to various modifications such as acetylation, methylation, phosphorylation, ubiquitination and SUMOylation of arginine (R) and lysine (K) residues, resulting in changes in DNA function and transcription (9). Histone lysine methylation can be an activator or suppressor of transcription depending on the histone residue that is methylated. For example, methylation of H3K4, H3K36 and H3K79 is associated with activation of transcription, whereas methylation of H3K9, H3K27 and H4K20 is associated with silencing of transcription (9, 10).
Histone lysine methylation is regulated by histone lysine methyltransferases (KMTs) and demethylases (KMDs). One of the earliest KMTs to be characterized was SUV39H. SUV39H belongs to the SET domain family and specifically methylates H3K9(11). Subsequently, H3K9 may be the most extensively studied site of histone methylation (12). Trimethylation of H3K9 (H3K9me3) is associated with pericentric heterochromatin formation, transcriptional repression and DNA methylation (13–15).
Very little is known about H3K9 modifications in glioma pathogenesis. H3K9me3 is increased in cells expressing mutant isocitrate dehydrogenase 1 (IDH1 R132H) (4, 5, 16). The NADP-positive dependent enzyme IDH is mutated in ~70% of World Health Organization (WHO) grade II and grade III gliomas and secondary glioblastomas (17–19). Mutations in IDH result in neomorphic activity of the enzyme catalyzing the production of the oncometabolite 2-hydroxyglutarate (2-HG) from α-ketoglutarate (α-KG) (20, 21). 2-HG is a structural analogue of α-KG and can inhibit several α-KG-dependent oxygenases (22, 23). The α-KG-dependent oxygenases form a large family of enzymes that influence various cellular functions, such as collagen modifications, carnitine synthesis, hypoxic sensing and histone and DNA demethylation (24). For example, KDM4C (belonging to the Jumonji C family of KDMs) uses α-KG, oxygen and Fe(II) as cofactors to demethylate histone lysine residues. Immortalized human astrocytes or murine neurosphere cultures overexpressing IDH1 R132H show increases in H3K9me3 compared to cells overexpressing wild type IDH1 (4, 16)l, which is mediated in part by the inhibition of KDM4C by 2-HG (4). Similarly, heterozygous knock-in of IDH1R132H in HCT116 colon cancer cell lines results in increased H3K9me3 (5).
Based on these observations we hypothesized that IDH mutant gliomas would show increased H3K9me3. The goals of this study were to evaluate the relationship between H3K9me3 and IDH mutations in gliomas of different subtypes and grades, and to determine if H3K9me3 had any prognostic significance in gliomas. The results suggest that the effect of IDH mutations on histone methylation may differ between oligodendrogliomas and high-grade astrocytomas and may provide insight into the role played by mutant IDH in the pathogenesis of these tumors.
MATERIALS AND METHODS
Case Material
Cases were obtained from the University of California San Francisco, the University of Pennsylvania, the University of Pittsburgh, Children’s Hospital of Philadelphia and Memorial Sloan-Kettering Cancer Center (MSKCC) following approval from the respective institutional review boards. All cases were de-identified prior to analysis and included both whole tissue and tissue microarray sections (Supplemental Digital Content 1, Methods, http://links.lww.com/NEN/A432).
The cohort demographics consisted of a total of 284 gliomas (Table 1). The tumors consisted of 263 adult gliomas (53 WHO grade II oligodendrogliomas, 47 grade III anaplastic oligodendrogliomas, 6 grade I pilocytic astrocytomas (supratentorial), 28 grade II diffuse astrocytomas, 43 grade III anaplastic astrocytomas and 86 grade IV glioblastomas) and 21 grade I pediatric pilocytic astrocytomas (14 cerebellar and 8 supratentorial). Each case was reviewed individually by a neuropathologist at each institution and then confirmed by a neuropathologist at MSKCC. Loss of heterozygosity (LOH) data for 1p and 19q loci was available in 51/53 of grade II and 40/47 grade III oligodendrogliomas. In oligodendrogliomas, 84% (43/51) of grade II and 75% (30/40) of grade III contained co-deletions of chromosomal arms 1p and 19q.
Table 1.
