Abstract
The resistance of hypoxic cells to radiotherapy and chemotherapy is a major problem in the treatment of cancer. Recently, an additional mode of hypoxia-inducible factor (HIF)-dependent transcriptional regulation, involving modulation of a specific set of micro RNAs (miRNAs), including miR-210, has emerged. We have recently shown that HIF-1 induction of miR-210 also stabilizes HIF-1 through a positive regulatory loop. Therefore, we hypothesized that by stabilizing HIF-1 in normoxia, miR-210 may protect cancer cells from radiation. We developed a non-small cell lung carcinoma (NSCLC)-derived cell line (A549) stably expressing miR-210 (pmiR-210) or a control miRNA (pmiR-Ctl). The miR-210-expressing cells showed a significant stabilization of HIF-1 associated with mitochondrial defects and a glycolytic phenotype. Cells were subjected to radiation levels ranging from 0 to 10 Gy in normoxia and hypoxia. Cells expressing miR-210 in normoxia had the same level of radioresistance as control cells in hypoxia. Under hypoxia, pmiR-210 cells showed a low mortality rate owing to a decrease in apoptosis, with an ability to grow even at 10 Gy. This miR-210 phenotype was reproduced in another NSCLC cell line (H1975) and in HeLa cells. We have established that radioresistance was independent of p53 and cell cycle status. In addition, we have shown that genomic double-strand breaks (DSBs) foci disappear faster in pmiR-210 than in pmiR-Ctl cells, suggesting that miR-210 expression promotes a more efficient DSB repair. Finally, HIF-1 invalidation in pmiR-210 cells removed the radioresistant phenotype, showing that this mechanism is dependent on HIF-1. In conclusion, miR-210 appears to be a component of the radioresistance of hypoxic cancer cells. Given the high stability of most miRNAs, this advantage could be used by tumor cells in conditions where reoxygenation has occurred and suggests that strategies targeting miR-210 could enhance tumor radiosensitization.
Keywords: apoptosis, hypoxia, microRNA, non-small cell lung cancer, radiotherapy
It has been well established that hypoxic cells are more resistant to radiation therapy than are normoxic cells, as first demonstrated in 1953 by Gray et al.1 Part of this resistance, called the ‘oxygen effect', can be explained by a reduction in production capacity and reaction of free radicals of water radiolysis.2 However, the master transcriptional activator of the hypoxic response, the hypoxia-inducible factor 1 (HIF-1),3 has also been widely linked to the radiation resistance of hypoxic cells.4 HIF-1 affects numerous processes, such as proliferation, migration or cellular metabolism, to activate survival pathways in response to radiation, and has been shown to be highly associated with poor prognosis and resistance to anticancer radiotherapy in many cancers.5, 6, 7, 8 In addition, targeting HIF-1 in tumor and stromal cells using different strategies can resensitize tumors to radiotherapy.8, 9, 10, 11
MicroRNAs (miRNAs)12 are new factors implicated in the regulation of gene expression, and over the past decade they have been found to have key roles in cancers,13, 14, 15, 16 including lung cancer.17 These molecules typically inhibit translation and/or reduce the stability of mRNAs of genes involved in tumorigenic processes, such as inflammation, cell cycle regulation, stress response, differentiation, apoptosis and invasion. Of note, recent studies have suggested a link between expression of some miRNAs and radiotherapy, particularly in lung cancer.18, 19, 20, 21 Alterations in miRNAs expression can be caused by various mechanisms, including deletions, amplifications or mutations, epigenetic silencing, or dysregulation of transcription factors.16 Notably, it has been demonstrated that a specific set of miRNAs are upregulated by hypoxia.22 Among these hypoxia-regulated miRNAs, the HIF-1-responsive miR-210 appears as a unique evolutionarily conserved and ubiquitously expressed ‘hypoxamir'.22, 23 MiR-210 involvement in a wide-range of processes, including cell proliferation, migration, apoptosis, differentiation, DNA repair and cell metabolism,22, 23 and more recently in the regulation of antitumor immune responses,24 underline the role of this miRNA as a central master regulator of the hypoxic response. Among this wide range of cellular effects, miR-210 has been shown to repress mitochondrial metabolism by targeting a number of proteins that are crucial for normal tricarboxylic acid cycle and electron transport chain (ETC) activity. These include an iron–sulfur cluster assembly homolog 1/2 (ISCU1/2),25, 26, 27 cytochrome c oxidase assembly protein (COX10),27 NADH dehydrogenase (ubiquinone) 1α subcomplex 4 (NDUFA4), a subunit of ETC complex I, and succinate dehydrogenase complex, subunit D (SDHD), a subunit of the ETC complex II.28 Recently, we demonstrated that miR-210 was overexpressed in the most aggressive stages in human non-small cell lung carcinomas (NSCLCs) and correlated with a hypoxic signature.28 We demonstrated that transient overexpression of miR-210 induced mitochondrial dysfunction and silenced the expression of SDHD, resulting in increased stabilization of HIF-1α, even in normoxia. We, thus, hypothesized that overexpression of miR-210 could stabilize HIF-1 and induce a hypoxic response to affect the sensitivity of cancer cells to radiotherapy in normoxia. To answer these questions, we developed NSCLC-derived cell lines stably expressing miR-210 or a miR control (miR-Ctl), and compared their resistance to high doses of radiation (0–10 Gy).
Results
Generation and characterization of an A549-derived cell line stably expressing miR-210
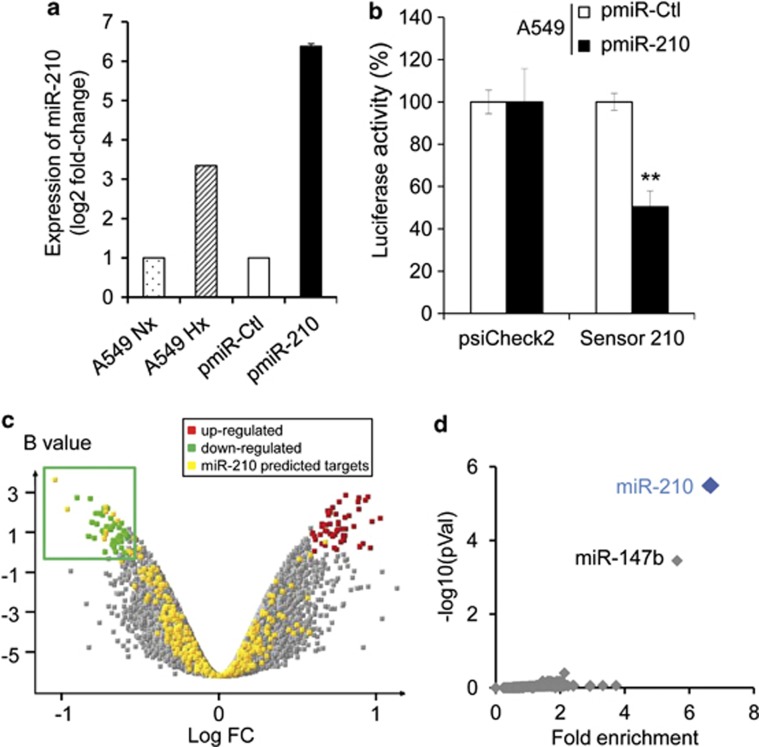
Using lentiviral vectors, we first established A549 cells that stably overexpressed hsa-miR-210 or a control ce-miR-67, named pmiR-210 and pmiR-Ctl, respectively. A stable and moderate overexpression of miR-210 was observed in pmiR-210 cells in comparison with miR-Ctl cells (Figure 1a). Luciferase activity of a miR-210 sensor construct29 was decreased in miR-210 cells compared with miR-Ctl cells, confirming that miR-210 activity was elevated in the pmiR-210 cells (Figure 1b). We then performed a transcriptomic analysis using human whole-genome expression microarrays. The comparison between miR-210 and miR-Ctl cells showed a modulation of 150 annotated transcripts, corresponding to 66 up- and 84 downregulated genes in the pmiR-210 cell line (Figure 1c). Several miR-210 predicted targets were found in the set of downregulated transcripts (Figure 1c). We next analyzed this set using our web tool ‘MiRonTop'30 to better characterize this effect. The results indicated an overrepresentation of miR-210-predicted targets in the set of downregulated transcripts (enrichment factor=6.55; P<0.0001), according to the ‘TargetScan' algorithm with 16 predicted targets (Figure 1d and Table 1). Gene ontology analysis of these 16 genes showed an association with the ‘ETC', confirming the downregulation of two ETC subunits, SDHD and NDUFA4, which we had previously identified.28 Moreover, validated miR-210 targets ISCU1/225 and GPD1L,31, 32 two genes associated with the ability of miR-210 to downregulate mitochondrial respiration or promote glycolysis, were confirmed. We, therefore, established a stable population expressing hypoxic physiopathological levels of miR-210, which are able to constitutively repress genes associated with mitochondria metabolism.
