Abstract
Rhabdomyosarcoma, an aggressive malignant neoplasm that shows features of skeletal muscle, is the most common soft tissue tumor of childhood. In children, the major subtypes are embryonal and alveolar. Although localized disease responds to a multimodal treatment, the prognosis for patients with high-risk features and metastasis remains dismal. Several in vivo models of rhabdomyosarcoma have been developed in mouse, human xenografts, zebrafish and Drosophila to better understand the underlying mechanisms governing malignancy. The findings so far have indicated the potential role of skeletal muscle precursor cells in malignant transformation. To better understand histogenesis and different aspects of tumorigenesis in rhabdomyosarcoma, we have previously developed a robust zebrafish model of kRAS-induced rhabdomyosarcoma, which shares morphologic and immunophenotypic features with the human counterpart. Cross-species mircroarray comparisons confirm that conserved genetic pathways drive rhabdomyosarcoma growth. The ease for ex vivo manipulation allows development of different transgenic and co-injection strategies to induce tumor formation in zebrafish. In contrast to other vertebrate model systems, the tumor onset in zebrafish is short, allowing for efficient study of different tumor processes including tumor growth, self-renewal, and maintenance.
Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue tumor in the pediatric population and displays phenotypic and biological features of embryonic skeletal muscle. RMS occurs in 4.6/million U.S. children under 15 years of age (Gurney et al., 1996). It falls into two major groups in children - embryonal and alveolar. Embryonal rhabdomyosarcoma constitutes the most common subtype, accounting for about 60% of childhood cases. Treatment for rhabdomyosarcoma is multimodal including surgical resection, chemotherapy and radiation. Alveolar rhabdomyosarcoma is often more aggressive than the embryonal subtype; however, the prognosis for patients with high-risk features or metastasis remains dismal regardless of subtypes.
Alveolar rhabdomyosarcoma (ARMS) is characterized by a (2;13) translocation in the majority of cases and a (1;13) translocation in a smaller subset of cases. These translocations juxtapose the 5′ DNA-binding domains of PAX3 or PAX7 genes on chromosomes 2 and 1, respectively, with the transactivation domain at the 3′ portion of FKHR gene on chromosome 13. Translocations generate chimeric PAX3/FKHR and PAX7/FKHR fusion genes that act as novel oncogenic transcription factors (Barr et al., 1992, Barr et al., 1993, Galili et al., 1993; Davis et al., 1994). Fusion gene-negative alveolar rhabdomyosarcoma comprise approximately 15% of the ARMS subtype and are clinically and molecular indistinguishable from embryonal rhabdomyosarcoma (Williamson et al., 2010). Embryonal rhabdomyosarcoma (ERMS) has frequent allelic loss in chromosomal region 11p15 (Koufos et al., 1985; Scrable et al., 1987), with this genetic interval harboring a number of imprinted genes implicated in oncogenesis, such H19, IGF2 and p57 kip2. Activating mutations of RAS genes are common in embryonal rhabdomyosarcoma with ~25% of patients harboring activating point mutations in either H-RAS, K-RAS, or N-RAS (Stratton et al., 1989; Chen et al., 2006). Moreover, cross-species comparisons of human and RAS-induced ERMS in zebrafish revealed that the RAS pathways is active in a majority of human ERMS, but the mechanism leading to constitutive RAS signaling has yet to be elucidated (Langenau et al, 2007). Activated RAS signaling has also been shown to block myogenic differentiation through down regulation of myogenic factors such as MyoD and myogenin (Lassar et al., 1989; Konieczny et al., 1989). Identification of germ-line mutations of H-RAS in Costello syndrome, a genetic disorder that predisposes affected individuals to embryonic tumors including embryonal rhabdomyosarcoma further supports the role of H-RAS in the pathogenesis of rhabdomyosarcoma. Other significant genetic alterations identified in both embryonal and alveolar rhabdomyosarcoma include p53 mutations and N-myc amplification (Mulligan et al., 1990; Stratton et al., 1990; Felix et al., 1992;). Taken together, it is clear that RAS pathway activation is important for tumor onset in ERMS while other genetic pathways likely aid in the multi-step progression to full malignancy.
Several murine models of RMS have been reported in the literature (Table 1). In one murine model, a combination of Her2/neu oncogene activation and p53 inactivation leads to the development of embryonal rhabdomyosarcoma (Nanni et al, 2003). In another murine model, simultaneous loss of Ink4a/Arf function and activation of c-Met signaling in mice transgenic for hepatocyte growth factor/scatter factor (HGF/SF) induces embryonal rhabdomyosarcoma (Sharp et al., 2002). Keller et al (2004) used the conditional knock-in strategy to generate a murine alveolar RMS model, by inducing expression of PAX3-FKHR gene in the skeletal muscle using a Myf6-Cre mouse line. However, the expression of PAX3-FKHR alone did not induce malignancy, and additional inactivation of the Ink4a/Arf and Trp53 pathways was necessary to induce ARMS. These models indicate that functional impairment of p53 is essential for development of rhabdomyosarcoma because Arf acts via p53, although additional genetic hits are necessary for tumorigenesis. Tsumura et al. (2006) generated a knock-in mouse line with oncogenic K-ras, conditionally activated by Cre/LoxP system in either heterozygous or homozygous for p53 knockout animals. Electroporation of Cre expression vector in skeletal muscle resulted in the generation of RMS in adults with tumor incidences of 100% at 10 weeks and 40% at 15 weeks, in p53 −/− and p53 −/+ backgrounds, respectively. The tumor has the morphologic features of pleomorphic rhabdomyosarcoma with haphazardly arranged large, round and pleomorphic cells and positive expression of myogenic markers on immunohistochemistry. Thus, K-ras and p53 deficiency cooperate to induce the development of pleomorphic rhabdomyosarcoma when targeted to late stage myoblast populations. Finally, the Hedgehog/Patched signaling pathway has also been shown to play a role in the pathogenesis of rhabdomyosarcoma. Rhabdomyosarcoma has been shown to develop in a subset of Pathced1 (Ptch) heterozygous mutant mice (Calzada-Wack et al., 2002). The study by Kapler et al. (2004) showed that RMS developed in Ptch1 heterozygous mutant mice are less aggressive and more differentiated than when compared to RMS developing from p53 heterozygous mutant mice. In addition, cDNA microarray analysis revealed distinct expression profiles in two groups of tumors, indicating that different causative mutations can lead to distinct gene expressions correlating with morphologic variations in RMS. In addition, it has also been shown that heterozygous Ptch1 mutation and p53 deficiency collaborate to hasten the onset to RMS formation in mice (Lee et al., 2007).
