Summary
Standardized studies examining environmental microbial exposure in populations at risk for asthma are necessary to improve our understanding of the role this factor plays in disease development. Here we describe studies aimed at developing guidelines for high-resolution culture-independent microbiome profiling, using a phylogenetic microarray (PhyloChip), of house dust samples in a cohort collected as part of the NIH-funded Inner City Asthma Consortium (ICAC). We demonstrate that though extracted DNA concentrations varied across dust samples, the majority produced sufficient 16S rRNA to be profiled by the array. Comparison of array and 454-pyrosequencing performed in parallel on a subset of samples, illustrated that increasingly deeper sequencing efforts validated greater numbers of array-detected taxa. Community composition agreement across samples exhibited a hierarchy in concordance, with the highest level of agreement in replicate array profiles followed by samples collected from adjacent 1×1 m2 sites in the same room, adjacent sites with different sized sampling quadrants (1×1 and 2×2 m2), different sites within homes (living and bedroom) to lowest in living room samples collected from different homes. The guidelines for sample collection and processing in this pilot study extend beyond PhyloChip based studies of house-associated microbiota, and bear relevance for other microbiome profiling approaches such as next-generation sequencing.
Keywords: Dust Microbiome, Standardized sampling, Phylogenetic microarray, 454-pyrosequencing
Introduction
Asthma prevalence in the US has increased substantially from 2001 to 2009; currently, about 10% of children in the United States suffer from this airway disease (Centers for Disease Control and Prevention 2011). Inner-city children, who live in urban census tracts with a high proportion of low-income families (Busse 2010), suffer from asthma prevalence and severity at a disproportionately high rate (Joseph et al. 2006). The Inner-City Asthma Consortium (ICAC) is a multi-site collaborative initiative to investigate possible causes of asthma and asthma morbidity in an attempt to reduce the occurrence and severity of the disease for inner-city pediatric populations.
One aim of ICAC is to investigate whether exposure to specific types of microbial communities (and to particular species within these assemblages), associated with a child's immediate indoor environment (i.e. house dust) may be related to asthma development outcomes. To address this issue and provide a broad, high-resolution profile of the bacterial communities present in house dust, we plan to use a culture-independent phylogenetic microarray, the 16S rRNA PhyloChip, a high-density array capable of detecting ~8,500 (G2; (Brodie et al. 2007) or ~60,000 (G3; (Hazen et al. 2010) bacterial taxa in a single economical assay. Much like gene expression arrays, data generated by the PhyloChip is normalized across the entire cohort of profiled samples. Thus, data can be used to identify species that exhibit the greatest differences in relative abundance across treatment groups (e.g. case and control or exposure groups). Data may also be used to perform multivariate analyses using associated study metadata (e.g. dog ownership or number of siblings) to identify key aspects of community structure and specific microbial members most highly associated with these variables. Species identified by such statistical approaches are then typically targeted for further study in mammalian models to confirm their contributory role, as we have previously described (Ivanov et al. 2009). The parallel nature of the array also affords the advantage of detecting lower abundance species as readily as the dominant species in a given community (Flanagan et al. 2007; Saulnier et al. 2011), thus providing a high-resolution profile of the bacterial community members present in a given sample and increasing the likelihood of detecting relevant target organisms for subsequent study.
Because the PhyloChip is highly sensitive, it is imperative to optimize and define the standards for sample collection and processing, for use in future planned studies. Therefore the study reported here aimed to evaluate the optimal weight of house dust necessary to produce sufficient DNA for PhyloChip microbiome profiling, the concentration of PCR product necessary to detect the breadth of diversity present, and the reproducibility of this tool across technical replicates. The study also examined concordance between this microbiota profiling approach and next-generation 454-sequencing. Following these initial optimization efforts, we applied the developed protocol to test for spatial heterogeneity and specific taxa characteristics of house dust samples from sites within and across homes, in an attempt to examine associated community variability and identify the optimal site for sample collection for these studies.
Experimental Procedures
Dust sample collection
Dustream® collectors (Indoor Biotechnologies, Charlottesville, VA) were used to collect separate vacuumed dust samples from homes for the time specified using a standardized protocol. In the case where a Dustream® collector was filled before the specified time was complete, the timer was stopped and a new collection tube was used to complete the remaining time. Dust from both collection tubes was used for DNA extraction.
For initial optimization studies, eighteen available banked dust samples (samples S1-18; Table 1), collected from the kitchen, TV/living room, and bedroom floors of homes from the greater Baltimore area were used. Ten of these samples were from inner-city homes, and eight from suburban homes. Sample quadrants (1×1 meter) had been vacuumed for 2 minutes prior to sieving of dust samples through a ~400 μm filter. These sieved dust samples had been stored at – 80 °C for 7-13 years. Samples were shipped to UCSF on dry ice and stored at -80 °C until processed for ba cterial community profiling.
Table 1.
Samples analyzed.
| Sample Type | Quantity (Sample number) |
|---|---|
| Banked Samples: | |
| Initial optimization study | 18 (S1-18) |
| Prospectively collected samples: | |
| Bed | 10 (S19-28) |
| Bedroom floor original (1×1 m2) | 11 (S29-39) |
| Bedroom floor adjacent (1×1 m2) | 9 (S40-48) |
| Bedroom floor adjacent (2×2 m2) | 1 (S49) |
| TV/Living room floor original (1×1 m2) | 9 (S50-58) |
| TV/Living room floor adjacent (1×1 m2) | 1 (S59) |
| TV/Living room floor adjacent (2×2 m2) | 8 (S60-67) |
For the subsequent spatial heterogeneity study, 49 dust samples (samples S19-68) were collected prospectively by trained staff from the bedroom and TV/living room floors and bed (Gruchalla et al. 2005) of 12 inner-city Baltimore homes (Table 1). Rooms were vacuumed for 2 minutes for each square meter within a defined quadrant (2 minutes for 1×1 m2 or 8 minutes for 2 × 2 m2) yielding between 10 mg and 400 mg dust. Dust samples were shipped to UCSF on dry ice, and stored at -80 °C until processed for bacterial community composition profiling.
