Abstract
Previously, TX5179, a disruption mutant of the enterococcal polysaccharide antigen (epa) gene cluster of Enterococcus faecalis strain OG1RF was shown to be attenuated in translocation, biofilm mouse peritonitis and was more susceptible to polymorphonuclear leukocyte phagocytic killing. Here, wild-type E. faecalis OG1RF and TX5179 strains were tested in a mixed-infection (inoculum, ~1:1) mouse urinary tract infection model. Wild-type OG1RF outnumbered TX5179 in the kidneys (P < .001) and bladder (P < .001). In conclusion, the epa locus of E. faecalis OG1RF contributes to murine urinary tract infection and is the firs such enterococcal polysaccharide locus shown to be important in this site.
Enterococci are the most common organisms among gram-positive bacteria to cause urinary tract infections (UTIs) and can be found as a single organism or as part of a polymicrobial infection [1]. In the hospital setting, identified risk factors for enterococcal UTI include stay in the intensive care unit, use of urinary catheters, immunosuppression, and use of broad-spectrum antibiotics [1]. However, the pathogenesis of enterococcal UTIs has not been clearly elucidated.
Polysaccharides on bacterial surfaces are known to interact with the human host and to play important roles in bacterial pathogenesis, including for enterococci [2–5]. We previously described a large enterococcal polysaccharide antigen (epa) gene cluster of E. faecalis and showed that a mutant of OG1RF, TX5179, with a disrupted epaB (formerly orfde4) gene, showed attenuation in a mouse peritonitis model [2], susceptibility to neutrophil phagocytic killing [3], and reduced biofilm formation [3, 6], compared with the wild-type E. faecalis strain OG1RF. Here, we studied the TX5179 mutant versus wild-type OG1RF in a murine UTI model.
Materials and methods
E. faecalis OG1RF [7] and TX5179 [2] have been described previously. Growth curve experiments of test bacteria were performed in Bacto Brain-Heart Infusion broth (Becton Dickinson) medium with 40% horse serum (Sigma; BHIS). The cultures were grown at 37°C with gentle agitation. A reading of optical density at 600 nm was determined every hour from 0 h to 8 h and then at 14 and 24 h. At intervals of 0 h, 6 h, and 24 h, the number of colony-forming units (cfu) per milliliter were also determined for both strains by plating serial dilutions on brain-heart infusion agar plates.
For inocula preparation, bacterial strains were grown for ~10 h with gentle agitation at 37°C in BHIS medium. The cells were pelleted for 10 minutes (10,000 rpm at 10°C) and resuspended in 10 mL of 0.9% saline. For the mouse UTI model, white, female Imprinting Control Region mice (Harlan Sprague Dawley) with a mean weight of 25 g were used. Methods for mouse catheterization, inoculation, organ recovery, and tissue homogenization for bacterial recovery were the same as those published elsewhere [8]. In brief, isoflurane-anesthetized mice were infected via intraurethral catheterization using 200 μL of the bacterial suspension consisting of an ~1:1 ratio of wild-type E. faecalis OG1RF: TX5179 (ie, 3.2 ×105 geometric mean [GM] cfu of OG1RF to 4.1 ×105 GM cfu of TX5179). The urethral catheter was removed soon after injection of the bacteria, and all animals had free access to food and water throughout the course of study. Mice were euthanized by CO2 inhalation at 48 h after transurethral challenge. The urinary bladder and kidney pair were excised, weighed, and homogenized in 1 mL and 5 mL of saline, respectively; dilutions were plated onto brain-heart infusion agar, with or without antibiotics, as appropriate. The minimum detection limit of bacteria in this experiment was 102 cfu/g of tissue homogenate. Mice with sterile cultures of kidney and urinary bladder homogenates were considered to have no UTI. Identity of the recovered test bacteria from infected organs was confirmed by verifying their appropriate antibiotic resistance markers. All experiments involving mice were performed in accordance with guidelines stipulated by the animal care committee, University of Texas Health Science Center at Houston.
The log10 cfu/g of TX5179 and OG1RF in the inocula and tissue of each animal (kidney or bladder) from the mixed-infection model were analyzed for statistical significance by the paired t test. The Fisher exact test was used to compare total infection of kidneys versus that of bladders (combining data for all animals) during mixed infection with OG1RF:TX5179. Graph Pad Prism, version 4.0 for Windows (GraphPad Software), was used for statistical analysis.