Figure 1.
Identification of a miR-210 transcriptional signature in pmiR-210 A549 cells. (a) MiR-210 expression was analyzed by qRT-PCR on A549 cells cultivated in normoxia or hypoxia (1% O2) and on A549-derived pmiR-Ctrl and pmiR-210 cells. Mean±S.E.M. is representative of three independent experiments carried out in duplicate. (b) Activity of miR-210 was assayed in pmiR-Ctl and pmiR-210 cell lines with a 2 × perfect sensor construct, and were normalized using psiCheck2. Mean±S.E.M. is representative of three independent experiments carried out in triplicate. **P<0.05. (c) Volcano plot showing the distribution of differentially expressed genes between pmiR-210 versus pmiR-Ctl A549 cells. The green window focuses on significantly downregulated transcripts, containing a high proportion of miR-210-predicted targets (yellow dots). (d) Overrepresentation of miR-210-predicted targets in the pmiR-210 cell line according to TargetScan algorithm. Representation of miRNA predicted targets in the set of downregulated genes was compared with the set of all expressed genes. For each miRNA, a fold enrichment value (horizontal axis) and an associated P-value (vertical axis) were calculated. The best enrichment score and P-value are obtained for miR-210. A significant score is also obtained for miR-147b, confirming the close functional proximity of miR-210 and miR-147b11
Table 1. List of the top miR-210 predicted targets significantly downregulated in the pmiR-210 A549 cell line.
| Symbol | Gene ID | AVEEXPR | LOGFC | Description |
|---|---|---|---|---|
| CORO1C | 23603 | 11.68 | −0.44 | Coronin, actin binding protein, 1C |
| DIMT1L | 27292 | 12.13 | −0.89 | DIM1 dimethyladenosine transferase 1-like |
| ELFN2 | 114794 | 9.27 | −0.61 | Extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| ENOX2 | 10495 | 9.65 | −0.54 | Ecto-NOX disulfide-thiol exchanger 2 |
| FGFRL1 | 53834 | 8.56 | −0.75 | Fibroblast growth factor receptor-like 1 |
| GPD1L | 23171 | 9.58 | −0.55 | Glycerol-3-phosphate dehydrogenase 1-like |
| INF2 | 64423 | 10.78 | −0.62 | Inverted formin, FH2 and WH2 domain containing |
| ISCU1/2 | 23479 | 13.06 | −0.58 | Iron-sulfur cluster scaffold homolog |
| MITF | 4286 | 9.84 | −0.52 | Microphthalmia-associated transcription factor |
| NDUFA4 | 4697 | 14.87 | −0.58 | NADH dehydrogenase (ubiquinone) 1αsubcomplex, 4, 9 kDa |
| SDF2 | 6388 | 10.63 | −0.61 | Stromal cell-derived factor 2 |
| SDHD | 6392 | 11.06 | −0.46 | Succinate dehydrogenase complex, subunit D, integral membrane protein |
| ST6GAL2 | 84620 | 10.20 | −0.57 | ST6 β-galactosamide α-2,6-sialyltranferase 2 |
| TMEM20 | 159371 | 7.98 | −0.49 | Transmembrane protein 20 |
| TNPO1 | 3842 | 9.69 | −0.41 | Transportin 1 |
| VAMP7 | 6845 | 11.21 | −0.78 | Vesicle-associated membrane protein 7 |
Sixteen of the miR-210 predicted transcripts selected using the bioinformatics tool miRonTop (http://www.microarray.fr:8080/miRonTop/index) are listed. The Target Prediction Tool used was TargetScan. Cutoff values for selection are equal to 7 for logarithm (base 2) of the average intensity (AveExpr), to 0.4 for the logarithm (base 2) of the ratio (pmiR-210/pmiR-Ctr) (logFC) and 0 for the B-value. Gene ID: corresponds to NCBI Gene ID (http://www.ncbi.nlm.nih.gov/gene)
MiR-210 alters mitochondrial function in A549 cells
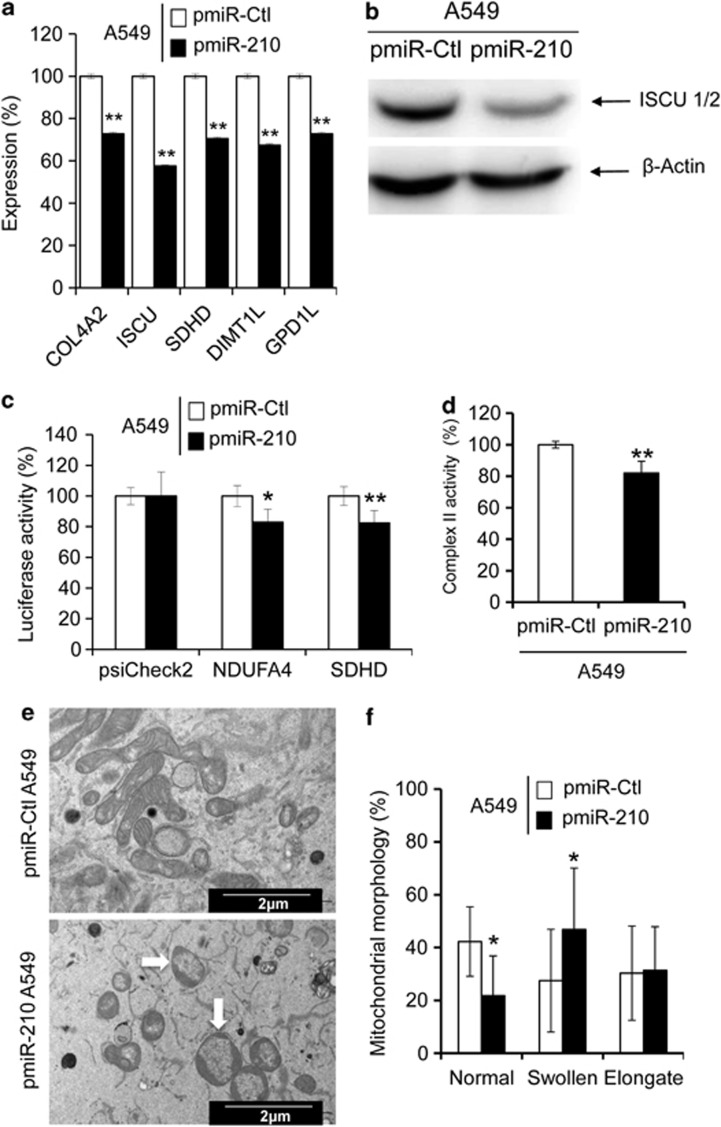
We next analyzed the expression level of a selection of validated targets of miR-210 by qRT-PCR. A reduction of mRNA levels was observed for five targets of miR-210 in pmiR-210 cells, including COL4A2, ISCU1/2, SDHD, DIMTL and GPDL1 (Figure 2a). A 50% decrease in the protein expression of ISCU1/2 confirmed the effective reduction in mRNA levels (Figure 2b). Transfection of NDUFA4 and SDHD 3′-untranslated region (UTR) pSiCheck-2 constructs showed a reproducible reduction of luciferase activity in pmiR-210 cells compared with pmiR-Ctl cells, confirming that both genes are direct miR-210 targets (Figure 2c). A significant reduction of SDH complex II activity was also noticed in pmiR-210 cells (Figure 2d). Finally, electron microscopy performed on pmiR-210 cells revealed enlarged mitochondria with rearranged cristae in comparison with the normal mitochondria morphology in pmiR-Ctl cells (Figures 2e and f). Overall, these results confirmed the strong link between miR-210 and mitochondrial functions.