DNA extraction
For the initial eighteen banked dust samples, DNA was extracted from 0.1 g, 0.2 g, and 0.4 g of each sample using a modified cetyl trimethylammonium bromide (CTAB) buffer and polyethylene (PEG) method as described previously (DeAngelis et al. 2009). Briefly, 0.5 ml of modified CTAB extraction buffer (1:1 10% CTAB in 1 M NaCl to 0.5 M phosphate buffer (pH 7.5 – 8) in 1 M NaCl) was added to dust samples in Lysing Matrix E tubes (MP Biomedicals, Solon, OH), followed by 500 μl of phenol:chloroform:isoamyl alchohol (25:24:1). Each sample was vortexed and placed on ice until all samples were processed. Samples were bead-beaten using MPBio FastPrep-24 (MP Biomedicals, Solon, OH) for 30 sec at 5.5 m/s prior to centrifugation for 5 min at 16k × g at 4 °C and transfer of the supernatant to heavy phase-lock gel tubes (5 Prime, Gaithersburg, MD). One volume of chloroform was added to each sample prior to inversion and centrifugation for 5 min at 12k × g at 4 °C. An add itional 0.5 ml of CTAB modified extraction buffer was added to each lysing matrix tube to maximize recovery of nucleic acid from each sample. One μl of linear acrylamide was added to the extracted supernatant followed by two volumes of PEG. Following a 2-hour incubation at room temperature, samples were washed with ice-cold 70% ethanol and resuspended in 30 μl of H2O. Extracted nucleic acids from each sample were pooled prior to application to the DNA column of the Qiagen AllPrep DNA/RNA extraction kit (Valencia, CA); DNA was extracted according to manufacturer's protocol. For samples (n=6) that had insufficient dust for extraction of all three weights, 0.4 g extractions were omitted. For the subsequent prospectively collected samples, the same extraction protocol was applied to 0.2 g of house dust.
PhyloChip Processing
The G2 PhyloChip is a high-density phylogenetic DNA microarray (16S rRNA microarray) containing 500,000 oligonucleotide probes. Each taxon is represented by a set of perfect match (PM) and mismatch (MM) probe pairs allowing the parallel detection of about 10,000 microbial taxonomic units based on 16S rRNA gene sequence variations. PhyloChip design and the selection of 25-mer PM and MM oligonucleotide probes is described in more detail elsewhere (Brodie et al. 2006; DeSantis et al. 2007).
PCR reactions for PhyloChip analyses were performed in 25 μl reactions using 0.02 U μl-1 Takara ExTaq (Takara Mirus Bio Inc, Madison, WI), 1X Takara buffer with MgCl2, 0.3 pmol μl-1 of 27F and 1492R primers (Lane 1991), 0.8 mg ml-1 bovine serum albumin (Roche Applied Science, Indianapolis, IN), and 30 ng of DNA template. A total of twelve reactions per sample were performed in an Eppendorf Mastercycler gradient thermocycler (Eppendorf, Westbury, NY) across a gradient (48-58.4 °C) of annealing temperatures to maximize recovered diversity. Reaction conditions were as follows: initial denaturation (95 °C for 3 min) followed by 25 cycles of 95 °C (30 s), annealing (30 s), and extension at 72 °C (2 min), and a final ext ension of 72 °C (10 min). 16S rRNA amplification was verified using a 1% TBE agarose gel.
PCR products for each sample were pooled and purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). Three concentrations (125 ng, 250 ng, and 500 ng) of 16S rRNA amplicon were applied to the G2 PhyloChip to determine whether application of higher concentrations of PCR product impacted the breadth of diversity detected by the array. The samples were prepared for PhyloChip analysis as previously described (Brodie et al. 2006). Briefly, purified 16S rRNA PCR products along with a mixture of amplicons of known concentrations (spike in control) were combined and fragmented with DNase I (Life Technologies, Carlsbad, CA) in 1X One-Phor-All buffer (Affymetrix, Santa Clara, CA) to 150-250 bp. The 3’ ends of fragments were biotin-labeled prior to application to the chip using GeneChip DNA Labeling reagent (Affymetrix, Santa Clara, CA) and Terminal Deoxynucleotidyl Transferase (Promega, Fitchburg, WI) following instructions from the Affymetrix technical manual. Labeled DNA was denatured at 99°C for 5 min and hybridized to the microarray at 48°C for at least 16 h at 60 rpm. Hybridized PhyloChips were washed and stained in a three-step process first with ImmunoPure Streptavidin (Thermo Scientific Pierce, Rockford, IL), then with Biotinylated Anti-Streptavidin antibody (Vector Labs, Burlingame, CA) and finally with a Streptavidin, R-phycoerythrin conjugate (SAPE; Life Technologies, Carlsbad, CA) in an Affymetrix GeneChip Fluidics Station 450 according to standard Affymetrix protocols (Masuda and Church 2002). Fluorescence of hybridized probes was measured using the Affymetrix GeneChip Scanner GCS 3000, and intensity data was acquired using the Affymetrix GCOS software. Array data were conservatively filtered, with taxa deemed present if ≥ 90% of probes per probe set were positive as previously described (Brodie et al. 2006). This was followed by a two-step normalization procedure based on spike in control oligonucleotides that corrects for technical variation (Ivanov et al. 2009). To test for assay reproducibility, six samples out of 15 yielding a sufficient amount of amplified 16S rRNA gene product were randomly chosen for triplicate PhyloChip profiling (S1, S7, S8, S11, S14, S18).