Table 1.
Animal models of rhabdomyosarcoma
| Species | Approach | Histologic Subtype |
Tumor Onset | References |
|---|---|---|---|---|
| Zebrafish | Mosaic Transgenic Approach (rag2- kRASG12D injection at 1-cell stage) |
Embryonal | 10 days post- fertilization |
Langenau et al., 2007 |
| Zebrafish | Heat Shock-inducible Cre-Lox approach (beta-actin-LoxP- EGFP-pA-LoxP- kRASG12D line × hsp70-Cre line) |
Embryonal | Heatshocked: Average 35 days Non heatshocked: Average 65 days |
Le et al., 2008 |
| Zebrafish | Tol2 gene trap system with transgene expressing constitutively active human H-RASV12 |
Costello syndrome |
60 to 365 days | Santoriello et al., 2009 |
| Mouse | Conditional Pax3:Fkhr knock-in/Myf6- specific Cre driver(Pax3:Fkhr heterozygous lines) Pax3:Fkhr homozygous lines × Trp53 −/+ × Trp 53 −/− × Ink4a/Arf −/− |
Alveolar --------------→ --------------→ --------------→ |
383 days 202 days 75-1 days 56-89 days |
Keller et al., 2004 |
| Mouse | HGF/SF-transgenic x Ink4a/Arf −/− |
Embryonal | ~90 days | Sharp et al., 2002 |
| Mouse | Cre-Loxp conditional krasG12V knock in x p53 −/− |
Pleomorphic | ~30-70 days | Tsumura et al., 2006 |
| Human Tumor Xenografts in Mice |
Cell lines derived from normal human skeletal muscle precursor cells (SkMCs) or human myoblasts (HSMM) stably expressing SV40 T/t-Ag (T), hTERT, H-Ras (V12G); transformed tumor cells were applied to xenograft assays |
Embryonal | SkMC: ~30-77 days (SQ injection) HSMM: 14 days (SQ); 56 days (SQ) |
Linardic et al., 2005 |
| Mouse | Conditionally regulated expression of an activated allele (Smoothened (R26- SMOM2)) × Cre transgene (CAGGS- CreER) induced by tamoxifen |
Embryonal | 35 days | Mao et al., 2006 |
Linardic et al. generated a genetic model of human rhabdomyosarcoma (2005). Two types of primary human skeletal muscle cells, primitive fetal skeletal muscle precursors (SkMC) and human postnatal skeletal myoblasts (HSMM), were transformed with defined genetic elements to deregulate the p53, Rb, Myc, telomerase and Ras pathways. The study showed that SkMC-derived tumors showed a range of phenotypes with most developing sarcomas having unclassified morphology based on H&E analysis with some evidence of skeletal muscle differentiation based on positive expression of myogenic markers. In contrast, HSMM-derived tumors showed morphologic and immunophenotypic features resembling the embryonal-type rhabdomyosarcoma. Electron microscopy also demonstrated evidence of skeletal muscle differentiation with formation of myofilaments. The study further showed the invasive and metastatic nature of the tumor. These data suggest that ERMS requires multiple genetic pathways to be disrupted to convert normal muscle into RMS including those regulating the p53, RB, Myc, telomerase and Ras pathways. HSMMs can be induced to undergo malignant transformation into rhabdomyosarcoma, supporting the utility of this genetic model in dissecting the molecular mechanisms underlying human disease.
Generation of murine models is expensive, time consuming, requires complex breeding schemes requiring multiple generations to develop reproducible models of RMS, multiple genetic perturbations are often required, and RMS commonly develops only after a long latency (Table 1). In contrast, zebrafish are amenable to large-scale genetic screens due to their fecundity, easy ex vivo manipulation from embryonic stage to adulthood, short latency for tumor development, and biological and pathologic similarity to human malignancies. We have developed a robust zebrafish transgenic model of kRAS-induced rhabdomyosarcoma (Langenau et al., 2007). Morphologically, zebrafish rhabdomyosarcoma resembles features of the human counterpart, showing spindle cell morphology and occasional nests of primitive round blue cells, recapitulating different stages of embryonic muscle development. The presence of cross striations in some tumor cells further supports that ERMS cells are blocked in myogenic differentiation. Similar to human rhabdomyosarcoma, zebrafish tumors express myogenic markers that are commonly used in to diagnose human disease including desmin, myoD and myogenin. This chapter gives an overview of different approaches utilized to induce zebrafish rhabdomyosarcoma, to identify tumor-propagating ERMS cell subpopulations, and to uncover conserved pathways in cancer using genomic resources and bioinformatics approaches.
Rationale
As in most types of human sarcomas, the cell of origin remains to be determined in rhabdomyosarcoma. As animal models have indicated that cells involved in skeletal muscle development play a role in the pathogenesis of rhabdomyosarcoma, the advantages of the zebrafish models of rhabdomyosarcoma can be utilized to investigate the histogenesis and molecular mechanisms underlying the disease. This chapter gives an overview of different approaches to generate rhabdomyosarcoma. Zebrafish rhabdomyosarcoma is highly similar to human embryonal rhabdomyosarcoma and using fluorescent transgenic approaches, we can visualize tumor onset, image tumor growth over time, and study the tumor self-renewal processes.
Material and Methods
I. Generation of Zebrafish Rhabdomyosarcoma
Mosaic Transgenic Approach
The rag2 promoter is expressed in T- and B- lymphoid progenitor cells, olfactory rosettes and sperm (Jesse et al., 2001; Langenau et al., 2003). A transgenic model using the rag2 promoter to drive expression of the mouse c-Myc gene results in the development of T-cell acute lymphoblastic leukemia (Langenau et al., 2003). We have subsequently shown that rag2 is also expressed in the mononuclear component of the skeletal muscle, which is composed of satellite cells and early myoblasts (Langenau et al., 2007). The rag2 promoter is not active in terminally differentiated multinucleated muscle fibers.