Figure 2.
Mitochondrial functions are altered in pmiR-210 A549 cells. (a) Expression levels of five miR-210 targets (predicted and confirmed) were measured by qRT-PCR in pmiR-Ctl and pmiR-210 A549 cells. Mean±S.E.M. is representative of three independent experiments carried out in triplicate. (b) Western blotting showing downregulation of ISCU1/2 in pmiR-210 cells. Representative blot out of three independent experiments is shown. (c) Effect of miR-210 on NDUFA4 and SDHD was assessed using luciferase reporter assay. NDUFA4 3′-UTR, SDHD 3′-UTR construct or the control construct were transfected into pmiR-Ctl and pmiR-210 A549 cells and assayed for luciferase activity. Renilla luciferase values were normalized to Firefly luciferase activity. Mean±S.E.M. is representative of three independent experiments carried out in triplicate. (d) Complex II activity assays were performed on pmiR-Ctl and pmiR-210 A549 cells 48 h after plating. Mean±S.E.M. is representative of three independent experiments carried out in duplicate. (e) Electron micrograph showing morphology of mitochondria in pmiR-Ctl and pmiR-210 A549 cells. (f) Quantification of mitochondrial morphology. The quantification was done on thirty cells for each cell line. *P<0.01 and **P<0.05
Stable expression of miR-210 stabilizes HIF and promotes a more glycolytic phenotype
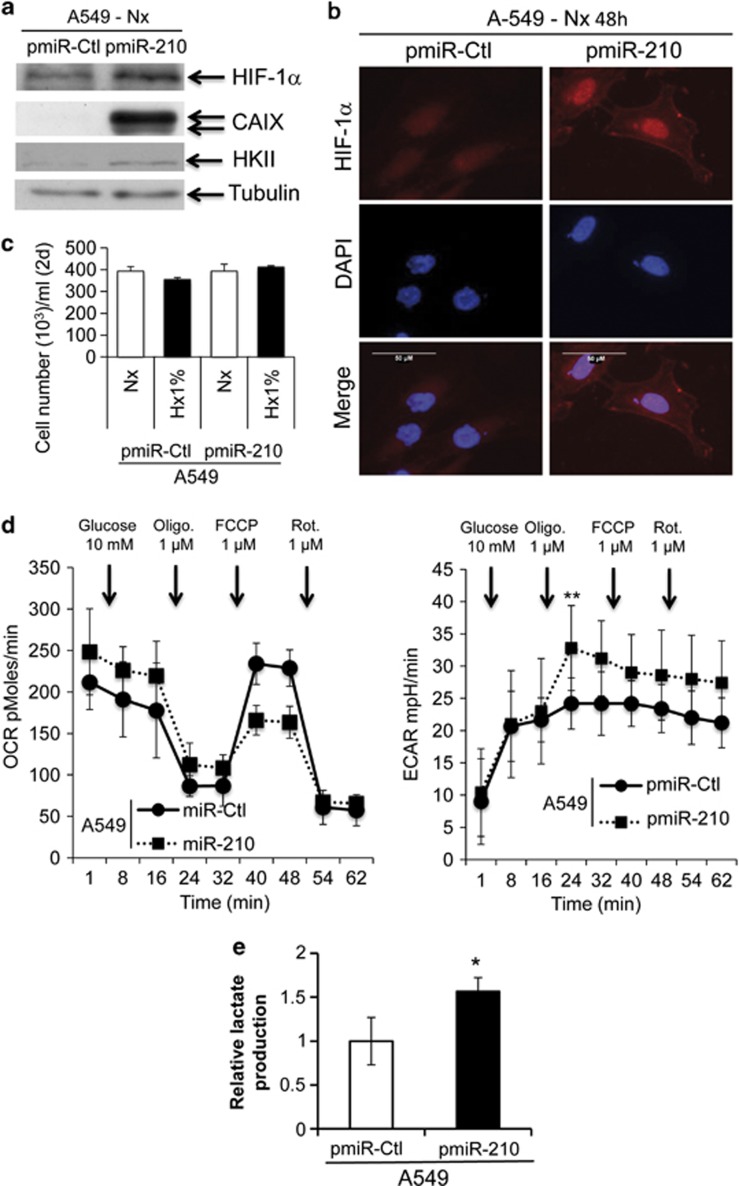
We investigated whether miR-210 was able to promote the stabilization of HIF-1 in pmiR-210 cells. We first monitored the expression of a sensitive target of HIF-1, carbonic anhydrase IX (CAIX, Figure 3a). Immunofluorescence confirmed that indeed HIF-1 was stabilized in pmiR-210 cells in normoxia in close relationship with the expression of CAIX (Figure 3b). No differences were observed between the proliferation rates of pmiR-Ctl and pmiR-210 cells (Figure 3c), strongly suggesting that the stabilization of HIF-1 was mainly due to miR-210-mediated metabolic gene targeting and not via an indirect mechanism mediated by an increased cell proliferation.33
Figure 3.
Metabolic characterization of pmiR-Ctl and pmiR-210 A549 cells. (a) Immunoblotting of HIF-1α, CAIX, HKII and tubulin in pmiR-Ctl and pmiR-210 A549 cells in normoxia. (b) Immunofluorescence of HIF-1α in pmiR-Ctl and pmiR-210 A549 cells in normoxia; scale bar, 50 μm. (c) Characterization of the growth of pmiR-Ctl and pmiR-210 A549 cells incubated in normoxia (Nx) or in hypoxia 1% O2 (Hx1%) for 2 days (2d). Mean±S.E.M. is representative of two independent experiments carried out in duplicate. (d) OCR (left panel) and ECAR (right panel) were measured in real time in a Seahorse XF bioenergetic assay. A total of 2 × 104 pmiR-Ctl and pmiR-210 A549 cells were seeded for 2d. Average OCR and ECAR were calculated from at least four measurements during the treatment of each compound (glucose, oligomycin, FCCP, rotenone) at the concentration as indicated. Mean±S.E.M. is representative of six independent experiments carried out in tetraplicate. **P<0.01 shows significant difference from the miR-Ctl after addition of oligomycin. (e) After 2d of culture, cells were lysed in assay buffer by sonication. Then, lactate was quantified in cellular extracts. Lactate quantity is expressed in relative lactate production. Mean±S.E.M. is representative of three independent experiments carried out in duplicate. *P<0.05 shows significant difference from the miR-Ctl
To compare the metabolic profiles of pmiR-Ctl and pmiR-210 cells, we used the SeaHorse XF bioenergetic system to quantify the mitochondrial respiration in which OXPHOS is measured by oxygen consumption rate (OCR) and glycolysis is measured by the generation of lactate as monitored by the consequent extracellular acidification rate (ECAR). Cells were deprived of glucose for 1 h, after which the effects of successive addition of glucose, oligomycin, FCCP and rotenone were measured for OXPHOS and glycolysis rates in real time. The maximum capacity of pmiR-Ctl cell respiration measured from the baseline in the presence of oligomycin and then FCCP was much larger than that obtained in the pmiR-210 cells, suggesting pmiR-210 cells have a lower respiratory capacity (Figure 3d, left panel). Moreover, the addition of oligomycin, which blocks mitochondrial ATP production, showed that pmiR-210 cells have an increased glycolytic rate compared with pmiR-Ctl (Figure 3d, right panel). Finally, pmiR-210 cells showed greater lactate production compared with pmiR-Ctl cells (Figure 3e), confirming the increase in glycolytic flux.