454-pyrosequencing of the 16S rRNA V4-V5 region
Four bedroom dust samples (S29 – S31) with sufficient DNA for further profiling, were chosen to compare bacterial community compositional profiles, generated in parallel from extracted DNA, using PhyloChip and 454-pyrosequencing. PCR for 454-pyrosequencing analyses were performed in 25 μl reactions in triplicate using 1x HotMasterMix (5 Prime, Gaithersburg, MD), 0.3 pmol μl-1 each of primer pair 515f (5’-GTGCCAGCMGCCGCGG TAA-3’; (Bergmann et al. 2011) and 926r (5’-CCGYCAATTYMTTTRAGTTT-3’; variant of (Liu et al. 1997), and 20 ng of DNA. The forward primer included a 12 nt error-correcting Golay barcode (Fierer et al. 2008) unique for each sample and a GA linker. The reverse primer included an AG linker. Reactions were performed under the following conditions: initial denaturation (95 °C for 2 min) followed by 30 cycles of 95 °C (20 s), anneali ng at 50 °C (20 s), and extension at 65 °C (1 min), and a final extension of 65 °C (10 m in). A 2-step purification was performed on the amplified products, which included Qiaquick Gel Extraction (Qiagen, Valencia, CA) followed by QIAquick PCR purification (Qiagen, Valencia, CA), according to the manufacturer's instructions. Sample concentrations were adjusted to 10 ng μl-1 and pooled prior to final dilution to achieve a 1 × 107 mol μl-1 solution according to manufacturer's instructions and submitted to the Research Technology Support Facility at Michigan State University (http://rtsf.msu.edu/) for sequencing using GS FLX titanium chemistry.
Raw sequence data were processed by trimming barcode tags and primer sequences and removing low quality sequences through the Ribosomal Database Project's (RDP) pyrosequencing pipeline – initial process (http://pyro.cme.msu.edu/index.jsp). Default filter parameters (Forward primer max edit distance = 2; Reverse primer max edit distance = 2; Max number of N's = 0; Minimum Average Exp Quality Score = 20) were used with the exception that the minimal sequence length was set at 150 bp. Taxonomy was assigned using the RDP classifier (Wang et al. 2007) prior to alignment.
Raw sequence data were processed by trimming barcode tags and primer sequences and removing low quality sequences through the Ribosomal Database Project's (RDP) pyrosequencing pipeline – initial process (http://pyro.cme.msu.edu/index.jsp). Default filter parameters (Forward primer max edit distance = 2; Reverse primer max edit distance = 2; Max number of N's = 0; Minimum Average Exp Quality Score = 20) were used with the exception that the minimal sequence length was set at 150 bp. Taxonomy was assigned using the RDP classifier (Wang et al. 2007) prior to alignment.
Comparison of 454-pyrosequencing and PhyloChip data
Of the ~8,500 taxa represented on the G2 PhyloChip, a total of 6,195 taxon-representative 16S rRNA bacterial sequences are available on Greengenes (http://greengenes.lbl.gov). These sequences were aligned and downloaded prior to extraction of the V4-V5 region from the alignment and provision of this sequence region to the RDP classifier for classification. This approach ensured that taxa detected by the array and by 454 sequencing were assigned identity using the same classification scheme. Rarefaction curves were plotted for sequence data from each sample using Analytic Rarefaction 1.3 software (www.huntmountainsoftware.com). Comparisons, using a simple binary output for presence or absence of community members, were made at both upper and lower levels of classification i.e phylum and family level respectively between 454 and parallel PhyloChip data generated from the same extracted DNA. To examine whether the additional PhyloChip findings were due to improved community coverage by the parallel nature of the array, we performed resampling of the sample with the greatest sequence reads (S29) at different read depths (1,000, 5,000, 10,000, 20,000 and 31,982 sequence reads). Classified genera at each sequencing effort level were compared to the 419 classified genera derived from representative sequences of the taxa detected by PhyloChip.
Data analyses
Bacterial community diversity (Inverse Simpson's index) was calculated using the Vegan (Oksanen 2008) package in the R statistical environment (R Development Core Team 2011). Between-group differences (e.g. bedroom vs TV/Living room floor) in taxon relative abundance was determined by two-tailed t-testing and adjusted for false discovery using q-values, as we have previously described (Cox et al. 2010; Fujimura et al. 2010; Huang et al. 2011). Fluorescence intensity variance components were estimated using restricted maximum likelihood via linear mixed effects models. The generalized logarithmic transformation (R package vsn) (Huber et al. 2002) was first applied to the intensity values, and the transformed data were fit using R package lme4 (Bates et al. 2011) with random effects specified for taxa, sample, and their interaction. Different models were fit to estimate the variability attributed to replicate samples (S1, S7, S8, S11, S14, S18), samples from adjacent areas, samples from adjacent areas of different sizes, and samples from different locations (bed, bedroom, and living room) in order to determine the effect of replication, area location and size, and site on sample collection. Fluorescence intensity values were then simulated using these models, and the concordance correlation coefficient (R package epiR) was used to assess the agreement of the intensity values. Normality and homoscedasticity of the models were inspected using residual plots, and approximate 95% percentile confidence intervals for the variance components were estimated using 5000 parametric bootstrap samples.