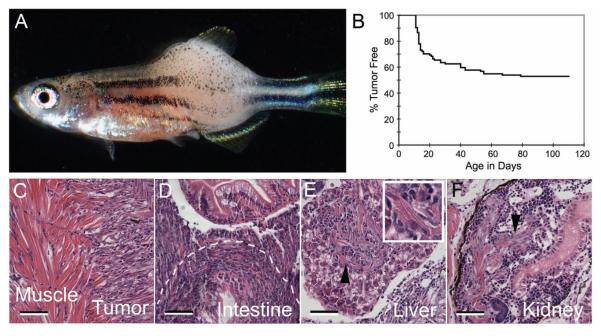
Mosaic transgenic zebrafish that are predisposed to developing RAS-induced ERMS can be created using the following method. First, the human kRASG12D is amplified by PCR and cloned into the rag2-GFP vector (Jessen et al., 2001; Langenau et al., 2003). The rag2- kRASG12D construct was linearized, phenol/chloroform extracted and ethanol precipitated. The DNA was resuspended in 0.5x Tris-EDTA buffer + 100 mM KCl and was injected into wild-type embryos (i.e. AB-strain) at the one-cell stage of development. For most applications, 60-120 ng/microliter of DNA can be microinjected. Animals were monitored for tumor onset beginning at 10 days of life and a subset of mosaic zebrafish containing the rag2-kRASG12D transgene develop externally visible tumors (Figure 1A). In our experience, animals begin to develop ERMS by 10 days post-fertilization (dpf, Figure 1B) and nearly 40% of mosaic transgenic zebrafish develop tumors by 80 dpf (dpf). ERMS can also be labeled with green fluorescence by coinjection of rag2-kRASG12D and rag2-GFP. In this instance, the two transgenes co-segregate and co-express in the developing tumor; allowing ERMS cells to be tracked by fluorescent imaging using a dissecting microscope (Figure 1). Finally, ERMS is the major type of tumor detected in mosaic transgenic animals. Only one mosaic animal has been identified that developed a lymphoid hyperplasia/leukemia following injection of rag2-kRASG12D into one- cell stage animals, with >500 animals analyzed to date.
Figure 1.
Mosaic transgenic approaches to develop RAS-induced embryonal rhabdomyosarcoma. rag2-kRASG12D was microinjected into one cell stage fish and animals were monitored for tumor onset over time. (A) Bright-field image of a 30-day-old zebrafish with ERMS. (B) Tumor onset in AB strain fish injected with rag2-kRASG12D. (C-F) ERMS cells invade into adjacent muscle (C), intestine (D), liver (E), and kidney (F). Scale bars are 50 microns.
Generation of Stable Transgenic Lines
Costello syndrome is caused by germ-line mutations in the H-Ras gene and is characterized by craniofacial dysmorphology, cardiac defects and tendency to develop various types of neoplasia, including rhabdomyosarcoma and neuroblastoma. To model Costello syndrome in zebrafish, Santoriello et al (2009) utilized the Tol2 gene trap system (Kawakami et al., 2000) to generate a stable transgenic line carrying a constitutively active form of human Ras, HRASV12, tagged with GFP. The authors observed that fish carrying the integrated transgene showed several features of Costello syndrome including craniofacial dysmorphology, short stature, and development of tumor. In older fish (5-12 months old), the transgenic group showed a higher frequency of tumor formation (n = 12/200) compared with the wild-type controls (1 in 500). Tumor types observed include rhabdomyosarcoma (n = 1), hepatocellular carcinoma (n = 2), metastatic melanoma (n = 2) and gut carcinoma (n = 1). Taken together, this zebrafish model of Costello syndrome is powerful tool for studying the constellation of abnormities associated with this syndrome. However, as other tumor types can develop in this model and that RMS develops only after a long-latency with low penetrance, it may be difficult to discover RMS-specific pathways associated with tumor onset using these transgenic lines.
Heat Shock-inducible Cre-Lox approaches to induce rhabdomyosarcoma in zebrafish
Le et al. (2007) developed a heat shock-inducible Cre-Lox approach to conditionally induce the expression of activated human kRASG12D. Briefly, to express activated human kRASG12D in various tissue types, a zebrafish beta-actin promoter was used to drive expression of a floxed transgene containing enhanced green fluorescent protein (EGFP) cassette that is followed by a transcriptional stop signal and located upstream of the constitutively active human kRASG12D. B-actin-LoxP-EGFP-pA-LoxP-kRASG12D stable transgenic lines were bred to the heat shock inducible Cre (hsp70-Cre) line. Because the hsp70 promoter is leaky during early larval development, a portion of cells recombine the locus and of those that develop tumors, ~40% develop ERMS while numerous other lesions are also seen. By contrast, embryos heat shocked at 37O C from 4 to 5 hpf or 24-25 hpf exhibited high levels of Cre expression without compromising embryonic development and genomic recombination was significantly enhanced when compared to non-heatshocked sibling controls. 80% tumor-bearing animals developed rhabdomyosarcoma in the heat-shocked cohort. Overall, the heat-shocked group showed increased tumor incidence and earlier tumor onset compared to the non-heat shocked group. Although this model provides a nice methodology to rapidly induce RMS in stable transgenic animals, these tumors do not morphologically resemble human RMS, nor have detailed analysis been undertaken to assess the molecular similarity to human disease. Moreover, other tumor cell types are evident in double transgenic animals potentially confounding analysis of RMS in these animals.