We next used another NSCLC cell line, H1975, along with HeLa cells stably expressing luciferase under a hypoxia responsive element (HRE) named HeLa–HRE Luc.34 The two cell lines were infected with pre-miR-210 lentiviral vectors, leading to a stable and moderate expression of mature miR-210 (Supplementary Figure S1a). As expected, miR-210 overexpression led to a decrease in miR-210 sensor luciferase activity and NDUFA4 or SDHD 3′-UTR pSiCheck-2 constructs in H1975 cells, as well as a significant downregulation of ISCU1/2 protein levels in the two cell lines (Supplementary Figures S1b and c). As miR-210 acts on several mitochondrial targets that could mediate apoptosis,28 we next measured caspase-3 activity in the different cell line models (Supplementary Figures S1d–f). We could detect a moderate but significant induction of caspase-3 in the pmiR-210 population compared with pmiR-Ctl cells for H1975 and HeLa–HRE Luc cell lines, whereas no significant differences could be detected for A549 cells. However, no significant differences in cell growth and viability could be observed between pmiR-210 and pmiR-Ctl conditions for the three cell lines (data not shown). Immunofluorescence indicated that HIF-1 was also stabilized in the two additional pmiR-210 cell types in normoxia compared with pmiR-Ctl cells (Supplementary Figures S2a and b). Moreover, although luciferase activities measured from the two subpopulations of HeLa–HRE Luc were similar in hypoxia, a significantly higher activity was found in pmiR-210 cells in normoxia (Supplementary Figure S2c). These data were confirmed by lactate production, with the two pmiR-210 cells showing greater lactate production compared with control cells (Supplementary Figure S2d). Overall, these results confirm and extend to other cell lines our previous data,28 showing that miR-210 alone can partially mimic hypoxic conditions.
MiR-210 confers resistance to radiation
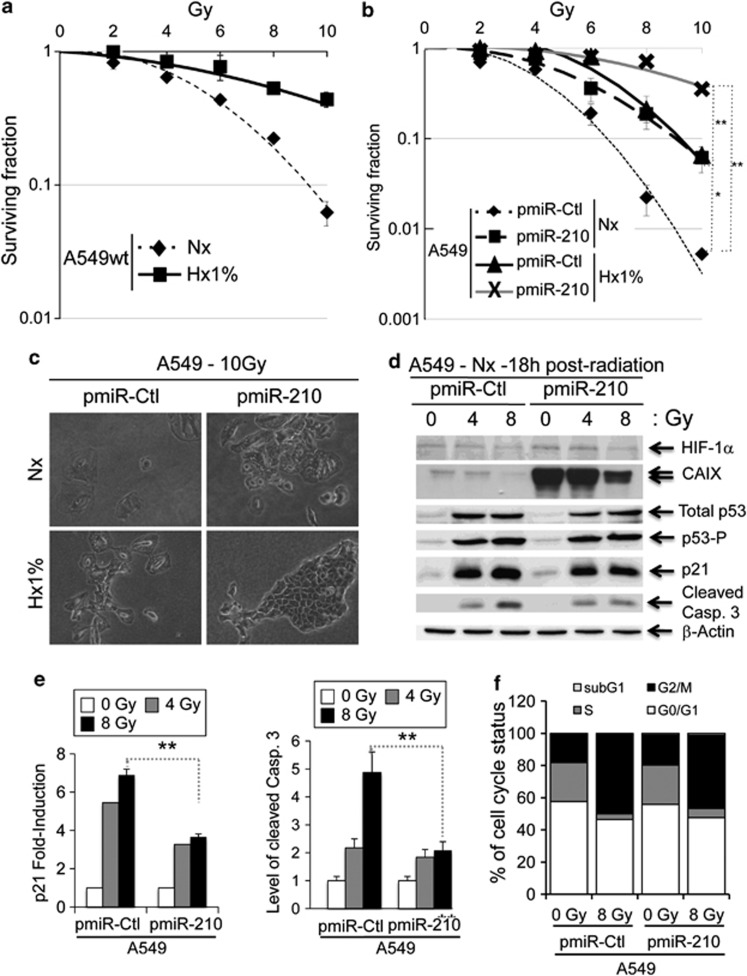
We next analyzed the impact of miR-210 expression levels on the sensitivity to radiation. As a control, we first confirmed the protective effect of hypoxia on A549 parental cells. Figure 4a illustrates the fraction of clonogenic survival as a function of radiation dose in A549 wt cells in normoxia and hypoxia 1% O2 (Hx1%), confirming that hypoxic A549 cells are more radioresistant. We then tested radiosensitivity of pmiR-Ctl and pmiR-210 A549 cells in either normoxia or hypoxia (Figure 4b and Supplementary Figure S3a). Interestingly, pmiR-210 A549 cells in normoxia exhibited a radioresistance similar to that of pmiR-Ctl cells in hypoxia. Moreover, hypoxia was able to increase this resistance. The morphology of the different colonies obtained 7 days after irradiation (Figure 4c) confirmed the protective effect associated with miR-210 expression. Importantly, pmiR-210 H1975 as well as pmiR-210 HeLa–HRE Luc cells also presented a significant radioresistance in normoxia compared with pmiR-Ctl (Supplementary Figures S3b and c).
Figure 4.
Radioresistance of pmiR-210 A549 cells. (a) Radioresistance of wild-type A549 (A549wt) cells cultured for 2 days in normoxia (Nx) and in hypoxia 1% O2 (Hx1%), and treated with the indicated dose of radiation. Cells were then subjected to a clonogenic cell survival assay. In x axis is the dose of X-radiation. In y axis is the surviving fraction. Mean±S.E.M. is representative of two independent experiments carried out in duplicate. (b) Radioresistance of both pmiR-Ctl and pmiR-210 A549 cells cultured for 2 days in normoxia (Nx) and hypoxia 1% O2 (Hx1%). The cells were then subjected to a clonogenic cell survival assay. In x axis is the dose of X-radiation. In y axis is the surviving fraction. Mean±S.E.M. is representative of three independent experiments carried out in duplicate for a+b. (c) Cell morphology from microscope images of pmiR-Ctl and pmiR-210 A549 cells cultured for 2 days in normoxia (Nx) and hypoxia 1% O2 (Hx1%), treated with 10 Gy and subject to clonogenic cell survival. (d) Immunoblotting of HIF-1α, CAIX, total p53, p53-P, p21, cleaved caspase-3 (cleaved Casp.3) and β-actin in pmiR-Ctl and pmiR-210 A549 cells in normoxia (Nx) 18 h after irradiations (0, 4 and 8 Gy). (e) Histograms show the quantification of the p21 and the cleaved caspase-3 bands. Mean±S.E.M. is representative of three independent experiments. **P<0.01. (f) Eighteen hours after the indicated dose of irradiation, cells were fixed and labeled with propidium iodide for analysis by flow cytometry. Histograms represent the percentage of cells in each phase of the cell cycle (subG1, G0/G1, S and G2/M). Representative results are shown from three independent experiments
We next monitored the p53 status between pmiR-Ctl and pmiR-210 A549 cells following radiation. We observed a rapid and prolonged induction of p53 expression at both 4 and 8 Gy radiation at the protein level in both pmiR-Ctl and pmiR-210 A549 cells (Figure 4d and Supplementary Figure S4). Expression level of p21, a direct target of p53, followed p53 activity after radiation but remained reproducibly at a lower level in pmiR-210 A549 cells (Figures 4d and e and Supplementary Figure S5). Consistent with the miR-210-mediated resistance to radiation, caspase-3 activity was 60% lower in pmiR-210 A549 cells subjected to 8 Gy than in pmiR-Ctl cells (Figures 4d and e). However, analysis of the cell cycle showed no significant differences between pmiR-210 and pmiR-Ctl A549 cells, either before or after radiation (Figure 4f).