Results
DNA extraction from dust samples
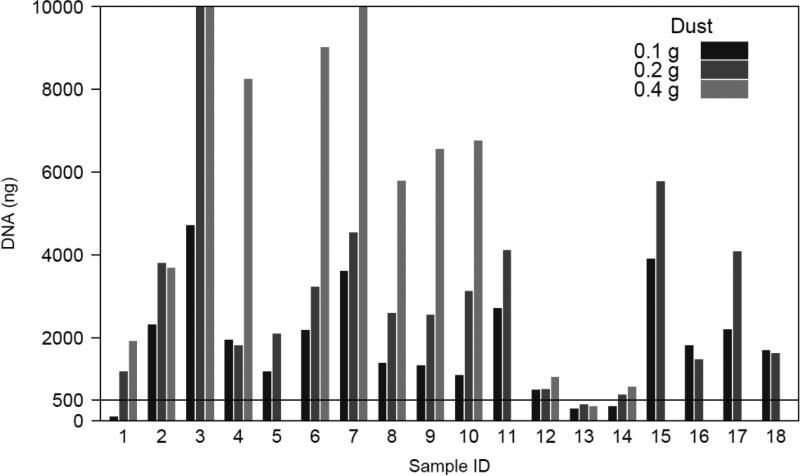
The standardized PCR conditions used to amplify the 16S rRNA for PhyloChip analysis typically require 360 ng of total DNA (30 ng per reaction). In addition, DNA is typically required for subsequent Q-PCR analyses to confirm the presence and relative abundance of detected taxa of interest. Hence a total DNA concentration of ≥500 ng is considered sufficient for profiling using this approach. A total of 15 of the 18 banked samples (83%) yielded at least 500 ng of total DNA from 0.1 g of dust (Figure 1). Extraction of DNA from 0.2 g of dust recovered ≥500 ng from 17 samples (94%). Extraction from 0.4 g of dust typically yielded higher concentrations of total DNA, though 0.4 g was too large for a single extraction tube and required the sample to be processed using two DNA isolation columns, thus doubling the costs and effort associated with extraction form this weight of dust. Only a single sample (sample 13) did not produce ≥500 ng DNA from 0.1 g, 0.2 or 0.4 g of dust. This suggests a low burden of total DNA in this sample. We noted that this sample also differed in physical appearance from all other samples and consisted primarily of human or animal hair.
Figure 1.
Total DNA isolated from eighteen banked dust samples used for optimization of array-based microbiota profiling protocol. The line indicates the minimal amount of DNA required for subsequent sample processing for PhyloChip microbiome profiling (500 ng).
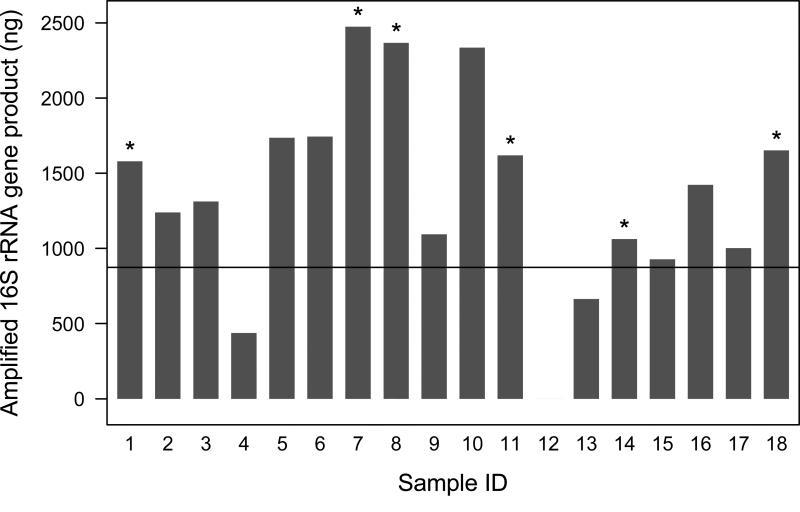
The concentration of total 16S rRNA amplified from the samples varied considerably, due presumably to wide variations in the burden of bacteria associated with these samples (Figure 2). However, the majority of samples produced substantial 16S rRNA PCR product (> 500 ng). Two exceptions included samples 4 and 12. Despite meeting our arbitrary ≥500 ng total DNA concentration, sample 4 produced only ~400 ng of 16S rRNA, while sample 12 consistently produced no PCR product. To confirm that the latter finding was not due to PCR inhibition, parallel spike-in control PCR reactions containing 100 ug of purified Pseudomonas aeruginosa genomic DNA were performed. These spiked control reactions were PCR product positive, indicating that the lack of 16S rRNA amplification in the original reactions was not due to PCR inhibition, but to a lack of bacteria in the sample. Though bacterial burden is sample specific, for the majority of samples, 0.2 g of house dust yielded sufficient DNA (≥500 ng) to produce in excess of 500 ng of amplified 16S rRNA PCR product.
Figure 2.
Total 16S rRNA concentration amplified from banked dust samples used for optimization of array-based microbiota profiling protocol. Replicate PhyloChip analyses using 250 ng was performed on samples indicated with an asterisk. The line indicates the concentration of amplified 16S rRNA needed to run 125ng, 250ng, and 500ng on the PhyloChip.
Optimal amount of 16S rRNA for PhyloChip analysis
We applied three concentrations of 16S rRNA gene product (125 ng, 250 ng, 500 ng) from each sample to the PhyloChip to determine the optimal conditions for detection of bacterial diversity present. The total concentration of amplified 16S rRNA product required for this approach was 875 ng per sample. This threshold was met by 15 out of 18 samples (83%; Figure 2). Though sample S13 produced low concentrations of total DNA, 16S rRNA amplification from this sample under standardized conditions produced sufficient PCR product to apply 125 ng and 250 ng to the PhyloChip. Sample S4 only produced sufficient amplicon to be analyzed at 125 and 250 ng, while as previously mentioned, sample S12 failed to produce any PCR product despite repeated attempts.