Patient Demographics and Number of Cases Positive for Trimethylation of Histone 3 Lysine 9 or Bearing Isocitrate Dehydrogenase Mutations in Gliomas
| Tumor type and World Health Organization Grade | Total | Age | Sex | H3K9me3 | IDH mutant | ||
|---|---|---|---|---|---|---|---|
| n | Mean | Median | M | F | Total positive n (%) | Total positive n (%) | |
| Oligodendroglioma II | 53 | 44 | 42 | 33 | 20 | 44 (83) | 46 (87) |
| Oligodendroglioma III | 47 | 46 | 45 | 24 | 23 | 37 (79) | 41 (87) |
| Pilocytic Astrocytoma I | 27 | 14 | 11 | 15 | 12 | 13 (48) | 0 (0) |
| Diffuse Astrocytoma II | 28 | 48 | 44 | 14 | 14 | 22 (78) | 20 (71) |
| Astrocytoma III | 43 | 49 | 45 | 25 | 18 | 29 (67) | 24 (56) |
| Primary Glioblastoma IV | 70 | 58 | 59 | 44 | 26 | 55 (78) | 0 (0) |
| Secondary Glioblastoma IV | 16 | 41 | 40 | 10 | 6 | 13 (81) | 16 (100) |
Patients were stratified according to H3K9me3 positivity (n indicates number of H3K9me3 positive (+) or negative (−) cases for each tumor subtype). The log-rank (Mantel-Cox) test with 95% confidence intervals was used to determine differences in overall survival.
M = male; F= female. IDH, isocitrate dehydrogenase; H3K9me3, trimethylation of histone 3 lysine 9.
Immunohistochemistry and Scoring
Immunohistochemical studies were performed as previously published (4, 25, 26). In brief, immunohistochemical detection was performed using the Discovery XT processor (Ventana Medical Systems, Tucson, AZ). Tissue sections were blocked for 30 minutes in 10% normal goat serum in 2% BSA in PBS. Sections were incubated for 5 hours with 0.1 μg/ml of the rabbit polyclonal anti-H3K9me3 (ab8898, Abcam, Cambridge, MA) or mouse monoclonal anti-IDH1 R132H (DIA H09, Dianova, Hamburg, Germany; 1:30) antibodies. Tissue sections were then incubated for 60 minutes with biotinylated goat anti-rabbit IgG (PK6101, Vector Laboratories, Burlingame, CA) or goat anti-mouse IgG (Vector, BA9200) at 1:200 dilution. Blocker D, Streptavidin- HRP and DAB detection kit (Ventana Medical Systems) were used according to the manufacturer instructions.
Whole mount mouse embryos were used as positive controls for H2K9me3 (27). H3K9me3 was also assessed in normal cortical adult post mortem brain tissue obtained from 4 cases. H3K9me3 in all of those cases was negative in glia and in endothelial cells but showed scattered nuclear staining in neurons (Figure, part A, Supplemental Digital Content 2, http://links.lww.com/NEN/A433). Two individuals scored H3K9me3 and IDH1 R132H as positive or negative in a blinded manner. Only nuclear staining for H3K9me3 in tumor cells was considered positive. Preservation of antigenicity in tumor sections that were negative for H3K9me3 was confirmed by retained neuronal nuclear staining in surrounding brain tissues as an internal control when available for evaluation (Figure, part C, Supplemental Digital Content 2, http://links.lww.com/NEN/A433). Sections stained for IDH1 R132H were scored positive or negative based on described criteria (25, 28, 29).
IDH1/2 Mutational Analyses
Genomic DNA was extracted from formalin-fixed paraffin-embedded blocks from 2 10-μm-thick slices using the Formapure kit (Agencourt, Beverly, MA) in a 96-well format, using a modified version of the manufacturers’ method, and in a semi-automated fashion. IDH1/2 mutational analyses were performed on cases that were IDH1 R132H-negative using previously described protocols (26, 30). Due to limitations of tissue availability, cases from the tissue microarray containing glioblastomas and pilocytic astrocytomas were not available for mutational analyses.
Statistical Analysis
Statistical analyses were performed in consultation with the MSKCC biostatistics core. SPSS (version 21, IBM, Chicago, IL) and Prism (version 5, La Jolla, CA) were used to analyze data. Fisher exact test was used to determine the odds ratios to evaluate the strength of association between IDH mutational status and H3K9me3 positivity or the association between 1p-19q deletion and H3K9me3 positivity in oligodendrogliomas. The log-rank (Mantel-Cox) test was used to examine the association of H3K9me3 positivity on overall survival within each tumor subtype. Multivariate survival analysis was performed using the Cox proportional hazards model. Differences were considered significant when p < 0.05 (95% confidence intervals).