MiR-210 cells rapidly repair double-strand breaks (DSBs) after radiation
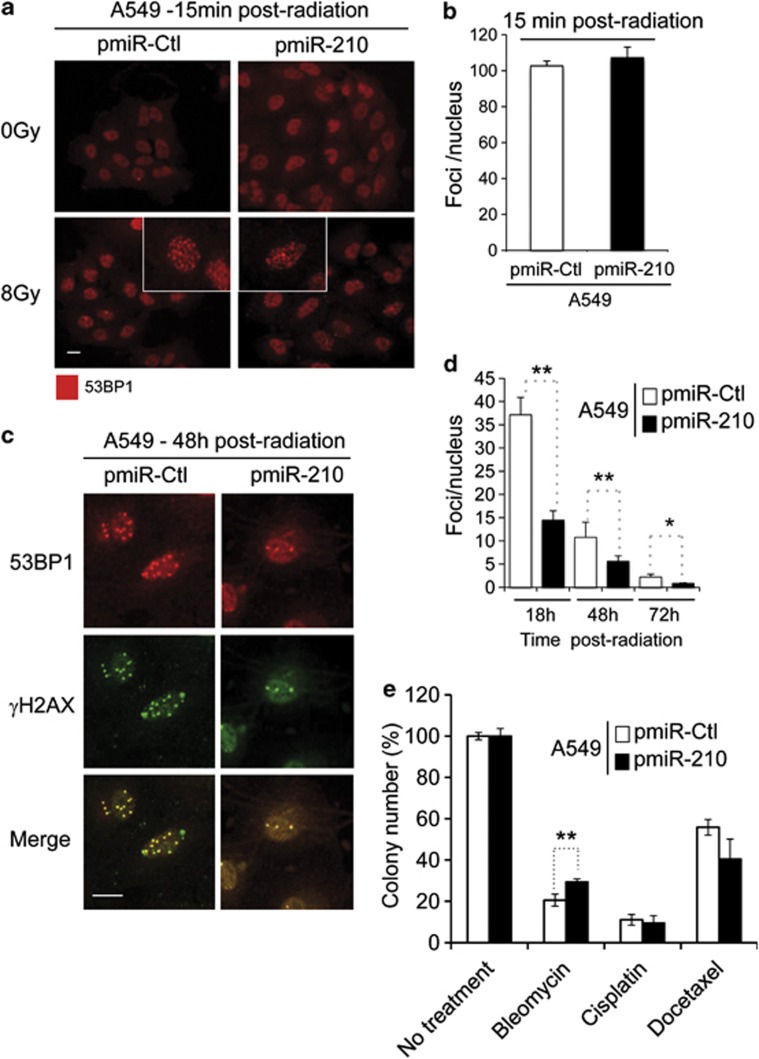
We followed the expression of two markers of DSB-induced DNA repair, γH2AX and tumor suppressor p53-binding protein 1 (53BP1), by immunofluorescence in A549 cells. Neither hypoxia nor overexpression of miR-210 was able to activate the expression of γH2AX and/or 53BP1 (Supplementary Figure S6). As expected, radiation (8 Gy) induced DSBs as indicated by the expression of 53BP1 at 15 min post radiation in both the pmiR-Ctl and pmiR-210 A549 cells (Figure 5a). Total 53BP1 foci numbers were similar (∼100 foci per nucleus) in both cell types at this time point (Figure 5b). However, after 48 h post radiation we clearly observed a decrease in foci for pmiR-210 A549 cells compared with pmiR-Ctl cells (5.6±1.2 versus 10.8±3.3; Figure 5c). The number of foci and the location of the DSBs merged perfectly between γH2AX and 53BP1. This difference in the number of foci was also observed at early (18 h, 37.1±3.8 versus 14.5±2.0) and late (72 h, 2.2±0.6 versus 0.8±0.1) time points post radiation (Figure 5d, Supplementary Figures S7 and S8). Counting apoptotic nuclei after staining with DAPI revealed that pmiR-210 A549 cells had significantly fewer apoptotic nuclei (7.4%) than pmiR-Ctl cells (13.2% Supplementary Figure S9). Increased mitosis was also found in pmiR-210 cells (Figure S7, yellow arrow) after 18 h post radiation. Finally, we investigated whether miR-210 expression could confer resistance to other genotoxic treatments. We tested three compounds used in chemotherapy in several cancers, including NSCLC: (i) bleomycin, which generates DSBs and DNA single-strand breaks, (ii) cisplatin, classified as an alkylating agent and (iii) docetaxel, a spindle poison. We observed that pmiR-210 A549 cells (29.4%±1.5) showed an enhanced resistance to bleomycin compared with pmiR-Ctl (20.6%±2.9), whereas no differences were observed for cisplatin and docetaxel (Figure 5e and Supplementary Figure S10).
Figure 5.