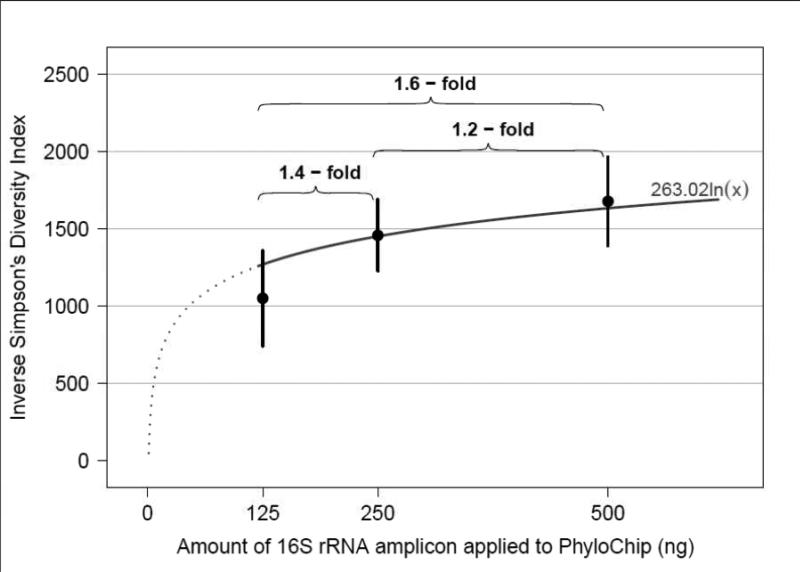
Community diversity was calculated for each sample at each of the 16S rRNA concentrations applied to the array and demonstrated expected increases in diversity with application of increased concentrations of 16S rRNA amplicon (Figure 3). While a dose-dependent response was observed, the increase in diversity was not linear, as the curve approached an asymptote at concentrations of 500 ng, suggesting that application of higher concentrations would not significantly increase the community diversity detected.
Fig. 3.
Observed Simpsons diversity (a metric based on community richness i.e. the number of taxa present, and evenness i.e. the relative distribution of those taxa within a community) in samples for which 125, 250 and 500 ng concentrations of amplified, purified 16S rRNA were applied to the PhyloChip. 125ng was significantly different (p 0.0003) from 250ng and 500ng; 250ng was not significantly different from 500ng (p 0.76)
We also examined reproducibility of microbiome profiles generated by the PhyloChip through analysis of variation across triplicate technical replicates. Following spike in control and total array intensity normalization, deviation in reported taxon fluorescence intensity across replicate datasets was examined and compared to variation in fluorescence intensity for taxa detected in distinct samples, and for all taxa detected within a single sample. As expected, variation in fluorescence intensity reported for individual taxon across replicate arrays varied the least (0.6% of total variability), followed by individual taxon across different samples (0.7%), then taxa across different samples (9.0%), and finally variation was greatest across taxa in a given single sample (89.8%; which is clearly a function of community composition). The fact that the variability in fluorescence intensity across distinct taxa was 157 times larger compared to the variability in fluorescence intensity per taxon across replicate arrays indicates the high reproducibility across replicate arrays.
Comparison of PhyloChip and 454-Pyrosequencing bacterial community profiles
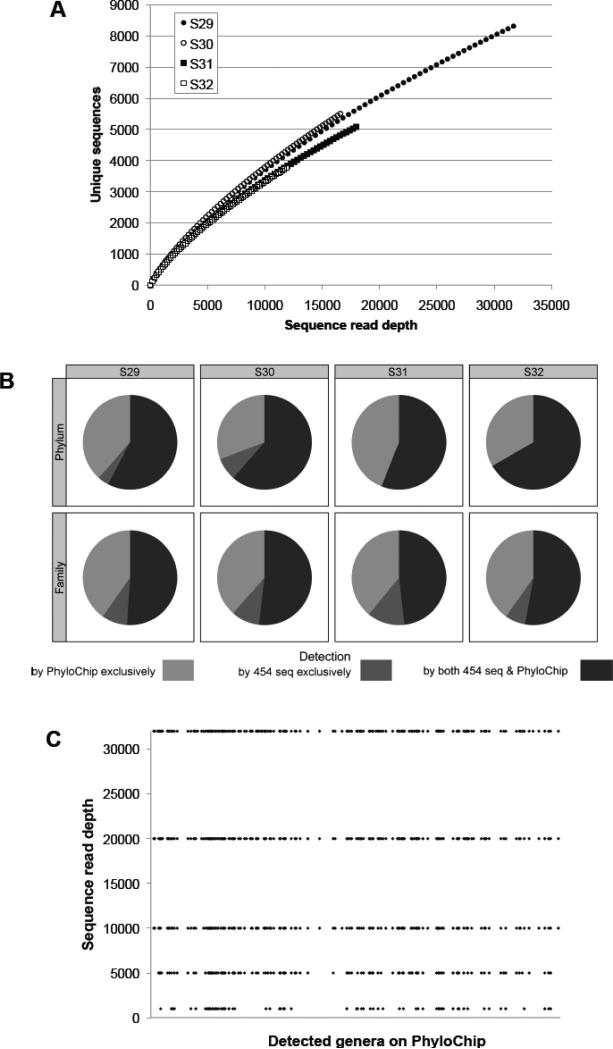
Using quality-filtered 454-sequence reads ranging from 11,994-31,982 per sample, rarefaction curves were constructed for each of four samples chosen for this component of the study (see materials and methods; Figure 4A) to determine community coverage at the sequence depths performed. All samples exhibited exponential curves with steep slopes, indicating that a large fraction of the species diversity remained to be discovered in these samples and that the depth of sequence performed for these samples, though substantial by published standards, permitted sampling of only a subset of the community members present.
Fig. 4.
A. Rarefaction curve at 0.03 distance clustering showing exponential increase in number of new sequence clusters detected (even at ~30,000 sequences), indicating highly rich microbiota. B. Plots illustrating at the Phylum and family level of classification, those community members detected by both profiling approaches (black) or exclusively by sequencing (dark gray) or PhyloChip (light gray). C. Increasing sequence read depth from S29 (which produced the greatest number of sequence reads) results in identification of increasingly greater numbers of genera detected by the PhyloChip (each line plot represents the number of genera detected by both methods).