RESULTS
H3K9me3 Is Associated with IDH Mutations in Oligodendrogliomas
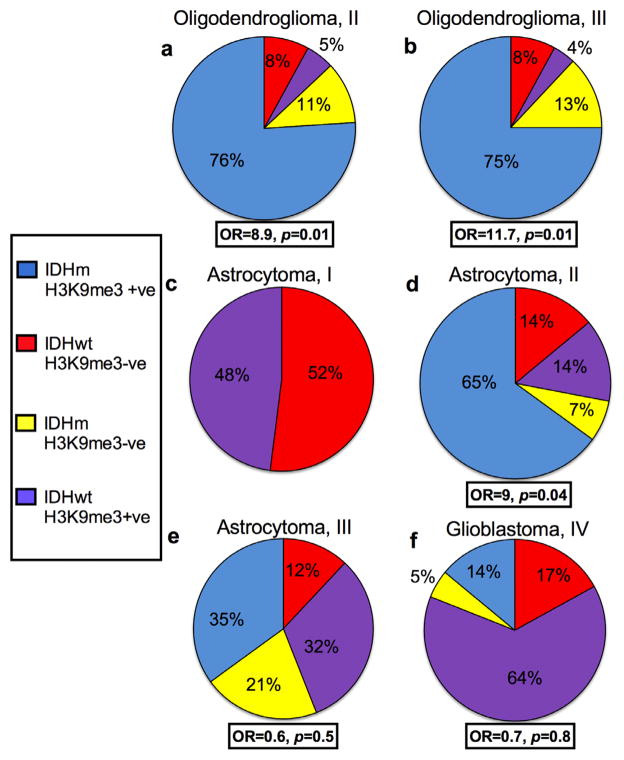
Positive H3K9me3 nuclear staining in gliomas was seen in 83% (44/53) of grade II oligodendrogliomas and 79% (37/47) of grade III oligodendrogliomas (Table 1; Fig. 1). There was a significant relationship between H3K9me3 positivity and IDH mutations in grade II oligodendrogliomas (76% [40/53] of cases with IDH mutations and positive for H3K9me3, odds ratio 8.9, p = 0.01), and grade III oligodendrogliomas (75% [35/47] of cases with IDH mutations and positive for H3K9me3, odds ratio 11.7, p = 0.01) (Figs. 1, 2).
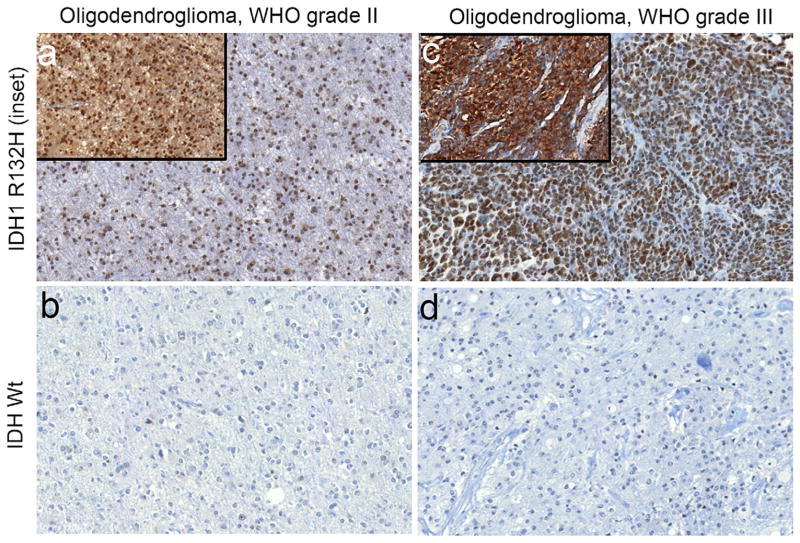
Figure 1.
Histone 3 lysine 9 trimethylation (H3K9me3) is associated with isocitrate dehydrogenase 1/2 (IDH1/2) mutations in oligodendrogliomas. H3K9me3 immunohistochemistry was assessed in World Health Organization (WHO) grade II oligodendrogliomas and grade III anaplastic oligodendrogliomas. IDH1 R132H mutational status was assessed using immunohistochemistry (insets) in a blinded manner and compared to H3K9me3 staining in each case. Cases that were negative for IDH1 R132H were sequenced for other IDH1/2 mutations. (a) Representative images from an IDH1R132H mutant (inset, 20x) grade II oligodendroglioma that was positive for H3K9me3 (10X). (b) Representative image from an IDH wild type grade II oligodendroglioma that was negative for H3K9me3 (10x). (c) Representative images from an IDH1R132H (inset, 20x) mutant grade III oligodendroglioma that was positive for H3K9me3 (10X). (d) Representative image from an IDH wild type grade III oligodendroglioma that was negative for H3K9me3 (10x).
Figure 2.