II. Identification of genetic modifiers of cancer initiation and maintenance
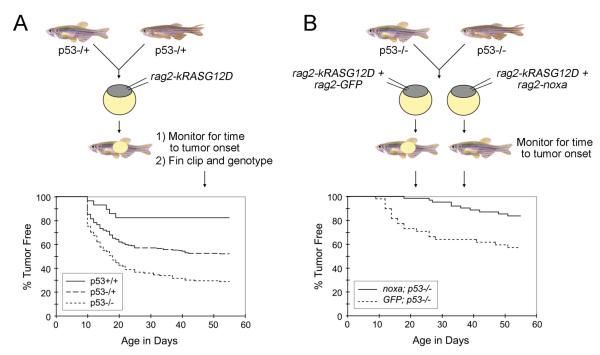
P53 pathway disruption is common in ERMS. To determine whether disruption of this genetic pathway can affect tumor initiation, p53-/+ heterozygous mutant zebrafish were uncrossed (Berghmans et al., 2005) and the resulting one-cell stage animals injected with rag2- kRASG12D (Langenau et al. 2007). Animals were monitored for externally visible tumor masses beginning at day 10 of life and a subset of animals were sacrificed, fixed, and sectioned to confirm tumor morphology. All animals were genotyped at the end of the experiment using an allele specific genomic PCR assay (Berghmans et al., 2005) and time to tumor onset was plotted by the method of Kaplan-Meier. Heterozygous and homozygous p53 loss-of-function fish displayed a marked increase in tumor incidence compared with wild-type injected siblings (Figure 2A). In addition, homozygous p53 loss-of-function animals developed more tumors compared with heterozygous p53 siblings. These findings indicate that p53 loss-of-function collaborates with kRASG12D to induce rhabdomyosarcoma and establishes that coinjection approaches can be used to rapidly identify tumor-modifying events that are relevant to human disease. Importantly, these methods do not require multiple generations to introduce your transgenic model into a various genetic backgrounds, but instead the modifying gene effects can be assessed directly by microinjection into various mutant lines.
Figure 2.
Assessing collaborating genetic events that modify time to ERMS onset in mosaic transgenic zebrafish. A) Schematic demonstrating injection of oncogenic rag2-kRASG12D into embryos with p53 alterations. P53 loss accelerates time to tumor onset and overall numbers of animals that develop ERMS. B) Schematic demonstrating injection strategy to determine the effect of noxa on p53 pathway in kRAS-induced zebrafish rhabdomyosarcoma. In these experiments, tumor onset is assessed by the development of externally visible tumors and then animals from each genotype are compared by Kaplan-Meier survival analysis (bottom). Noxa suppresses tumor onset in p53-deficient animals.
The co-injection technique can also be utilized to identify genetic modifiers of tumor initiation. Langenau et al. (2008) previously showed that noxa, a pro-apoptotic BH3-only protein and a direct transcriptional target of p53, is a potent suppressor of tumor initiation. Specifically, wild-type AB-strain animals injected with rag2-kRASG12D and rag2-noxa showed longer latency in tumor onset compared to animals injected with rag2-kRASG12D and rag2- dsRED2. Tumor onset was also assessed in p53-LOF animals co-injected with rag2-kRASG12D and either rag2-noxa or rag2-GFP. Again, noxa functioned as a potent genetic suppressor as tumor onset was more delayed in the group co-injected with rag2-kRASG12D and rag2-noxa compared to the group co-injected with rag2-kRASG12D and rag2-GFP (Figure 2B). This transgenic coinjection methodology establishes a rapid assay to identify enhancers and suppressors of tumor initiation and growth.
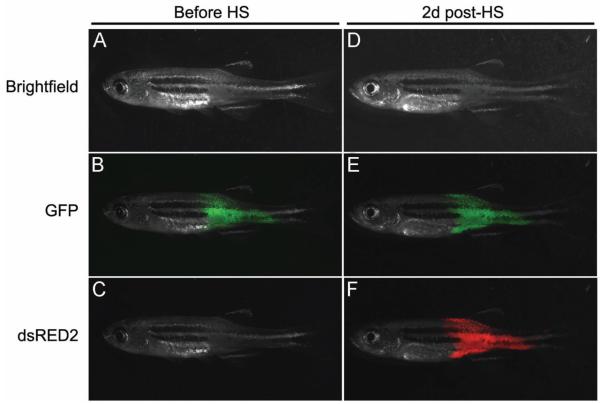
The coinjection strategy may also prove useful for assessing pathways required for tumor maintenance (Figure 3). For example, coinjection of rag2-kRASG12D + rag2-GFP + heat shock-inducible-dsRED (hsp70-dsREDexpress) into one cell stage animals led to the development of GFP+/red-negative tumors. Following heatshock at 37OC for 45 minutes, ERMS become dsRED-labeled two days latter and persisted until 10 days. Engineering heatshock- inducible constructs that deregulate pathways in established cancers will likely provide a unique and rapid assay for identifying pathways required for tumor maintenance and stopping tumor growth.
Figure 3.
Co-injection approaches to selectively induce gene expression in RAS-induced ERMS. The rag2-kRASG12D transgene was co-injected with rag2-GFP and hsp70-dsRED2 into AB- strain embryos at one-cell stage. Animals develop GFP-labeled tumors that do not express high levels of dsRED2 (A-C, Before HS). ERMS-effected animals were heat shocked at 37OC degrees for 45 minutes. Two days post-heat-shock (2d post-HS), ERMS express both GFP and dsRED2 in surviving animals (D-F). Bright field (A and D), GFP (B and E) and dsRED2 (C and F).