Application of PhyloChip to these samples identified a total of 1,474 ± 242 (SD) taxa. Representative sequences of taxa detected by the array were classified in RDP and compared to those detected by 454-pyrosequencing of the same samples. A total of 25 phyla were detected by the combination of PhyloChip and 454-sequencing; 60.5% of these phyla were detected by both methods, 36.6% were detected exclusively by PhyloChip, and 2.9% exclusively by 454-sequencing (Figure 4B). Those exclusively detected by PhyloChip included Aquificae, Chlorobi, Thermodesulfobacteria, BRC1, and OP10, putatively representing minor phyla undetected by sequencing efforts in the house dust communities profiled. Phyla detected exclusively by 454-pyrosequencing included the Lentisphaerae and SR1, the former is represented by three taxa on the array (all of which were detected at levels that did not pass our threshold for presence), the latter SR1, is not represented by any taxa on the array. When the data was examined at the family level of phylogenetic classification, the trend towards greater detection of community members by PhyloChip was again evident. About 194 families were detected by the combination of the two methods; 50.9% were detected by both methods, 39.5% exclusively by PhyloChip, and 9.6% exclusively 454-sequencing. Hence comparative analysis of parallel PhyloChip and 454-pyrosequencing microbiome profiles generated in parallel from the same extracted DNA, revealed that while the large majority of organisms detected by 454-sequencing were also detected by PhyloChip, in all samples the array consistently detected substantially more community members (Figure 4B). It should also be noted that, because a fraction of taxa on the G2 array (~30%) do not have an associated representative 16S rRNA sequence (typically because of insufficient sequence length or suspicion of a chimeric sequence), the community members detected exclusively by 454-pyrosequencing may be inflated. Indeed these phyla or families may well have been identified by PhyloChip, but because their representative species 16S rRNA sequences are not included in the comparative analyses, they are not considered detected by the array.
To further illustrate that the improved detection afforded by the array is due to increased community coverage, we performed resampling at different sequencing depths (1,000, 5,000, 10,000, 20,000 and 31,982 reads) from the sample with the greatest number of sequence reads (S29). The number of classified genera detected at each defined depth of sequence reads was compared to the 419 classified genera detected by PhyloChip in this sample. This approach demonstrated that increasingly deeper sequencing efforts validated more of the PhyloChip detected genera (Figure 4C; see supplemental table for the complete list of detected genera), suggesting that for comparative analyses of highly diverse and complex communities, this tool represents an economical approach to provide a relatively high-resolution profile of bacterial community composition.
Spatial heterogeneity of microbial flora
To test our optimized protocol, we examined heterogeneity in dust-associated bacterial communities within and across homes. Of the 49 dust samples collected, 40 samples from 9 homes with sufficient material for microbiome profiling were used to examine relationships between bacterial community diversity detected and both the area of floor-covering sampled as well as the site of sample collection (i.e. TV/living room floor, child's bedroom floor, and bed). The residual variances contributed by samples from different floor positions (1×1 m2 quadrant vs an adjacent 1×1 m2), different sized sampling quadrants (1×1 m2 quadrant vs an adjacent 2×2 m2), and different locations (bed vs bedroom floor vs TV/living room floor) are presented in Table 2. Variability between samples again exhibited a hierarchy. It was smallest between adjacent 1×1 m2 sampling quadrants, marginally greater between samples from different sized quadrants in the same room, even greater between samples from different locations in the same home, while variability across samples from the same site in different homes exhibited the greatest variability (Figure 5). It should be noted that sampling a 2×2 m2 quadrant did not substantially increase detected sample diversity; due to the added effort to obtain and process such samples it was determined that a 1×1 m2 sampling quadrant provided sufficient material for such studies.
Table 2.
Variability in fluorescence intensity due to size of area and location where dust sample is collected.
| Adjacent Area Same Size 1×1 vs. 1×1m2 | Adjacent Area Different Sizes 1×1 vs. 2×2m2 | Location Bed, BRa Floor, LRb Floor | |
|---|---|---|---|
| Variance* | 0.0073 | 0.0093 | 0.0125 |
| 95% Confidence Intervalc | 0.0072 to 0.0075 | 0.0091 to 0.0094 | 0.0124 to 0.0126 |
| % of Total Variabilityd | 3.6% | 4.5% | 6.1% |
BR, bedroom
LR, TV/Living room
Residual variance component estimate in the linear mixed effects model after accounting for taxa-to-taxa variability, home-to-home variability, and their interaction.
95% percentile confidence intervals based on 5000 parametric bootstrap samples
(Residual variance / Total variance) × 100%
Fig. 5.
Agreement in taxa fluorescence intensity between i) technical replicates of the same sample (provided for reference), ii) adjacent 1×1 m2 bedroom samples, iii) adjacent 1×1 m2 and 2×2 m2 living room samples, iv) 1×1 m2 samples from the living room, bedroom, or bed in a home, and v) 1×1 m2 samples from the living room, bedroom, or bed in different homes. Shown in each plot is the concordance correlation coefficient (95% confidence interval) superimposed on a bivariate kernel-density estimate of the data. The outermost 100 observations on the distribution of the kernel-density are also plotted.
Finally we performed comparative taxon-level analyses on the bedroom and living room samples to determine which site represented the most appropriate room for sample collection. We hypothesized that that due to relevant activities in each room, bedroom dust microbiota would be predominantly composed of skin-associated microbiota (self-microbial exposures) while living room would be associated predominantly with environmental species. With this small sample, not unexpectedly, none of the findings passed our false discovery criteria for significance; however, initial indications suggested that bedroom dust microbiota possessed significantly higher abundance of Streptococcaceae, Prevotellaceae, and Staphylococcaceae (p < 0.05), bacterial families synonymous with human colonization. In comparison species detected in significantly higher abundance in living room dusts were members, typically associated with environmental origin, of the Comamonadaceae, Desulfomicrobiaceae, Actinomycetaceae, Synthorphobacteriaceae and Flavobacteriaceae.