Increased kinetics of DSBs repair after radiation in pmiR-210 A549 cells. (a) Induction of 53BP1 foci in pmiR-Ctl and pmiR-210 A549 cells following 0–8 Gy at 15 min; scale bar, 7.3 μm. (b) Histograms show the quantification of the average foci per nucleus from at least three independent measurements. (c) 53BP1 colocalizes with the canonical DSB marker γH2AX in pmiR-Ctl and pmiR-210 A549 cells at 48 h following 8 Gy; scale bar, 7.3 μm. (d) Histograms show the quantification of the foci per nucleus on average from at least three independent measurements in a time-responsive manner (18, 48 and 72 h after irradiations). (e) A549 cells, pmiR-Ctl and pmiR-210 were cultured for 2 days, and at this point, treated with cisplatin (1 μg/ml) for 24 h or docetaxel (25 nM) for 4 h, or bleomycin for 2 h (10 μg/ml). Then, cells were trypsinized and plated in triplicate (1000 cells per well in 6-well plates) for clonogenic survival. Cells were allowed to form colonies during 10 days. At this time, they were fixed and stained with crystal violet and counted using ImageJ software. Data represent the average of two independent experiments. *P<0.05 and **P<0.01
MiR-210 confers radioresistance to cancer cells through HIF-1
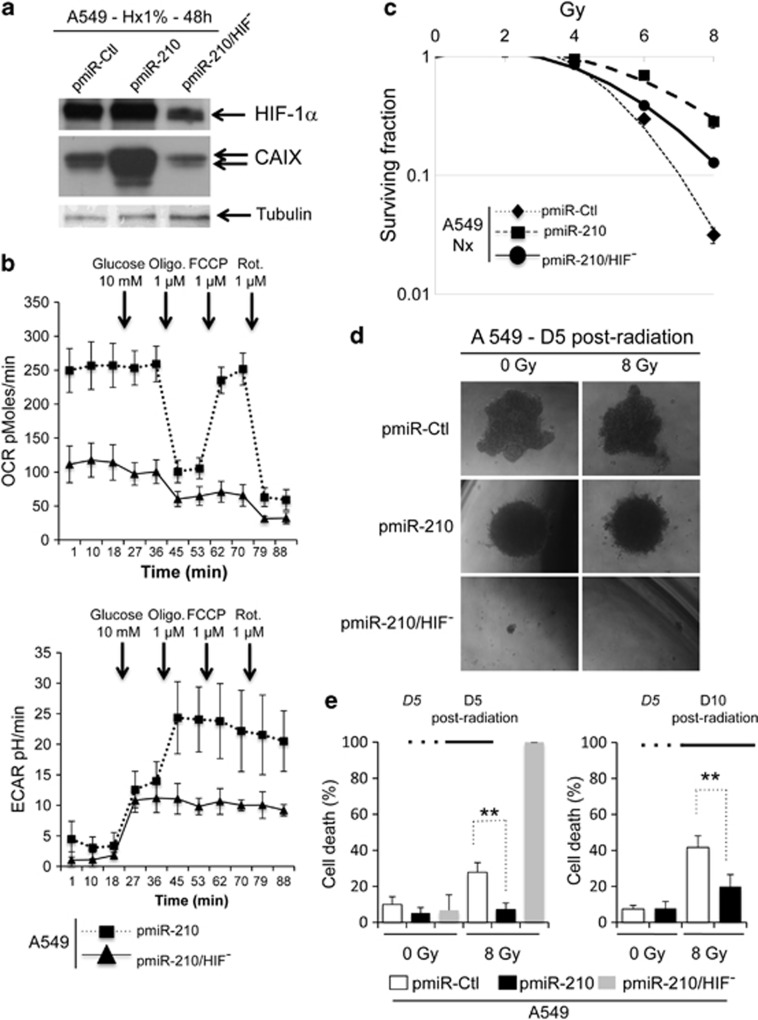
We established new stable pmiR-210 A549 clones, in which HIF-1α was knocked down (pmiR-210/HIF−) using the pTerHIF-1 constructs already described35 (Figure 6a). Surprisingly, we observed no difference in growth between pmiR-Ctl, pmiR-210 and pmiR-210/HIF− in normoxia or hypoxia (Supplementary Figure S11a). We next compared the metabolism of both pmiR-210 and pmiR-210/HIF− A549 cells by real-time monitoring of OCR and ECAR, following the same protocol described above. The invalidation of HIF-1α in pmiR-210 A549 cells had a dramatic impact on mitochondrial respiration (Figure 6b, top panel). In parallel, the addition of oligomycin had no effect on the glucose-stimulated ECAR, indicating that all of the available glucose is utilized for glycolysis (Figure 6b, bottom panel) and that pmiR-210/HIF− A549 cells had lost their ability to use OXPHOS. Radioresistance of pmiR-210/HIF− A549 cells was then tested. We observed that pmiR-210/HIF− cells were less resistant to radiation than pmiR-210, but still slightly more resistant than pmiR-Ctl (Figure 6c). We finally explored the effect of modulation of miR-210 and HIF-1α expression in tumor spheroids containing a natural oxygen gradient in vitro36 (Figure 6d). Comparing first the morphology of spheroids, we observed that pmiR-Ctl A549 spheroids showed irregular contours unlike the uniform pmiR-210 spheroids obtained with the same amount of cells (Supplementary Figure S11a). Although cell growth in 2D did not differ between the different cells, spheroids obtained with pmiR-210/HIF− were 10 times smaller than control spheroids owing to a strong defect in cell growth (Supplementary Figure S11b). Moreover, 5 days after radiation (8 Gy), cells in the pmiR-210 spheroids survived better than pmiR-Ctl spheroids (7.1%±3.6 versus 27.8%±5.4 cell death, respectively; Figure 6e, left panel). All A549 cells from pmiR-210/HIF− spheroids were dead after 5 days. The difference between pmiR-210 and pmiR-Ctl spheroids was still maintained 10 days after radiation (19.8%±6.7 versus 41.7%±6.5 cell death, respectively; Figure 6e, right panel).
Figure 6.
Involvement of HIF-1α in miR-210 radioresistance. (a) Immunoblot of HIF-1α, CAIX and tubulin in pmiR-Ctl, pmiR-210 and pmiR-210/HIF− A549 cells in hypoxia 1% O2 (Hx1%). (b) OCR (top panel) and ECAR (bottom panel) were measured in real time in a Seahorse XF bioenergetic assay. A total of 2 × 104 pmiR-210 and pmiR-210/HIF− A549 cells were seeded for 2 days. Average OCR and ECAR were calculated from at least four measurements during the treatment of each compound (glucose, oligomycin, FCCP, rotenone) at the concentration as indicated. Mean±S.E.M. is representative of three independent experiments carried out in tetraplicate. (c) Radioresistance of both pmiR-210 and pmiR-210/HIF− A549 cells cultured for 2 days in normoxia (Nx). The cells were then subjected to a clonogenic cell survival assay. In x axis is the dose of X-radiation. In y axis is the surviving fraction. Mean±S.E.M. is representative of three independent experiments carried out in duplicate. (d and e) A549 cells, pmiR-Ctl, pmiR-210 and pmiR-210/HIF−, were cultured as 3D spheroids for 5 days (D5), subjected to 0 or 8 Gy, and then cultured for 5 (D5) or 10 (D10) days. Spheroids were then subjected to accutase dissociation, and individualized live cells and dead cells were counted using Trypan blue. Data represent the average of three independent experiments. **P<0.01
These data indicated that an in vitro 3D system, which approximates spatial conditions observed in vivo, confirmed the miR-210-mediated radioresistance. Moreover, inactivation of HIF-1 in cells expressing miR-210 significantly reduces their radioresistance, confirming the strong molecular link between miR-210 and HIF-1.
Discussion
We show here for the first time that several normoxic cancer cell lines expressing the ‘hypoxamir' miR-210 exhibited a radioresistance similar to that found in hypoxic control cells. Our data shows that miR-210 stable expression mimics hypoxia-induced metabolic changes (CAIX and hexokinase II (HKII) expression, lactate production) associated with a slight but significant stabilization of HIF-1α. This information, combined with a strong reduction of radioresistance following HIF-1 silencing, reinforces the central role of HIF-1 in the resistance to radiotherapy. The molecular mechanism responsible for this radioresistance is not fully understood and is likely mediated by a complex network of miR-210 targets involved in a wide set of biological functions, including cell cycle control, survival, DNA repair and cell metabolism.22, 23 The precise contribution of these specific pathways in this phenotype is described below.
MiR-210 and cell cycle
Stimulation of cell proliferation may promote HIF-1 stabilization, and we have previously shown that a high cell density consumes large amounts of oxygen to reproduce hypoxic conditions.33 MiR-210 has been previously shown to repress or stimulate cell proliferation, depending on the cellular model. It targets proteins that are crucial for the cell cycle, such as E2F3,37, 38 FGFRL139, 40 or HOXA1,40 to inhibit cell growth. On the other hand, it also targets the myc-antagonist MNT41 promoting cell cycle progression in some cancer cells. Lung adenocarcinoma (A549 or H1975) cells stably expressing miR-210 did not show any modulation regarding their proliferation rate and are, therefore, unlikely to create such O2 local deprivation. We also focused on the cell cycle, as cells in S-phase are less radiosensitive;42 however, we observed no differences between pmiR-210 and pmiR-Ctl in 2D conditions (Figure 4f). This lack of effect on cell cycle may be explained by the fact that miR-210 function has so far been studied within hours or days following transient transfection using relatively high concentration of miRNAs mimics, whereas in our conditions cells have been adapted to a stable and moderate miR-210 concentration. Finally, we also looked at the induction of p53, which is well known to coordinate DNA repair with cell cycle progression and apoptosis.42 Similar expression levels of both total p53 and p53-P were induced in the two cell lines following radiation in a dose-dependent manner (Figures 4d and e), confirming our previous results. Surprisingly, expression of one of the main p53 effectors, the potent cyclin-dependent kinase inhibitor p21/WAF1, was almost 50% lower after radiation in cells expressing miR-210. Despite this p21 expression level, it is likely that there were sufficient p21 levels in pmiR-210 cells to induce growth arrest as shown in cell cycle analysis (Figure 4f).