III. Co-injection strategies to label ERMS cell subpopulations
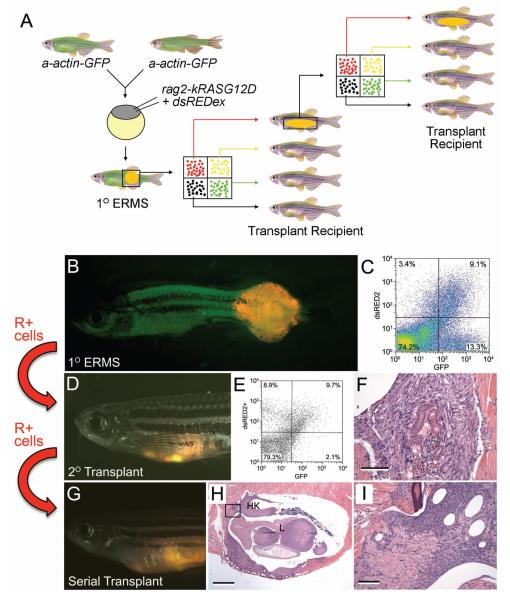
Co-injection of two constructs into one-cell stage embryos leads to co-segregation and co-expression of multiple transgenes within developing ERMS and T-cell leukemia (Langenau et al., 2008). In order to differentially label ERMS cells based on differentiation status, rag2- kRASG12D transgenic construct was co-injected with rag2-dsRED into alpha-actin-GFP transgenic fish (Figure 4A). rag2-dsRED is expressed in satellite cells and early myoblasts while alpha-actin-GFP is expressed in more mature muscle cells. To verify that heterogeneous ERMS cell populations could be differentially labeled with fluorescent proteins, ERMS was isolated from zebrafish by surgical resection and minced in 5 ml of 0.9X PBS/ 5% FBS (fetal bovine serum). To facilitate dissociation of tumor cells, the minced contents can be repeated pipetted up and down using a 10 ml pipette. Samples were filtered through a 40 ump cell strainer (#352340, BD Falcon), centrifuged at 1000g for 5-10 minutes and resuspended in 500 ul of 0.9x PBS × 5% FBS containing propidium iodide at a concentration of 1 ug/ml to exclude dead cells or debris. Fluorescence activated cell sorting (FACS) was performed based on forward and side scatter, green and dsRed fluorescence using FACSVantage flow cytometer (Beckton Dickinson). FACS analysis confirmed that the mononuclear component of the tumor was composed of four populations of cells (double negative, GFP+/dsRED-, dsRED+/GFP- and double positive, Figure 4C). Sorted cell populations were FACS sorted twice to optimize cell purity and assessed for morphology and gene expression. All four populations expressed human kRASG12D, while the R+ population expressed satellite cell marker expression (cMet+, m-cadherin+, and myf5+) and lower or undetectable levels of myoblast and mature muscle markers (myod, myogenin, muscle creatine kinase and myosin light chain-2 (mylz2)) when compared with R+G+ and G+ populations. Microarray profiling confirmed that these cell populations were molecularly distinct. Taken together, our data showed that coinjection strategies can be a powerful tool to label tumor cell populations within the tumor and may be amenable to other zebrafish tumor models. Experiments outlined below assessed if a specific subpopulation of ERMS cells was capable of self-renewal.
Figure 4.
Mosaic transgenic approaches to identify distinct cell populations within zebrafish ERMS and cell transplantation approaches to identify tumor-propagating cell populations. A) Schematic: rag2-kRASG12D and rag2-dsREDex are co-injected into one-cell stage a-actin-GFP stable transgenic animals. The primary ERMS was subjected to FACS to sort out distinct populations of fluorescent tumor cells. Each population of cells was transplanted into recipient animals. For subsequent serial transplantation, each round of transplantation was preceded by FACS-based cell sorting. B). Merged image from dsREDex fluorescence image and GFP fluorescence image of a primary ERMS. C). FACS profile of a-actin-GFP transgenic animals injected at the one-cell stage with rag2-dsRED2 and rag2-G12D. D) Merged image of GFP fluorescent, dsRED2 fluorescent, and bright field images of primary transplanted tumors from a- actin-GFP+/rag2-dsRED2+ zebrafish (primary recipient). E). FACS analysis of primary recipient with engrafted ERMS. F). Morphology of the tumor from the primary recipient, showing mostly fascicles of spindle cells and rare nests of round, hyperchromatic round cells. G). Merged image of GFP fluorescent, dsRED2 fluorescent, and bright field images of 5° recipient. H. Hematoxylin and eosin-stained section showing engrafted tumor at low power, showing infiltration of kidney and liver. I. High-power magnification of (H) showing tumor infiltration of the liver.
IV. Identifying the serially transplantable ERMS-propagating cell population
Short-term irradiation assay
The ability to remake tumor in transplant animals is a hallmark of cancer. To establish that ERMS are fully malignant, ERMS cells were transplanted into irradiated recipients. Wild- type recipients were subjected to sub lethal radiation at 20-25 Gy, 2 days before transplant and transplanted with unsorted primary ERMS by intraperitoneally injection (2x104 cells/animal). Small bundles of fluorescent tumor cells were detected as early as 7 days post-transplant using a fluorescence dissecting microscope and large tumor masses developed near the site of injection by 14 days post-transplant.
To define the population of cells that contains the transplantable ERMS cell population, FACS-sorted cells are obtained from primary rag2-dsRED+/alpha-actin-GFP+ ERMS and injected into irradiated recipient fish at limiting dilution (2 × 10^4, 4 × 10^3, 200, 50 and 10 cells). Specifically, 5 microliters of cell suspension was injected into the peritoneal cavity of irradiated AB strain adult animals using a Hamilton syringe (#701, Fisher Scientific). Each labeled cell population was transplanted from lowest to highest concentration and the syringe was cleaned when a different subtraction of cells was injected into recipient animals. Cleaning of the syringe was completed by pipetting and aspirating 10 washes in 10% bleach in dH20, 10 washes in 100% ethanol, and 20 washes in 0.9X PBS. Following cell transplant of ERMS cells into recipient zebrafish, animals were analyzed for engraftment based on dsRED2 and GFP fluorescence at 7, 11, 14 and 18 days post-injection (Figure 4D-F). To confirm that R+ cell subpopulation contained the long-term self-renewing ERMS cell, FACS-sorted populations from serially transplanted tumors were also assessed for engraftment potential. Specifically, ERMS cells were isolated from animals engrafted with ERMS from the R+ cell subpopulation and re- introduced into recipient animals by intraperitoneal injection (Figure 4G-I). These experiments confirmed that rag2-dsRED+/alpha-actin-GFP-negative cell population was better at engrafting ERMS than other tumor subpopulations and was serially transplantable – a hallmark of self- renewing cells. Serial transplantation at limiting dilution showed that as few as 10 R+ cells were required for transplantation while other tumor subpopulations required a much higher number of cells to transplant and engraft. A subset of transplant fish were sacrificed in order to verify that fluorescence corresponded to areas of tumor formation, that heterogeneity of tumor cell types was similar between passage, and that fish scored negative for tumor by fluorescent microscopy were indeed negative for tumor. Fluorescence imaging and histological analysis of tumors yielded similar total numbers of engrafted animals and confirmed that the rag2-dsRED+/alpha- acting-neagitve cells are the tumor-propagating cancer stem cells in ERMS. The limiting dilution assay allows us to identify the transplantable population of cells in ERMS. Monitoring and tracking this population facilitates the investigation of self-renewal mechanisms in ERMS.