Discussion
Microbiome studies using culture-independent tools offer a powerful approach to examine, without the restrictions of culture, the microbial community members associated with samples from a variety of sources. Though this approach has been favored, employed, and largely developed by environmental microbial ecologists for several decades, recent application of these tools to samples collected from humans has revealed a previously hidden world of microbial inhabitants at a variety of sites traditionally thought of as sterile or, at best, colonized by a handful of species (Brodie et al. 2007; Cox et al. 2010; Huang et al. 2011). Microbiome profiling, particularly that which has targeted the gastrointestinal tract, has revealed the presence of a wide diversity of bacterial phyla in a number of human niches, and provided invaluable information on the role these complex communities play in both metabolic and immune functions of the human host (Ivanov et al. 2009; Kwon et al. 2010; Turnbaugh et al. 2009; Turnbaugh et al. 2006). However, it is becoming increasingly apparent that microbial colonization of the human host is influenced, in part, by environmental microbial exposures and that microbial ecology in human environments can be markedly affected by human behaviors, such as livestock- (Eduard et al. 2004) or pet-keeping (Fujimura et al. 2010) or habitation in geographically adjacent but geoclimatic, vegetatively, and developmentally distinct locales (Pakarinen et al. 2008). It has been suggested that the decreased risk for asthma incidence associated with exposure to livestock or pets may be related to the specific patterns of environmental microbial communities associated with these animals (Braun-Fahrlander 2003; Fujimura et al. 2010).
To address such hypotheses it is necessary to examine, at a high level of resolution, the microbiota associated with specific environmental samples. This is particularly true of studies linking environmental samples to human samples and human disorders. For example, rare, low abundance species found detected in dust samples may serve to inoculate a human niche such as the gastrointestinal tract, where they may become dominant microbiome members due to more favorable microniche conditions and selective pressures in the host. Hence it is critical for such studies to identify the lower abundance species in environmental samples so that relationships between microbial exposures, host microbiome composition, and host disease states may be made. As such, we have optimized an approach to provide a high-resolution microbiome profile for sampled dust using a phylogenetic microarray.
Using a banked cohort of frozen dust samples, we demonstrated variable total DNA concentrations in samples from which a standardized weight of dust had been extracted. However, the majority of samples produced sufficient DNA (minimum of 500 ng) from 0.2 g of dust to proceed to the 16S rRNA amplification stage. Given the additional costs and effort associated with DNA extraction from 0.4 g of dust, DNA extraction from 0.2 g would be sufficient to yield adequate total DNA for subsequent 16S rRNA amplification and is recommended as the minimum weight of dust necessary for subsequent studies.
As expected, the concentration of 16S rRNA PCR product amplified from the samples also varied considerably. Interestingly, a sample that barely produced sufficient total DNA for subsequent amplification, yielded relatively large concentrations of 16S rRNA (in excess of 500 ng), while another possessing ample total DNA, failed to yield a 16S product. This underscores that while guidelines may be drawn up defining optimal concentrations of nucleic acids necessary to proceed to the amplification step, the concentration of 16S rRNA product produced is entirely dependent on the bacterial burden in a given sample, which cannot be estimated from total DNA concentrations.
Examining the level of diversity detected from application of a variety of amplified 16S rRNA product to the array, demonstrated a dose-dependent response; for samples examined in this study, 500 ng appeared to represent the concentration at which the detected diversity approached an asymptote. Though we recognize that the concentration of amplified 16S rRNA product obtained is dependent on the sample and that some types of samples (e.g. healthy subject airway samples) typically house low bacterial burden, we recommend that, if possible, 500 ng of amplified 16S rRNA product should be applied to the array to maximize community diversity detected.
We choose the 16S rRNA PhyloChip in preference to next-generation sequencing since it provides broad coverage of the majority of known bacterial species and can detect lower abundance species with the same efficiency as the dominant members of a bacterial community using considerably fewer resources. A comparative analysis of PhyloChip and 454-sequencing performed in parallel from the same pool of total extracted DNA revealed that, as we have previously demonstrated (DeSantis et al. 2007; Flanagan et al. 2007), the array consistently detected substantially more microbial community members than the sequencing approach. These additional community members are considered by some to represent cross-hybridization on the array and dismissed as extraneous and irrelevant species. However, here we demonstrate that the depth of sequence reads performed on a given sample can dramatically influence the number of genera detected and that as deeper sequence efforts are leveraged, 454-pyrosequencing identified increasingly greater numbers of genera detected by the array. This is particularly pertinent since our rarefaction curve analysis of the sequence-based profiles clearly demonstrated that even at depths of >30,000 sequence reads, community coverage is relatively poor. This suggests that the additional detected taxa by the array represent the long tail of rare species in the typically log-normal distribution of microbial communities. An on-going debate exists in the field as to whether these rare community members hold any significance to community composition or function. However, recent data generated by the European MetaHit consortium (Arumugam et al. 2011), demonstrated that specific genes detected in high abundance in the GI tract of subjects examined in their study were contributed by rare members of the microbial community, suggesting that these lower abundance community members may contribute substantially to community function. In addition, a more recent study demonstrated that Prophyromonas gingivalis, while remaining a relatively low abundance species in communities associated with murine periodontal disease, leverages host immune response to dramatically restructure community composition (Honda 2011), thus representing a true keystone species (i.e. one which exerts an effect on the community disproportionate to its relative abundance in the assemblage). Hence the importance of low abundance species should not be underestimated, though it is clear that additional studies are necessary to determine whether rare species in the environment can be transferred and established in the human body and what their relevance is to human health. Nonetheless, application of economical profiling approaches that detect both low and high abundance community members provide not only a higher resolution profile of community composition, but also increases resource conservation, which, for epidemiological and clinical studies with their attendant large sample sizes (Arumugam et al. 2011), is critical.