Histone 3 lysine 9 trimethylation (H3K9me3) is associated with isocitrate dehydrogenase 1/2 (IDH1/2) mutant oligodendrogliomas but not high-grade astrocytic tumors. H3K9me3 positivity was compared with IDH1/2 mutational status. Data were analyzed using Fisher exact test with 95% confidence intervals to evaluate the odds ratios reflective of the strength of association between H3K9me3 and IDH mutational status in (a) World Health Organization grade II oligodendrogliomas, (b) grade III oligodendrogliomas, (c) pilocytic astrocytomas, (d) grade II astrocytomas, (e) grade III astrocytomas and (f) glioblastomas. OR = odds ratio; +ve = positive; −ve = negative; IDHm = mutant IDH; IDHwt = wild type IDH.
H3K9me3 in Astrocytic Tumors
H3K9me3 positivity was as follows in astrocytic gliomas: 48% (13/27) of pilocytic astrocytomas (Figure, part B, Supplemental Digital Content 2, http://links.lww.com/NEN/A433), 78% (22/28) of diffuse astrocytomas, 67% (29/43) of anaplastic astrocytomas, and 79% of glioblastomas (68/86) (Table 1; Figs. 2, 3). As expected, all pilocytic astrocytomas were IDH1 R132H-negative. H3K9me3 showed a significant association with IDH mutations in diffuse astrocytomas (65% (18/28) of cases with IDH mutations and positive for H3K9me3, odds ratio 9, p = 0.04 (Figs. 2, 3). No significant relationship between IDH mutations and H3K9me3 positivity was observed in anaplastic astrocytomas or glioblastomas (Figs. 2, 3).
Figure 3.
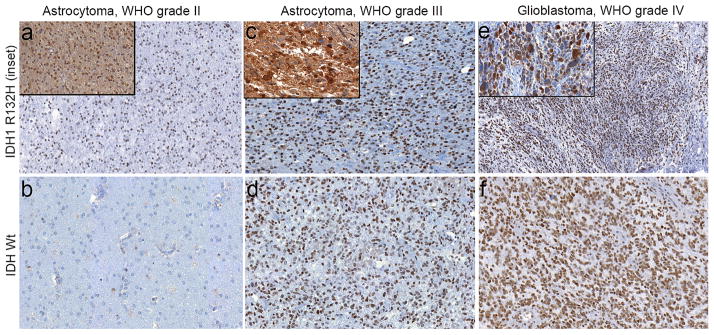
Histone 3 lysine 9 trimethylation (H3K9me3) immunostaining patterns in astrocytic tumors. H3K9me3 immunohistochemistry was assessed in World Health Organization (WHO) grade II astrocytomas, grade III astrocytomas, and glioblastomas. Isocitrate dIDH1R132H mutational status was assessed using immunohistochemistry in a blinded manner and compared to H3K9me3 staining in each case. Cases that were negative for IDH1R132H were sequenced for other IDH1/2 mutations. (a) Representative images from an IDH1R132H mutant (inset, 20x) grade II astrocytoma that was positive for H3K9me3 (10x). (b) Representative image from an IDH wild type grade II astrocytoma that was negative for H3K9me3 (10x). (c) Representative images from an IDH1R132H mutant (inset, 20x) grade III astrocytoma that was positive for H3K9me3 (10x). (d) Representative image from an IDH wild type grade III astrocytoma that was positive for H3K9me3 (10x). (e) Representative images from an IDH1R132H mutant (inset, 20x) glioblastoma that was positive for H3K9me3 (10x). (f) Representative image from an IDH wild type glioblastoma that was positive for H3K9me3 (10x).
H3K9me3 Is More Frequent but not Significantly Associated with LOH of Chromosomes 1p and 19q in Oligodendrogliomas
Loss of heterozygosity information for 1p-19q was available in 46/53 of grade II and 40/47 grade III oligodendrogliomas (Table, Supplemental Digital Content 3, http://links.lww.com/NEN/A434). 1p-19q co-deletions showed a strong correlation with IDH mutational status (grade II oligodendrogliomas: odds ratio = 41, p = 0.0001 and grade III oligodendrogliomas: odds ratio = 28.5, p = 0.01). Two grade II oligodendrogliomas had co-deletions but did not bear IDH mutations. All grade III anaplastic oligodendrogliomas with co-deletions bore IDH mutations. H3K9me3 positivity and 1p-19q co-deletions were noted in 72% (37/51) of grade II and 65% (26/40) grade III oligodendrogliomas. H3K9me3 was positive in 12% (6/51) of grade II and 17% (7/40) of grade III oligodendrogliomas with intact 1p-19q. H3K9me3 was negative in 14% (7/51) of grade II and 10% (4/40) of grade III oligodendrogliomas with 1p-19q co-deletions (Fig. 4; Table, Supplemental Digital Content 3, http://links.lww.com/NEN/A434). However, Fisher exact test did not reveal a statistically significant relationship between H3K9me3 and loss of 1p-19q in either grade II (odds ratio = 0.8, p = 1) or grade III (Odds ratio=2.8, p=0.3) oligodendrogliomas (Table, Supplemental Digital Content 4, http://links.lww.com/NEN/A435).