Transplanting into clonally syngeneic fish
One issue with transplanting tumor cells into an irradiated non immune- matched host is that immune suppression is temporary, and tumor can regress as the immune system recovers. The previous study by Smith et al. (2010) took advantage of the clonal syngeneic CG1 strain animals and compared the transplantation efficiency of T-cell acute lymphoblastic leukemia (T- ALL) cells to irradiated wild-type AB-strain animals. Leukemias were generated by coinjecting rag2-mouse-cMyc and rag2-dsRED express, rag2-zsYellow or rag2-Amcyan into one-cell stage CG1-strain embryos. Fluorescence-labeled T-ALL developed in a subset of animals and fluorescent blasts were isolated from primary leukemic fish by FACS and transplanted into non- irradiated CG1-strain animals, non-irradiated AB-strain animals and irradiated AB-strain animals (AB+IR). Compared to CG1 recipients, AB+IR recipients had fewer engrafted animals at 10 and 20 days post-transplant. In addition, irradiation also increased death rate in AB recipients. Most T-ALLs arising in AB+IR recipients regressed by 30 days post-transplant. However, tumors in CG1 recipients continued to grow. Limiting dilution analysis revealed that transplantation into AB+IR recipients underestimated the number of leukemia-initiating cells by 20-33 fold, secondary to inefficient ablation of the immune system and increased lethality from irradiation. The same study also showed that T-ALLs can also be transplanted into CG1 outcrossed animals. By third outcross to CG1 most tumors engrafted tumors arising in CG1 animals.
Our group has also shown that ERMS tumor cells derived from coinjection of CG1 strain animals with rag2-dsREDexpress and rag2-kRASG12D can engraft successfully into CG1 recipient animals. Moreover, a subset of RMS from AB strain animals that have been serially outcrossed to CG1 strain fish can be transplanted into CG1 recipient animals, suggesting that immune matching has occurred in a subset of animals. Serial transplantation in either experiment nicely demonstrated the existence of tumor-propagating cell population in ERMS. Although, the CG1 strain zebrafish have yet to be used in limiting dilution cell transplantation experiments in ERMS, our results from Myc-induced T-ALL models suggests that these experiments will identify long-term repopulating cell types in ERMS capable of remaking tumor.
V. Bioinformatics approaches to identify conserved pathways in zebrafish and human ERMS
Microarray analysis and gene set enrichment analysis (GSEA)
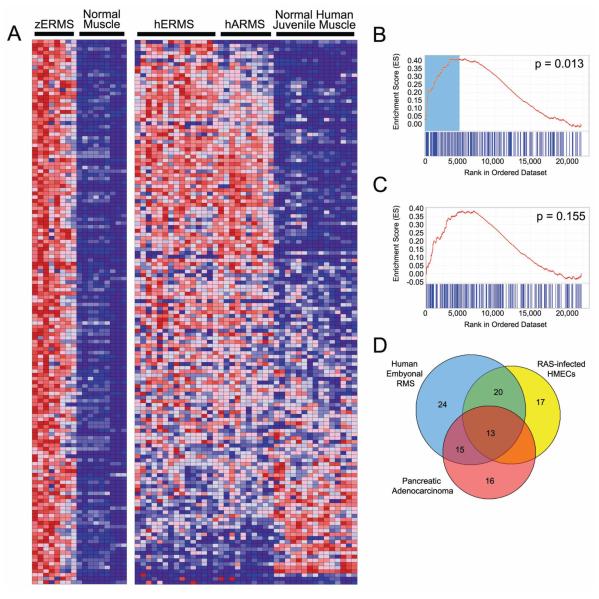
Gene Set Enrichment Analysis (GSEA) is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states (e.g. phenotypes). This method has been used in mouse and human to identify gene signatures associated with cancer and also in zebrafish to classify different types of tumor (Ramaswamy et al., 2001; Sweet-Cordero et al., 2005; Lam et al., 2005, Langenau et al., 2007). The study by Langenau et al. (2007) used GSEA to identify conserved pathways in both zebrafish and human RMS. The gene sets were determined by microarray analysis comparing zebrafish RAS-induced RMS (n=8) to normal control muscle (n=8) from AB-strain fish at 30 dpf. Several fold changes cut-offs were used in the GSEA analysis to verify that differences between disease and normal states were reproducible and not due to arbitrary assignment of gene lists. In the study, the zebrafish up-regulated gene sets were significantly associated with the human embryonal RMS data set at all five fold changes but never with the ARMS data set (Figure 5A-C). Unexpectedly, zebrafish ERMS up-regulated gene set was significantly associated with the human pancreatic adenocarcinoma. To test that the up-regulated zebrafish gene set comprised a RAS-specific signature, human mammary epithelial cells (HMECs) infected with activated RAS, MYC, SRC, B-CATENIN, or E2F3 were compared with cells infected with GFP using GSEA and the zebrafish ERM gene set. The upregulated zERMS gene set was determined to be significantly associated with RAS status. Further analysis of the gene set revealed several known transcriptional targets of RAS.
Figure 5.
Gene Set Enrichment Analysis (GSEA) identifies a conserved gene signature in both zebrafish and human ERMS. A) Heat map showing genes up-regulated in zebrafish ERMS when compared with normal muscle at 2.25-fold change (left) and juxtaposed to the corresponding human orthodox in ERMS, ARMS and normal juvenile muscle (right). Graphical representation of the rank-ordered gene lists found when comparing human ERMS (B) or translocation+ ARMS (C) to normal muscle. The up-regulated gene set identified in zebrafish ERMS is significantly enriched in human ERMS (B, p=0.013). (D) A Venn diagram illustrating the genes from the up-regulated zebrafish ERMS gene set that contribute maximally to the GSEA score in the pancreatic adenocarcinoma, human ERMS and RAS-infected human mammary epithelial cells (HMECs). Genes found in the non-overlapping blue portion are ERMS-specific and comprise the MYF5 transcription factor. By contrast, the other genes are likely RAS-targets and are found to be coordinately regulated in zebrafish RAS-induced ERMS and either RAS-infected human mammary epithelial cells or human pancreatic Aden carcinoma, of which >90% have activating mutations in kRAS.