MiR-210 and DSB repair
We could clearly demonstrate that miR-210 expression conferred a strong radioresistance to all of the three cell lines tested. Moreover, the number of 53BP1- and γH2AX-associated foci disappeared significantly faster in pmiR-210 A549 cells compared with pmiR-Ctl cells following radiation, suggesting that the process of DSB repair was more efficient in cells expressing miR-210. The source of radioresistance appeared to be specific for DSBs, as cells expressing miR-210 were also able to withstand a cytotoxic antibiotic treatment with bleomycin that generates DSBs. On the contrary, miR-210 has been implicated in the shutdown of DNA repair via targeting the enzyme RAD52.43 However, no modulation of RAD52 could be observed in our cellular model, which explains, in part, these conflicting data. Comparative genomic studies between pmiR-210 and pmiR-Ctl A549 cells did not reveal any modulation of a specific gene involved in DSB repair at the RNA level (data not shown). During preparation of this manuscript, Yang et al.44 reported that stable integration of an antisense sequence against miR-210 (iR-210) suppressed cell viability, induced cell arrest in the G(0)/G(1) phase, increased apoptotic rate and enhanced radiosensitivity in hypoxic human hepatoma. They proposed apoptosis-inducing factor, mitochondrion-associated, 3 (AIFM3) as a new miR-210 target involved in these processes. Although these results are consistent with our data, no significant expression of AIFM3 was found in our NSCLC cell line model. In contrast, another recent study focusing on prostate cancer has reported the absence of radiosensitization using a miR-210 inhibitor on prostate cancer cells in condition of anoxia, suggesting that miR-210 inhibition may depend on the type of cancer or on the degree and duration of hypoxia.45
MiR-210, cell metabolism and radioresistance
MiR-210 expression in normoxia resulted in a glycolysis-dominated metabolism (Figures 3d and e). By reducing the metabolism of mitochondria in normoxia, miR-210 would create a mitochondrial dysfunction that is known to lead to an increase in toxic reactive oxygen species (ROS).25 Adaptation of cells to an increased ROS production may be part of the mechanism that protects cells against radiation exposure. Moreover, it has been shown that lactate, the final product of glycolysis, has an antioxidant effect acting as a scavenger of free radicals.46, 47, 48 Therefore, an interaction between the increase in ROS production and increased lactate likely contributes to the resistance of the different pmiR-210 cell types analyzed in this study. Interestingly, hypoxia-induced miR-210 remains elevated even after reoxygenation,49 suggesting that miR-210 could sustain inhibition of mitochondria metabolism and HIF1 activation in the presence of oxygen. This mechanism may also occur in tumor cells in which HIF-1α expression is increased via hypoxia-independent pathways through transcriptional and/or translational regulation by oncogenes, loss of the tumor suppressor PTEN,50 or as suggested recently by systemic elevation of tissue inhibitor of metalloproteinases-1.51, 52 It might therefore be conceivable that miR-210 may be a marker of radioresistance in circumstances other than hypoxia.
MiR-210 as a marker of radioresistance?
MiR-210 overexpression has been found in numerous tumors,40, 53, 54, 55 including NSCLC.56 Whether miR-210 is linked to a poor prognosis and/or a poor response to treatment is not clearly established, although three studies previously reported miR-210 overexpression as a bad prognostic factor in a small cohort of patients with HNSCC,57 soft-tissue sarcoma58 and breast carcinoma.59 Interestingly, although a postoperative radiotherapy was delivered in the cohort of patients with HNSCC,57 no clear correlation could be found between miR-210 expression and oncogenic or DNA repair pathway signatures. It would be interesting to analyze in future patient cohorts the impact of miR-210 regarding tumor response after radiotherapy or chemotherapy for clinical outcome, especially in tumors presenting miR-210 overexpression, such as NSCLC.
In summary, we provide evidence that miR-210 can efficiently mediate radioresistance in the NSCLC cell line A549, suggesting that miR-210 is an attractive prognostic/predictive biomarker in this type of cancer. More knowledge is now required regarding the underlying molecular mechanisms, notably through knockdown strategies and in vivo models to evaluate whether targeting miR-210 may have potential value as an adjuvant therapy for tumor radiosensitization.
Materials and Methods
Cell culture
The lung adenocarcinoma A549 and H1975 cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in Ham's F-12K supplemented with penicillin, streptomycin and 10% heat-inactivated fetal calf serum (FCS) at 37 °C in a humidified 5% CO2 air atmosphere. The p3HRE-Δptk-LUC construct was described in Dayan et al.34 HeLa cells were cotransfected with p3HRE-Δptk-LUC and pBabepuro. Puromycin-resistant clones were tested in hypoxia for their ability to upregulate LUC activity (HeLa–HRE Luc). A Bug-Box anaerobic workstation (Ruskinn Technology, Bridgend, UK) set at 1% oxygen, 94% nitrogen and 5% carbon dioxide was used for hypoxic conditions.
Generation of stable miR-Ctl, miR-210 and miR-210/HIF-1− cell lines
A lentiviral miR-210 expression vector that expresses ∼400-base pre-miR-210 and empty lentiviral vector were purchased from System Biosciences (SBI, Mountain View, CA, USA). A pre-miR (ce-miR-67) from Caenorhabditis elegans was cloned into the lentiviral vector and used as a control. Virus production and cell transduction were performed following the manufacturer's protocol. The resulting pmiR-Ctl and pmiR-210 cells that stably overexpress miR-210 were selected by puromycin (1 μg/ml).
Several pmiR-210/HIF-1− A549 clones were obtained using two independent sequences of shRNA against HIF-1α.35 We selected one clone for each sequence, based on the decreased mRNA level of HIF-1α (>95% – data not shown) and the expression of HIF-1α and CAIX in hypoxia compared with the same conditions in pmiR-210 cells (Figure 6a). As both clones were then tested under the same conditions and gave the same results, we only show the results from one clone (sequence pTerHIF-1bis).
qRT-PCR analysis
miR-210 expression was evaluated using the TaqMan MicroRNA Assay (Applied Biosystems, Carlsbad, CA, USA) as specified in their protocol. Expression of target genes was assessed using TaqMan Gene Expression Assay (Applied Biosystems). Real-time qPCR was performed using TaqMan Universal Master Mix II, no UNG (Applied Biosystems) and a Lightcycler 480 (Roche, Meylan, France) real-time PCR machine. All reactions were performed in triplicate. Expression levels of miRNA and mRNAs were evaluated using comparative CT method (2−ΔCT). Transcript levels of RNU44 (for microRNA assay), β-actin and ubiquitin C (for gene expression assay) were used as endogenous control.
Luciferase assay
Transfection of 50 ng psiCHECK-2 constructs (Sensor 210 described in Yang et al.,29 and NDUFA4 and SDHD described in Puissegur et al.28) was carried out in 96-well plates with Lipofectamine 2000 (Invitrogen, Saint Aubin, France). The medium was replaced 8 h after transfection with fresh medium containing 10% FCS and supplemented with penicillin and streptomycin. At 48 h after transfection, firefly and Renilla luciferase activities were measured using the Dual-Glo luciferase assay (Promega, Charbonnieres, France). For experiments with HeLa–HRE Luc cells, cells were plated in six-well plates and incubated in 21% O2 or 1% O2 for 24 h. Cells were then lysed, firefly luciferase activity was measured and were normalized to total protein contents.