Figure 4.
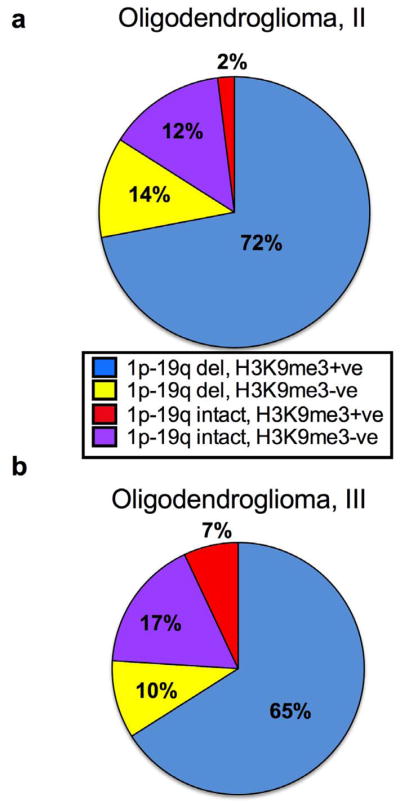
Histone 3 lysine 9 trimethylation (H3K9me3) is more frequent but not significantly associated with loss of heterozygosity of chromosomes 1p and 19q in oligodendrogliomas. H3K9me3 positivity was compared in oligodendrogliomas with and without 1p-19q loss of heterozygosity. (a) World Health Organization grade II oligodendrogliomas. (b) Grade III oligodendrogliomas. +ve = positive; −ve = negative.
H3K9me3 Is Associated with Improved Overall Survival in Grade II Oligodendrogliomas
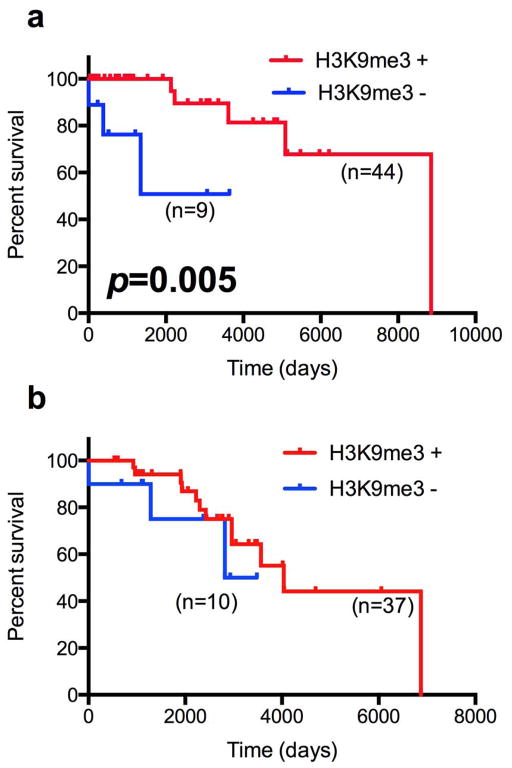
Overall survival was improved in H3K9me3 positive grade II oligodendrogliomas vs. their H3K9me3-negative counterparts (Log-rank (Mantel-Cox) test, hazards ratio = 5.9, p = 0.005) (Fig. 5a; Table 2). Multivariate analysis using the Cox proportional hazards model showed that H3K9me3 positivity was a significant prognostic factor (n = 53, p = 0.015), but age, sex, or IDH mutational status were not (Table, Supplemental Digital Content 4, http://links.lww.com/NEN/A435). 1p-19q LOH showed a trend towards improved overall survival (p = 0.052) (Table, Supplemental Digital Content 4, http://links.lww.com/NEN/A435). No significant difference in survival was noted in grade III oligodendrogliomas (Log-rank (Mantel-Cox) test, Hazards ratio = 1.6, p = 0.4, Fig. 5b; Table 2). No significant associations were observed in overall survival between H3K9me3-positive and -negative gliomas, regardless of grade in astrocytic tumors (Table 2).
Figure 5.
Histone 3 lysine 9 trimethylation (H3K9me3) is associated with improved overall survival in World Health Organization (WHO) grade II oligodendrogliomas. Patients with grade II or grade III anaplastic oligodendrogliomas were stratified according to H3K9me3 positivity (red = H3K9me3-positive, blue = H3K9me3-negative). Percent survival was graphed using Kaplan Meier curves and analyzed using the Log-rank (Mantel-Cox) test with 95% confidence intervals. (a) Grade II oligodendrogliomas. (b) Grade III anaplastic oligodendrogliomas.