To identify a set of genes involved in tumor-specific and tissue-restricted pathways associated with embryonal RMS, A subset of zebrafish ERMS up-regulated genes was further refined to include only genes that contribute maximally to the GSEA score in human ERMS but not pancreatic adenocarcinoma or human mammary epithelial cells (HMECs) transduced with RAS (Global Cancer Map data set from Ramaswamy et al. 2001). Specifically, a Venn diagram was used to identify the overlapping genes found in each of these human data sets (Figure 5D). Most genes are coordinately regulated in pancreatic adenocarcinoma or HMECs transduced with RAS, suggesting that these genes comprise a RAS specific signature. 24 genes including Muscle Regulatory Factor 5 (MYF5) were contained within only the human ERMS unregulated data set, identifying a tumor-specific and tissue-restricted signature associated with the ERMS phenotype. Myf5 is a myogenic transcription factor that is expressed in activated satellite cells (Cornelison and Wold, 1997; McKinnell et al., 2008) and is required along with MyoD to specify muscle in early embryogenesis (Weintraub et al., 1991; Kablar et al., 2003; Berkes and Tapscott, 2005). The presence of MYF5 expression in the ERMS-specific gene signature suggests that the gene likely participates in the lineage and stage-specific stage of ERMS cells.
Array CGH approach
Comparative Genomic Hybridization (CGH) measures DNA copy number differences between a test and reference genome. This technology scans the entire genome for variations in DNA copy number, hence can be applied to detect genetic imbalances present in a disease model. Array-based CGH is a high-resolution platform constructed from DNA fragments, PCR products or oligonucleotides that are mapped to the genome sequence. Array CGH has been an important tool in detecting genetic imbalances in human cancers and has also be utilized in various cancer models in other species such as the rodents, canines and even nematodes (refs). Freeman et al recently developed the first array CGH platform for use in zebrafish utilizing large genomic DNA fragments from zebrafish that were inserted into bacterial artificial chromosomes (BAC) (Freeman et al., 2010). 357 BAC clones were selected based on containing anthologies to human oncogenes from the CHORI-211 (zC), Danio Key (zK), and CHORI-73 (zH) genomic DNA libraries. BAC clones were selected based on containing orthologues to human oncogenes and tumor suppressor genes. Each clone was also assigned a precise cytogenetic location by fluorescent in situ hybridization.
The BAC array CGH was applied to three transgenic zebrafish cancer models: rhabdomyosarcoma, T-ALL, and melanoma (Freeman et al., 2010). For assessing chromosome alterations in ERMS, mosaic transgenic RAS-induced rhabdomyosarcomas were created by co- injection of rag2-kRASG12D and rag2-GFP into AB strain wild-type animals. GFP+ tumor tissue was identified using a fluorescent dissecting microscope and adjacent unlabeled normal tissue was used as the reference sample. Total genomic DNA was isolated from the test and the reference samples using the Gentra Puregene Genomic DNA Purification Kit (Qiagen) and differentially labeled with fluorescent molecules (e.g. Cy3, Cy5). Labeled DNA was competitively hybridized to the BAC array. A two-color scan of the arrays was conducted using the GenePix 4000B Scanner (Molecular Devices, Union City, CA) at a 5 μm resolution and the fluorescent intensities of Cy3 and Cy5 was quantified for each DNA spot. For a given genomic segment, gains or losses in copy number were detected based on deviation of Cy3 to Cy5 ratio from 1 to 1. Seven rhabdomyosarcoma specimens were analyzed on the custom zebrafish BAC- based array and observed to have 6 to 14 significant genomic imbalances per sample. Gains of multiple BAC clones were cytogenetically mapped to various regions on zebrafish chromosomes, indicating the presence of several genetic imbalances in zebrafish embryonal RMS. While zebrafish BAC-based array CGH has lower resolution compared to oligonucleotide array CGH platforms from other species, these experiments provide a nice proof-of-concept that initial genome-wide identification of chromosomal imbalances can be assessed by array CGH in zebrafish models of cancer. As the annotation of zebrafish genomic sequences nears completion, higher resolution platforms will be a useful resource for exploring changes in the zebrafish genome related to RMS.
Discussion
Studies have shown that only a limited number of cancer cells within the tumor mass has the ability for self-renewal and hence cause tumor formation (Reya et al., 2001). The same population of cells has also been implicated in treatment-refractory relapse. The tumor initiating cells or cancer stem cells have been characterized in some solid tumors such as brain, breast, colon and skin cancer (Alcantara et al., 2009; Todaro et al., 2010; Charafe-Jauffret et al., 2009; Banerjee and Eyden, 2008). However, for most malignancies including soft tissue sarcomas such as rhabdomyosarcoma, the mechanisms for self renewal remain to be elucidated. We have shown that kRAS-induced zebrafish rhabdomyosarcoma resembles human embryonal rhabdomyosarcoma. The discovery of a self-renewal population of tumor initiating cells in the zebrafish model of rhabdomyosarcoma opens the door for further investigation into the mechanism driving tumor self renewal. In addition, therapeutic targets aimed at genes involved in self renewal also promise better clinical management of cancer patients suffering from treatment refractory.
Zebrafish as a model for the study of human disease and development serves many advantages. The feasibility for large-screen genetic screens (forward or reverse) allows for rapid identification of essential genes in various biological and disease processes. The ability to identify genetically modifying events using approaches such as heat shock-inducible Cre-Lox transgenics and co-injection of transgenes also facilitates the understanding of pathways essential for different processes involved in tumorigenesis. Gene sets (“signatures”) identified from microarrays and GSEA will refine the conserved pathways in cancer and provide a tremendous opportunity for targeted therapy.
Acknowledgments
Eleanor Chen is supported by a T32 Training grant through Brigham and Woman’s Hospital (T32 HL007627). David Langenau is supported by NIH grants K01AR05562190-01A1 and K01AR05562190-S1, a new investigator grant from Alex Lemonade Stand, the Sarcoma Foundation of America, the Leukemia Research Foundation, and a seed grant from the Harvard Stem Cell Institute.