Results and discussion
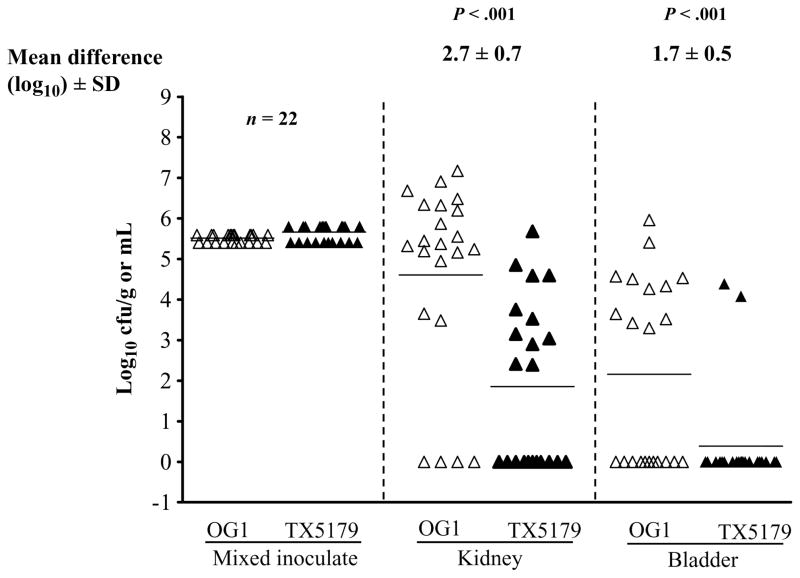
For the UTI model, a total of 22 mice were used (12 mice in one experiment and 10 in an independent experiment; the results were combined). In the kidneys, despite inoculating with slightly more of the mutant TX5179 (4.3 ×105 GM cfu) than the wild-type OG1RF (3.2 ×105 GM cfu), there was almost a 3-log10 cfu/g difference that favored wild-type OG1RF (6.7 × 104) over TX5179 (8.8 × 101 GM cfu; mean difference ± standard deviation, 2.7 ± 0.7 log10 cfu; P < .001). In the bladder, there was a mean (± standard deviation) difference of 1.7 ± 0.5 log10 cfu/g (P < .001) that favored wild-type OG1RF (8.2 ×102 GM cfu) over TX5179 (0.3 ×101 GM cfu) (figure 1). Kidneys—which have rich vascular supplies and which are made up of glomerular basement membrane consisting of a cross-linked meshwork of collagen (mostly type IV), laminin, polyanionic proteoglycans (mostly heparan sulfate), fibronectin, entactin/ nidogen, and several other glycoproteins [9]—appear to be preferred sites for colonization by enterococci, with 19 of 22 kidneys infected. Only 11 of 22 urinary bladders, where the mucosal surface has a relatively low vascular supply, were found to be infected. This preference for kidney colonization by E. faecalis is consistent with previous observations by us [8] and others [10]. Our previous study showed a role for ebp pili in upper tract infection in this model [8]. In contrast, in a previous study of Shankar et al. [11], in which an Esp (enterococcal surface protein, encoded by an acquired gene)–positive E. faecalis and its Esp-deficient mutant were compared, there was increased persistence of bacteria (log10 cfu) in the urinary bladders of mice, with no histological changes, by wild-type versus mutant; however, there was no difference in the bacterial log10 cfu level in kidneys. The wild-type OG1RF used in the present study lacks the Esp [7].
Figure 1.
Results for urinary tract infection. Mixed infection (competition assay) with wild-type Enterococcus faecalis OG1RF and TX5179 (epaB; formerly orfde4) gene disruption mutant) in the kidneys and urinary bladder of 22 mice. Data are expressed as log10 colony-forming units (cfu) of wild-type E. faecalis OG1RF or TX5179. Empty and solid triangles represent wild-type E. faecalis OG1RF and TX5179, respectively, from kidney and urinary bladder homogenates. Horizontal bars represent geometric mean titers. Mean fold difference in log10 cfu are given. Log10 cfu were compared for statistical significance using a paired t test. SD, standard deviation.