Table 2.
H3K9me3 Is Associated with Improved Overall Survival in World Health Organization Grade II Oligodendrogliomas
| Tumor type and World Health Organization grade | N | Median survival (d) | Log-rank test | ||
|---|---|---|---|---|---|
| H3K9me3-positive | H3K9me3-negative | H3K9me3-positive | H3K9me3-negative | p value | |
| Oligodendroglioma II | 44 | 9 | 8852 | Undefined** | 0.005 |
| Oligodendroglioma III | 37 | 10 | 4037 | 4846 | 0.4 |
| Astrocytoma II | 21 | 6 | 1613 | Undefined** | 0.2 |
| Astrocytoma III | 29 | 14 | 1430 | Undefined** | 0.3 |
| Glioblastoma IV | 67 | 19 | 517 | 332 | 0.4 |
Percent survival in this group at the end of the study was greater than 50% and was thus undefined by the statistical software.
DISCUSSION
IDH mutations are seen in ~70% of intermediate grade gliomas. How mutations in IDH contribute to brain tumor development is not known. Mutant IDH catalyzes the production of 2HG from α-KG (20, 21). 2HG is structurally similar to α-KG and inhibits α-KG dependent dioxygenases, such as the Jumonji C family of histone demethylases, resulting in increased histone methylation marks including H3K9me3 in vitro (4, 22, 23). We sought to determine the relationship between H3K9me3 and IDH mutations in gliomas. We found significant associations between the 2 factors in oligodendrogliomas and grade II astrocytomas. While 67% of anaplastic astrocytomas and 78% of glioblastomas showed H3K9me3 positivity, there was no significant association with IDH mutations. Finally, H3K9me3 showed a beneficial survival effect in grade II oligodendrogliomas but not in grade III oligodendrogliomas or in any grade of astrocytic tumors.
Because IDH mutations are closely associated with 1p-19q co-deletions (18, 31–33), we considered whether H3K9me3 was associated with 1p-19q deletions in oligodendrogliomas. Indeed, 72% of grade II and 65% of grade III oligodendrogliomas that contained 1p-19q deletions were H3K9me3-positive. This association was not, however, statistically significant, suggesting that 1p-19q deletions may not directly regulate histone methylation. Recurrent mutations in the CIC gene (homolog of the Drosophila gene capicua) on chromosome 19q and FUBP1 gene (FUSE binding protein 1) on chromosome 1p have been recently described in oligodendrogliomas (34, 35). These proteins are thought to influence the RAS/MAP-kinase pathway (CIC) and MYC activation (FUBP1) and thus may not directly methylate histones (34).
H3K9me3-positive grade II oligodendrogliomas, but not other glioma subtypes showed significantly improved overall survival. Future studies with a larger cohort are needed to confirm this observation but the prognostic significance of H3K9me3 needs to be interpreted with caution. While H3K9me3 showed a significant relation with IDH mutations in oligodendrogliomas and grade II astrocytomas, we did not observe a one-to-one correspondence. For example, 7% of diffuse astrocytomas, 11% of grade II oligodendrogliomas and 13% of anaplastic oligodendrogliomas were IDH mutant but H3K9me3-negative. Conversely, 14% of diffuse astrocytomas, 5% of grade II oligodendrogliomas and 4% of anaplastic oligodendrogliomas were H3K9me3-positive but IDH wild type. We thus did not expect the prognostic value of IDH mutations to entirely translate to H3K9me3. One of the limitations of this study is that we are restricted to assessing H3K9me3 by immunohistochemistry due to tissue availability. Nevertheless, these observations reflect the complexity of the pathogenesis of IDH mutant gliomas. Multiple mechanisms downstream of mutant IDH and 2-HG may contribute to the pathogenesis with H3K9me3 representing one piece of the puzzle (36). Nevertheless, the finding that trimethylation of H3K9 is differentially regulated in oligodendrogliomas and high-grade astrocytomas is a significant observation. It suggests that mutant IDH may have different roles in oligodendrogliomas vs. high-grade astrocytomas; thus, it may provide clues for understanding the pathogenesis of these tumors.