References
- Alcantara Laguna S, Chen J, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee SS, Eyden B. Divergent differentiation in malignant melanomas: a review. Histopathology. 2008;52(2):119–129. doi: 10.1111/j.1365-2559.2007.02823.x. [DOI] [PubMed] [Google Scholar]
- Barr FG, Galili N, et al. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;3(2):113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- Barr FG, Holick J, et al. Localization of the t(2;13) breakpoint of alveolar rhabdomyosarcoma on a physical map of chromosome 2. Genomics. 1992;13(4):1150–1156. doi: 10.1016/0888-7543(92)90030-v. [DOI] [PubMed] [Google Scholar]
- Berghmans S, Murphey RD, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A. 2005;102(2):407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16(4-5):585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Calzada-Wack J, Kappler R, et al. Unbalanced overexpression of the mutant allele in murine Patched mutants. Carcinogenesis. 2002;23(5):727–733. doi: 10.1093/carcin/23.5.727. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, et al. Breast cancer stem cells: tools and models to rely on. BMC Cancer. 2009;9:202. doi: 10.1186/1471-2407-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Takita J, et al. Mutations of the PTPN11 and RAS genes in rhabdomyosarcoma and pediatric hematological malignancies. Genes Chromosomes Cancer. 2006;45(6):583–591. doi: 10.1002/gcc.20322. [DOI] [PubMed] [Google Scholar]
- Cornelison DD, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol. 1997;191(2):270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- Davis RJ, D’Cruz CM, et al. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54(11):2869–2872. [PubMed] [Google Scholar]
- Felix CA, Kappel CC, et al. Frequency and diversity of p53 mutations in childhood rhabdomyosarcoma. Cancer Res. 1992;52(8):2243–2247. [PubMed] [Google Scholar]
- Freeman JL, Ceol C, et al. Construction and application of a zebrafish array comparative genomic hybridization platform. Genes Chromosomes Cancer. 2009;48(2):155–170. doi: 10.1002/gcc.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N, Davis RJ, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5(3):230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Gurney JG, Davis S, et al. Trends in cancer incidence among children in the U.S. Cancer. 1996;78(3):532–541. doi: 10.1002/(SICI)1097-0142(19960801)78:3<532::AID-CNCR22>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Jessen JR, Jessen TN, et al. Concurrent expression of recombination activating genes 1 and 2 in zebrafish olfactory sensory neurons. Genesis. 2001;29(4):156–162. doi: 10.1002/gene.1019. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, et al. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev Biol. 2003;258(2):307–318. doi: 10.1016/s0012-1606(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Kappler R, Bauer R, et al. Profiling the molecular difference between Patched- and p53- dependent rhabdomyosarcoma. Oncogene. 2004;23(54):8785–8795. doi: 10.1038/sj.onc.1208133. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A, et al. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97(21):11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, et al. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18(21):2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny SF, Drobes BL, et al. Inhibition of myogenic differentiation by the H-ras oncogene is associated with the down regulation of the MyoD1 gene. Oncogene. 1989;4(4):473–481. [PubMed] [Google Scholar]
- Koufos A, Hansen MF, et al. Loss of heterozygosity in three embryonal tumours suggests a common pathogenetic mechanism. Nature. 1985;316(6026):330–334. doi: 10.1038/316330a0. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21(11):1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SH, Wu YL, et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat Biotechnol. 2006;24(1):73–75. doi: 10.1038/nbt1169. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, et al. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic Zebrafish. Oncogene. 2008;27(30):4242–4248. doi: 10.1038/onc.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Traver D, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299(5608):887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- Lassar AB, Thayer MJ, et al. Transformation by activated ras or fos prevents myogenesis by inhibiting expression of MyoD1. Cell. 1989;58(4):659–667. doi: 10.1016/0092-8674(89)90101-3. [DOI] [PubMed] [Google Scholar]
- Le X, Langenau DM, et al. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc Natl Acad Sci U S A. 2007;104(22):9410–9415. doi: 10.1073/pnas.0611302104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kawagoe R, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26(44):6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- Linardic CM, Downie DL, et al. Genetic modeling of human rhabdomyosarcoma. Cancer Res. 2005;65(11):4490–4495. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- Mao J, Ligon KL, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66(20):10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10(1):77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan LM, Matlashewski GJ, et al. Mechanisms of p53 loss in human sarcomas. Proc Natl Acad Sci U S A. 1990;87(15):5863–5867. doi: 10.1073/pnas.87.15.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni P, Nicoletti G, et al. Development of rhabdomyosarcoma in HER-2/neu transgenic p53 mutant mice. Cancer Res. 2003;63(11):2728–2732. [PubMed] [Google Scholar]
- Ramaswamy S, Tamayo P, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci U S A. 2001;98(26):15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Santoriello C, Deflorian G, et al. Expression of H-RASV12 in a zebrafish model of Costello syndrome causes cellular senescence in adult proliferating cells. Dis Model Mech. 2009;2(1-2):56–67. doi: 10.1242/dmm.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrable HJ, Witte DP, et al. Chromosomal localization of the human rhabdomyosarcoma locus by mitotic recombination mapping. Nature. 1987;329(6140):645–647. doi: 10.1038/329645a0. [DOI] [PubMed] [Google Scholar]
- Sharp R, Recio JA, et al. Synergism between INK4a/ARF inactivation and aberrant HGF/SF signaling in rhabdomyosarcomagenesis. Nat Med. 2002;8(11):1276–1280. doi: 10.1038/nm787. [DOI] [PubMed] [Google Scholar]
- Smith AC, Raimondi AR, et al. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010;115(16):3296–3303. doi: 10.1182/blood-2009-10-246488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Fisher C, et al. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Res. 1989;49(22):6324–6327. [PubMed] [Google Scholar]
- Stratton MR, Moss S, et al. Mutation of the p53 gene in human soft tissue sarcomas: association with abnormalities of the RB1 gene. Oncogene. 1990;5(9):1297–1301. [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37(1):48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- Todaro M, Francipane MG, et al. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138(6):2151–2162. doi: 10.1053/j.gastro.2009.12.063. [DOI] [PubMed] [Google Scholar]
- Tsumura H, Yoshida T, et al. Cooperation of oncogenic K-ras and p53 deficiency in pleomorphic rhabdomyosarcoma development in adult mice. Oncogene. 2006;25(59):7673–7679. doi: 10.1038/sj.onc.1209749. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Dwarki VJ, et al. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991;5(8):1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- Williamson D, Missiaglia E, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28(13):2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]