To evaluate whether growth of TX5179 mutant differed from that of wild-type OG1RF in BHIS, the optical density at 600 nm and bacterial titers were measured. TX5179 did not show any apparent growth defect, compared with wild-type OG1RF, up to 14 h, with a slight decrease in optical density at 600 nm at 24 h, compared with wild-type OG1RF (figure 2A). However, the bacterial titers for both strains were comparable at all 3 time points: at 0 h, 5.4 ×107 cfu/mL and 4.6 ×107 cfu/mL; at 6 h, 2.8 ×109 cfu/mL and 2.6 ×109 cfu/mL; and at 24 h, 1.4 ×109 cfu/mL and 1.2 ×109 cfu/mL for TX5179 and wild-type OG1RF, respectively (figure 2B). Thus, the gene disruption in TX5179 did not cause a growth defect when grown in BHIS medium. Similarly, TX5179 BHIS-grown cells plated on brain-heart infusion agar plus kanamycin (2000 μg/mL), compared with brain-heart infusion agar plates from inoculum prepared for animal infection, demonstrated an almost equal number of colony-forming units per milliliter on plates with and without the antibiotic (data not shown), indicating in vitro stability of the disruption. We previously demonstrated in vivo stability of gene disruption of the TX5179 in a mouse peritonitis model [2].
Figure 2.
Growth curves and levels of colony-forming units (cfu). Empty squares, OG1RF; solid squares, TX5180. A, Comparison of wild-type OG1RF and TX5179 growth in Bacto Brain-Heart Infusion broth (Becton Dickinson) medium with 40% horse serum. Optical densities (OD600) were measured every hour until 8 h and then at 14 and 24 h. B, The level of cfu determined at 0 h, 6 h, and 24 h.
Various enterococcal polysaccharides (capsular or cell wall) and related genes and/or gene clusters [2, 3, 12] have been reported in the literature. In our previously published studies, we showed that TX5179 was attenuated in a mouse peritonitis model, displayed decreased resistance to phagocytosis and/or killing by polymorphonuclear leukocytes [3] and formed less biofilm than did wild-type OG1RF [6]. In the present report, we have shown that TX5179 is also significantly attenuated, compared with wild-type OG1RF, in a murine model of ascending UTI. TX5179, with a disrupted epaB gene (formerly orfde4), lacks or has modified a polysaccharide that is important for efficient colonization and/or infection of kidneys and urinary bladder, compared with wild-type OG1RF. Our previously published work used Western blots and a serum specimen obtained from a patient with E. faecalis endocarditis to demonstrate the presence of high-molecular-weight (smear) and low-molecular-weight bands in the polysaccharide extracts obtained from wild-type OG1RF that were absent in the polysaccharide extracts obtained from TX5179; periodate treatment and carbohydrate staining confirmed the polysaccharide nature of this material in wild-type OG1RF [3]. Our recent work has shown that the purified Epa polysaccharide from wild-type OG1RF is composed mainly of rhamnose and glucose, as well as of small amounts of galactose, GalNAc, and GlcNAc, whereas a new polysaccharide present in TX5179 mutant does not contain rhamnose but has other carbohydrate residues as listed for wild-type OG1RF (F. Teng, K. V. Singh, A. Bourgogne, J. Zeng, and B. E. Murray; unpublished data), suggesting the possibility that altered polysaccharide of TX5179 contributes to its attenuation in our murine UTI model. Our previously published work suggested that epaB (orfde4) of TX5179 was cotranscribed with the downstream gene epaC (orfde5)) [3], and our recent work shows that epaB (orfde4) is cotranscribed with epaC (orfde5) and epaD (orfde6) (F. Teng, K. V. Singh, A. Bourgogne, J. Zeng, and B. E. Murray; unpublished data), indicating that there is a polar effect on these downstream genes in TX5179 mutant. Our effort to test complemented TX5179 in the mouse UTI model were unsuccessful, because the vector pAT18 with cloned complementation fragment showed in vivo loss of ~2–3 log10 cells, although it did not show any in vitro loss when grown in BHIS medium for inoculum preparation. However, our intent was to show an effect of disruption within the epa cluster, which these data provide, rather than to associate a specific gene with the defect.
The importance of carbohydrate residues on E. faecalis cell surface and their adherence to different cell lines has been previously suggested by Guzman et al. [13] using carbohydrate inhibition experiments and sodium m–periodate treatment of cells; in these studies, E. faecalis carbohydrate residues (D-mannose and D-glucose) were noted to be expressed by strains of both UTI and endocarditis origin when the cells were grown in brain-heart infusion broth and mediated adherence to either urinary tract epithelial cells or the Girardi Heart cell line. Other carbohydrate residues (D-galactose and L-fucose) were present only on endocarditis strains that mainly adhered to Girardi Heart cells, but these ligands were also expressed by UTI isolates after growth in serum. In the current study, both wild-type OG1RF and TX5179 were grown in the presence of 40% serum, because in our preliminary experiments, wild-type OG1RF grown in brain-heart infusion broth was found to be less infective in the murine UTI model, compared with brain-heart infusion–40% serum–grown OG1RF (unpublished data). Recently, our group has shown the in vitro serum-elicited adherence of 43 diverse E. faecalis strains, which included 4 strains isolated from urine samples of patients and wild-type OG1RF, to extracellular matrix proteins (ie, fibonectin, fibrinogen, and collagen types I and IV) demonstrating that all strains, when grown in brain-heart infusion–40% serum versus brain-heart infusion alone, showed increased adherence to extracellular matrix proteins regardless of the source of isolation [4]. Serum is known to alter expression of some E. faecalis genes [14, 15] and is the subject of additional studies.