We found significant H3K9me3 positivity in anaplastic astrocytomas and glioblastomas without any significant associations with IDH mutations. Similarly, H3K9me3 exhibited no prognostic associations in these tumors. Methylation of H3K9 is regulated by a complex dynamic interplay involving the enzymatic activity of KMTs and KDMs. It is possible that the activity of these enzymes is deregulated in higher-grade astrocytomas and glioblastomas by means of a yet undefined process that is independent of the effect of mutant IDH. Further, DNA methylation can promote histone methylation and vice versa. IDH mutations result in DNA methylation and reprograming the methylome to the glioma-CpG island methylation phenotype (G-CIMP), which could in turn influence H3K9 methylation (16). DNA methyltransferases can also influence histone methylation (37–39). Because Dnmt1 and Dnmt3 are deregulated in glioblastomas (40, 41), it is possible that aberrant activity of these enzymes mediate trimethylation of H3K9 in an IDH mutant/ 2-HG independent manner. The data suggest the hypothesis that H3K9 can be trimethylated in high-grade astrocytomas and glioblastomas in both mutant IDH1-2HG dependent and independent manner. Furthermore, other epigenetic marks such as H3K27me3 may be altered in these tumors. Future experiments examining this question will address this issue.
A high frequency of positivity of H3K9me3 was observed in all subtypes or grades of gliomas including pilocytic astrocytomas. Pilocytic astrocytomas show alterations in BRAF (42), which is known to alter the methylome in melanomas and colon cancer (43, 44). Further, melanomas bearing the BRAF(V600E) mutation show amplification of SETDB1, a H3K9 KMT resulting in increased H3K9me3 (45). It is possible that BRAF alterations contribute to H3K9me3 in pilocytic astrocytomas. To date, few studies have examined H3K9 modifications in glioma pathogenesis or glial development. H3K9me3 is thought to play a role in differentiation (46–48). Neurosphere cultures overexpressing mutant IDH1 R132H show increased H3K9me3 accompanied by a decrease in glial fibrillary acidic protein expression compared to cells overexpressing wild type IDH1, suggesting that H3K9me3 may contribute to repressing glial differentiation in IDH mutant gliomas (4).
Histone methylation remains poorly understood in gliomas and may be vital to further our understanding of the pathology of gliomas. We hypothesize that repressive trimethylation of H3K9 can occur in gliomas in IDH mutant dependent or independent fashion. Repressive H3K9 trimethylation may be regulated differently in oligodendroglial and astrocytic tumors and may be associated with IDH mutations in oligodendrogliomas, but may be regulated by other, yet undefined mechanisms in astrocytic tumors. This difference in H3K9 trimethylation could represent one of the ways by which IDH mutations play different roles in oligodendrogliomas and astrocytomas. Moreover, IDH mutations show differential associations with other genetic alterations in oligodendrogliomas vs. astrocytomas. IDH mutations in oligodendrogliomas are associated with 1p19q codeletions, while in astrocytic tumors IDH mutations are closely associated with ATRX and TP53 mutations (8, 49–52). ATRX is a SWI/SNF chromatin remodeler and plays a role in deposition of histone variant H3.3 (53). How IDH mutations, histone methylation and ATRX mutations contribute to the pathogenesis of gliomas is not known. The interplay between these complex genetic alterations and histone methylation in oligodendrogliomas and astrocytomas are avenues for future experimentation.
Supplementary Material
Acknowledgments
We thank the MSKCC cytology core facility, the Geoffrey Bene Translational core and Jianan Zhang for expert technical assistance and Elyn Riedel from the MSKCC biostatistics core for help with data analysis. We thank Dennis Pozega, Patrick Ward and Tullia Lindsten for critical reading of the manuscript.
This work was supported in part by grants from the NCI and NIH to C.B.T. C.B.T is co-founder of Agios Pharmaceuticals and has financial interest in Agios.
Footnotes
The authors of this study declare no other potential conflicts of interest.