Examination of spatial heterogeneity in microbiota composition across adjacent similar sized quadrants, adjacent different sized quadrants, different rooms in a house, and the same site across different houses, demonstrated that taxonomically, these samples exhibited a hierarchical trend towards increased variability across these groups respectively (i.e. similar sized adjacent quadrants exhibited more similar community composition, compared with TV/living room floor communities from different houses). This suggests that the profiling approach used can detect taxonomic differences that characterize these sites and that using this approach could lead to identification of bacteria in house dust that are associated with diseases such as asthma. In addition, this study provides evidence that sampling a 1 ×1 meter quadrant is sufficient to provide enough materials for epidemiological research and that living room dust samples appear to contain a better representation of environmental- as opposed to human-origin species, when compared to bedroom dust samples. Hence, for similar studies we would recommend sampling a 1 ×1 meter quadrant in the TV/living room of houses to represent an optimal site for examining home environmental microbial exposures related to particular studies of human disease.
Limitations of this study include the small number of samples and that the samples for the optimization component of the study were from a frozen sample bank. In addition, these samples originated from a single geographic area and are not population-based. The study also reveals the limitations of current culture-independent microbial profiling approaches. Sequencing may, if insufficient sequence reads are performed per sample, miss large swathes of microbial diversity present, while the array based approach can only detected those taxa that are known and targeted by the array. Strengths include the ability of the phylogenetic microarray used in this study to produce highly reproducible microbiome profiles and to detect subtle differences in distinct sample types. Thus this investigation represents a proof of principle study and provides guidelines for performing subsequent studies of house dust microbiome related to asthma development.
Supplementary Material
Highlights.
Provide guidelines for standardized microbiome-based studies of house dust.
DNA yield, 16S rRNA dose response and assay reproducibility were tested.
Pilot study revealed hierarchy in community composition concordance.
Least community variability between technical replicates, greatest between rooms from different houses.
454-pyrosequencing verified bacteria detected by phylogenetic microarray.
Acknowledgements
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under Contract numbers NO1-AI-25496, NO1-AI-25482, HHSN272200900052C and HHSN2722010000521. Additional support was provided by the National Center for Research Resources, National Institutes of Health, under grants RR00052, M01RR00533, 1UL1RR025771, M01RR00071, 1UL1RR024156, and 5UL1RR024992-02.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B. lme4: Linear mixed-effects models using S4 classes. R package version 0.999375-41. 2011.
- Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43(7):1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun-Fahrlander C. Environmental exposure to endotoxin and other microbial products and the decreased risk of childhood atopy: evaluating developments since April 2002. Curr Opin Allergy Clin Immunol. 2003;3(5):325–329. doi: 10.1097/00130832-200310000-00001. [DOI] [PubMed] [Google Scholar]
- Brodie EL, Desantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72(9):6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie EL, DeSantis TZ, Parker JP, Zubietta IX, Piceno YM, Andersen GL. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci U S A. 2007;104(1):299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse WW. The National Institutes of Allergy and Infectious Diseases networks on asthma in inner-city children: an approach to improved care. J Allergy Clin Immunol. 2010;125(3):529–537. doi: 10.1016/j.jaci.2010.01.036. quiz 538-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention [August 7, 2012];Asthma in the US. 2011 Available: http://www.cdc.gov/vitalsigns/Asthma/
- Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, Daly RA, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5(6):e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, Firestone MK. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3(2):168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53(3):371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- Eduard W, Douwes J, Omenaas E, Heederik D. Do farming exposures cause or prevent asthma? Results from a study of adult Norwegian farmers. Thorax. 2004;59(5):381–386. doi: 10.1136/thx.2004.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105(46):17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JL, Brodie EL, Weng L, Lynch SV, Garcia O, Brown R, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol. 2007;45(6):1954–1962. doi: 10.1128/JCM.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura KE, Johnson CC, Ownby DR, Cox MJ, Brodie EL, Havstad SL, et al. Man's best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126(2):410–412. e411–413. doi: 10.1016/j.jaci.2010.05.042. 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruchalla RS, Pongracic J, Plaut M, Evans R, 3rd, Visness CM, Walter M, et al. Inner City Asthma Study: relationships among sensitivity, allergen exposure, and asthma morbidity. J Allergy Clin Immunol. 2005;115(3):478–485. doi: 10.1016/j.jaci.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330(6001):204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- Honda K. Porphyromonas gingivalis sinks teeth into the oral microbiota and periodontal disease. Cell Host Microbe. 2011;10(5):423–425. doi: 10.1016/j.chom.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127(2):372–381. e371–373. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Variance Stabilization Applied to Microarray Data Calibration and to the Quantification of Differential Expression. Bioinformatics. 2002;18:S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph CL, Williams LK, Ownby DR, Saltzgaber J, Johnson CC. Applying epidemiologic concepts of primary, secondary, and tertiary prevention to the elimination of racial disparities in asthma. J Allergy Clin Immunol. 2006;117(2):233–240. doi: 10.1016/j.jaci.2005.11.004. quiz 241-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HK, Lee CG, So JS, Chae CS, Hwang JS, Sahoo A, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci U S A. 2010;107(5):2159–2164. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63(11):4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Church GM. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J Bacteriol. 2002;184(22):6225–6234. doi: 10.1128/JB.184.22.6225-6234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, et al. Community Ecology Package. R Package version 114-9. 2008.
- Pakarinen J, Hyvarinen A, Salkinoja-Salonen M, Laitinen S, Nevalainen A, Makela MJ, et al. Predominance of Gram-positive bacteria in house dust in the low-allergy risk Russian Karelia. Environ Microbiol. 2008;10(12):3317–3325. doi: 10.1111/j.1462-2920.2008.01723.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; Austria: 2011. [Google Scholar]
- Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.