In conclusion, in a murine ascending UTI model, wild-type E. faecalis OG1RF significantly outnumbered its epaB (formerly orfde4) gene disruption mutant TX5179 in both kidneys and urinary bladders, indicating that the polysaccharide related to the epa gene cluster of wild-type OG1RF contributes to murine UTI.
Acknowledgments
We thank Karen Jacques-Palaz and L. Charlene Thomson for their technical assistance.
Financial support: National Institutes of Health (R37 AI47923 to B.E.M) from the Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases.
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Mathai D, Jones RN, Pfaller MA. Epidemiology and frequency of resistance among pathogens causing urinary tract infections in 1,510 hospitalized patients: a report from the SENTRY Antimicrobial Surveillance Program (North America) Diagn Microbiol Infect Dis. 2001;40:129–36. doi: 10.1016/s0732-8893(01)00254-1. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y, Singh KV, Qin X, Murray BE, Weinstock GM. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect Immun. 2000;68:815–23. doi: 10.1128/iai.68.2.815-823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teng F, Jacques-Palaz KD, Weinstock GM, Murray BE. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influence resistance to phagocytic killing of E. faecalis. Infect Immun. 2002;70:2010–5. doi: 10.1128/IAI.70.4.2010-2015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nallapareddy SR, Murray BE. Role played by serum, a biological cue, in the adherence of Enterococcus faecalis to extracellular matrix proteins, collagen, fibrinogen, and fibronectin. J Infect Dis. 2008;197:1728–36. doi: 10.1086/588143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huebner J, Quaas A, Krueger WA, Goldmann DA, Pier GB. Prophylactic and therapeutic efficacy of antibodies to a capsular polysaccharide shared among vancomycin-sensitive and -resistant enterococci. Infect Immun. 2000;68:4631–6. doi: 10.1128/iai.68.8.4631-4636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohamed JA, Huang W, Nallapareddy SR, Teng F, Murray BE. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72:3658–63. doi: 10.1128/IAI.72.6.3658-3663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgogne A, Garsin DA, Qin X, et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh KV, Nallapareddy SR, Murray BE. Importance of the ebp (endocarditis- and biofilm-associate pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis. 2007;195:1671–7. doi: 10.1086/517524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller U, Brändli AW. Cell adhesion molecules and extracellular-matrix constituents in kidney development and disease. J Cell Sci. 1999;112:3855–67. doi: 10.1242/jcs.112.22.3855. [DOI] [PubMed] [Google Scholar]
- 10.Kau AL, Martin SM, Lyon W, Hayes E, Caparon MG, Hultgren SJ. Enterococcus faecalis tropism for the kidneys in the urinary tract of C57BL/6J mice. Infect Immun. 2005;73:2461–8. doi: 10.1128/IAI.73.4.2461-2468.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun. 2001;69:4366–72. doi: 10.1128/IAI.69.7.4366-4372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hufnagel M, Carey VJ, Baldassarri L, Reinert RR, Huebner J. Distribution of four capsular serotypes of Enterococcus faecalis among clinical isolates from different geographical origins and infection sites. Infection. 2006;34:22–5. doi: 10.1007/s15010-006-4100-5. [DOI] [PubMed] [Google Scholar]
- 13.Guzman CA, Pruzzo C, Plate M, Guardati MC, Calegari L. Serum dependent expression of Enterococcus faecalis adhesins involved in the colonization of heart cells. Microb Pathog. 1991;11:399–409. doi: 10.1016/0882-4010(91)90036-a. [DOI] [PubMed] [Google Scholar]
- 14.Nallapareddy SR, Singh KV, Sillanpaa J, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepard BD, Gilmore MS. Differential expression of virulence-related genes in Enterococcus faecalis in response to biological cues in serum and urine. Infect Immun. 2002;70:4344–52. doi: 10.1128/IAI.70.8.4344-4352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]