References
- 1.Dubuc AM, Mack S, Unterberger A, et al. The epigenetics of brain tumors. Methods Mol Biol. 2012;863:139–53. doi: 10.1007/978-1-61779-612-8_8. [DOI] [PubMed] [Google Scholar]
- 2.Kreth S, Thon N, Kreth FW. Epigenetics in human gliomas. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.04.008. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Martinez R, Esteller M. The DNA methylome of glioblastoma multiforme. Neurobiol Dis. 2010;39:40–6. doi: 10.1016/j.nbd.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 4.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan CG, Barwick BG, Jin G, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 2012;22:2339–55. doi: 10.1101/gr.132738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W, Xia Y, Hawke D, et al. PKM2 Phosphorylates Histone H3 and Promotes Gene Transcription and Tumorigenesis. Cell. 2012;150:685–96. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu G, Broniscer A, McEachron TA, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–3. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 9.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 10.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rea S, Eisenhaber F, O’Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 12.Krauss V. Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica. 2008;133:93–106. doi: 10.1007/s10709-007-9184-z. [DOI] [PubMed] [Google Scholar]
- 13.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan S, Horowitz S, Trievel RC. Structure and function of histone H3 lysine 9 methyltransferases and demethylases. Chembiochem. 2011;12:254–63. doi: 10.1002/cbic.201000545. [DOI] [PubMed] [Google Scholar]
- 16.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleeker FE, Lamba S, Leenstra S, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 20.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhury R, Yeoh KK, Tian YM, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol. 2008;4:152–6. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 25.Capper D, Zentgraf H, Balss J, et al. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118:599–601. doi: 10.1007/s00401-009-0595-z. [DOI] [PubMed] [Google Scholar]
- 26.Gorovets D, Kannan K, Shen R, et al. IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res. 2012;18:2490–501. doi: 10.1158/1078-0432.CCR-11-2977. [DOI] [PubMed] [Google Scholar]
- 27.Henckel A, Nakabayashi K, Sanz LA, et al. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum Mol Genet. 2009;18:3375–83. doi: 10.1093/hmg/ddp277. [DOI] [PubMed] [Google Scholar]
- 28.Capper D, Sahm F, Hartmann C, et al. Application of mutant IDH1 antibody to differentiate diffuse glioma from nonneoplastic central nervous system lesions and therapy-induced changes. Am J Surg Pathol. 2010;34:1199–204. doi: 10.1097/PAS.0b013e3181e7740d. [DOI] [PubMed] [Google Scholar]
- 29.Capper D, Weissert S, Balss J, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20:245–54. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horbinski C, Kofler J, Kelly LM, et al. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues. J Neuropathol Exp Neurol. 2009;68:1319–25. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe T, Nobusawa S, Kleihues P, et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–53. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27:5743–50. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 33.van den Bent MJ, Dubbink HJ, Marie Y, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 34.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–5. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yip S, Butterfield YS, Morozova O, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol. 2012;226:7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih AH, Levine RL. IDH1 mutations disrupt blood, brain, and barriers. Cancer Cell. 2012;22:285–7. doi: 10.1016/j.ccr.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 37.Rai K, Nadauld LD, Chidester S, et al. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol Cell Biol. 2006;26:7077–85. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esteve PO, Chin HG, Smallwood A, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rai K, Jafri IF, Chidester S, et al. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J Biol Chem. 2010;285:4110–21. doi: 10.1074/jbc.M109.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fanelli M, Caprodossi S, Ricci-Vitiani L, et al. Loss of pericentromeric DNA methylation pattern in human glioblastoma is associated with altered DNA methyltransferases expression and involves the stem cell compartment. Oncogene. 2008;27:358–65. doi: 10.1038/sj.onc.1210642. [DOI] [PubMed] [Google Scholar]
- 41.Kreth S, Thon N, Eigenbrod S, et al. O-methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One. 2011;6:e17156. doi: 10.1371/journal.pone.0017156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horbinski C. To BRAF or Not to BRAF: Is That Even a Question Anymore? J Neuropathol Exp Neurol. 2013;72:2–7. doi: 10.1097/NEN.0b013e318279f3db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou P, Liu D, Dong J, et al. The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells. Cell Cycle. 2012;11:286–95. doi: 10.4161/cc.11.2.18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karpinski P, Ramsey D, Grzebieniak Z, et al. The CpG island methylator phenotype correlates with long-range epigenetic silencing in colorectal cancer. Mol Cancer Res. 2008;6:585–91. doi: 10.1158/1541-7786.MCR-07-2158. [DOI] [PubMed] [Google Scholar]
- 45.Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–7. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohmann F, Loureiro J, Su H, et al. KMT1E mediated H3K9 methylation is required for the maintenance of embryonic stem cells by repressing trophectoderm differentiation. Stem Cells. 2010;28:201–12. doi: 10.1002/stem.278. [DOI] [PubMed] [Google Scholar]
- 47.Strobl-Mazzulla PH, Sauka-Spengler T, Bronner-Fraser M. Histone demethylase JmjD2A regulates neural crest specification. Dev Cell. 2010;19:460–8. doi: 10.1016/j.devcel.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Liu H, Liu J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2012;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 49.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–22. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kannan K, Inagaki A, Silber J, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower-grade glioma. Oncotarget. 2012;3:1194–203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu XY, Gerges N, Korshunov A, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol. 2012;124:615–25. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen DN, Heaphy CM, de Wilde RF, et al. Molecular and morphologic correlates of the alternative lengthening of telomeres phenotype in high-grade astrocytomas. Brain Pathol. 2012 doi: 10.1111/j.1750-3639.2012.00630.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ratnakumar K, Bernstein E. ATRX: The case of a peculiar chromatin remodeler. Epigenetics. 2012;8 doi: 10.4161/epi.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.