Microarrays
Expression microarrays
SurePrint G3 Human Gene Expression 8 × 60 K Microarray from Agilent Technology (Loveland, CO, USA) were used. RNA were labeled and hybridized following the manufacturer's instructions. Two biological replicates were performed for each comparison. The experimental data and microarray design have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under series GSE43622.
Statistical analysis
Normalization was performed using the Limma package available from Bioconductor (http://www.bioconductor.org). Intra- and interslide normalization was performed using the Print Tip Loess and the quantile methods, respectively. Means of ratios from all comparisons were calculated and B-test analysis was performed. Differentially expressed genes were selected using a B-value>0.
Biological theme analysis
Data from expression microarrays were analyzed for enrichment in biological themes (Gene Ontology molecular function and biological process) using Ingenuity Pathway Analysis software (http://www.ingenuity.com/) and Mediante (http://www.microarray.fr:8080/merge/index).
miR-210 targets analysis was performed with MiRonTop (available at http://www.microarray.fr:8080/miRonTop/index) as previously described.30
Transmission electronic microscopy
Cells were fixed in situ with 1.6% glutaraldehyde in 0.1 M phosphate buffer at room temperature for at least 1 h and then conserved at 4 °C. Samples were rinsed in the same buffer and then postfixed with 1% osmium tetroxide and 1% potassium ferrocyanide in 0.1 M cacodylate buffer for 1 h at room temperature to enhance the staining of cytoplasmic membranes. Cells were rinsed with distilled water and embedded in epoxy resin. Embedded samples were then processed for thin sectioning and observed with a JEM1400 transmission electron microscope (Jeol, Tokyo, Japan) equipped with a Morada CCD camera (Olympus SIS, Rungis, France).
Western blot analysis
Cells were lysed in 1.5 × Laemmli buffer and the protein concentration was determined using the bicinchoninic acid assay. Forty microgram of protein of whole-cell extracts was resolved by SDS-PAGE and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Membranes were blocked in 5% non-fat milk in TN buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl) and incubated in the presence of the primary and then secondary antibodies in 5% non-fat milk in TN buffer. After washing in TN buffer containing 0.1% TWEEN-20 and then in TN buffer, immunoreactive bands were visualized with the ECL system (Amersham Biosciences, Orsay, France).
Antibodies
Anti-HIF-1α (antiserum 2087) was produced and characterized in our laboratory as described previously.34 Rabbit polyclonal antibodies to HKII, p53 and phospho-p53 (ser15), 53BP1 and p21 were purchased from Cell Signaling (Danvers, MA, USA), Novus Biologicals (Cambridge, UK) and Santa Cruz (Heidelberg, Germany), respectively. Mouse antibodies to CAIX and ISCU1/2 were purchased from Bayer HealthCare Co. (Berlin, Germany) and Novus Biologicals, respectively. Membranes were also blotted with the mouse monoclonal antibodies to tubulin (Santa Cruz) and β-actin (Cell Signaling) as loading controls.
Complex II activity
Complex II activity was assessed using Complex II Enzyme Activity Microplate Assay Kit from Mitosciences (Abcam, Paris, France) following the manufacturer's instructions. Briefly, cells were lysed and complex II from lysate was immobilized in the well by a capture antibody. Succinate, 2,6-dichloroindophenol and ubiquinone 2 were added to the well, and the decrease in absorbance at 600 nm was measured.
Caspase-3/7 assay
The activation of executioner caspase-3 and -7 in all cell types was determined using the Caspase-Glo 3/7 Assay kit (Promega), according to manufacturer's instructions. Cells were plated in triplicate in 96-well plates and treated as described above. Luminescence was quantified after 1 h of incubation with the caspase substrate.
Lactate quantification
Lactate quantification was done on cellular extracts using a Lactate Assay Kit (Biovision, Milpitas, CA, USA) according to the manufacturer's instructions.
Assessment of the cell cycle by flow cytometry
Eighteen hours after radiations, cells were washed, fixed in 70% ethanol and left for a minimum of 24 h at −20 °C. Next, cells were washed 2 × in phosphate buffered saline (PBS) and incubated in PBS buffer containing 50 μg/ml of propidium iodide and 50 μg/ml RNase A for 15 min at 37 °C. Cell distribution across the different phases of the cell cycle was analyzed with LSR Fortessa cytometer (Becton-Dickinson, Franklin Lakes, NJ, USA).
3D cell culture, radiation and cell death
To grow spheroids, 5000 pmiR-Ctl, pmiR-210 and pmiR-210/HIF1− A549 cells were seeded in 20 μl drops of F-12 media supplemented with 10% fetal bovine serum. After 5 days, spheroids were irradiated (0 and 8 Gy) as described below. Spheroids were then collected and transferred to polyhema-coated 96-well plates containing the same media for 5 days. Spheroids were dissociated in Accutase (Life Technologies, Saint Aubin, France), and live and dead cells were counted (Trypan blue uptake).
Cell radiations
Radiation of tumor cells was performed in 60-mm dishes. Cells were irradiated 100 cm from the source with a bolus of 1.1 cm (under dishes; water equivalent of 2 cm). Low-energy photons were used (6 MV), delivered by a clinical device (PRIMUS, Siemens, Concord, CA, USA) with a 40 × 40 cm posterior field. The debit of the PRIMUS was 300-monitor units/min and 2 Gy corresponded to 93 monitor units (18.6 s).
Cloning efficiency
One thousand or 5000 cells were plated in normoxic or hypoxic conditions in 60-mm dishes or flasks, subsequently irradiated at day 2 (2, 4, 6, 8 and 10 Gy), then immediately trypsinized and plated in triplicate for clonogenic survival. Cells were allowed to form colonies during 7 days. At this time, they were stained with GIEMSA and counted using ImageJ software. For chemotherapy sensitivity, 400 000 cells were plated in 25 cm2 flask and cultivated for 2 days. At this point, cells were treated for 24 h with cisplatin or 4 h with docetaxel or for 2 h with bleomycin, then immediately trypsinized and plated in triplicate (1000 cells per well in 6-well plates) for clonogenic survival. Cells were allowed to form colonies during 10 days. At this time, they were fixed and stained with crystal violet and counted using ImageJ software.
Confocal microscopy
Cells were grown on glass coverslips and then fixed in 3.3% paraformaldehyde for 30 min at RT and permeabilized with 0.2% Triton-X-100 for 5 min. Cells were blocked with PBS containing 0.2% gelatin and 2% bovine serum albumin for 30 min at RT, and incubated with anti-rabbit HIF-1α (described in the Immunoblotting section) and/or antimouse CAIX (Bayer), or antirabbit 53BP1 (Novus Biologicals) and anti-mouse γH2AX (Abcam) antibodies in PBS for 3 h at RT. After washing, cells were incubated in the presence of a biotinylated antimouse secondary antibody conjugated to Alexa 594 (Invitrogen, Saint Aubin, France) (1 : 400) or antirabbit secondary antibody conjugated to Alexa 488 (Amersham Biosciences, Munich, Germany) (1 : 400) for 1 h at RT. After washing, coverslips were mounted in Cytifluor (Amersham Biosciences), detection of the confocal images by fluorescence was performed with a Leica (Leica Microsystem, Zurich, Switzerland) TCS-SP1 microscope, and images were taken using a Nikon Coolpix 990 digital camera (Nikon, Champigny sur Marne, France).
Respirometry and extracellular acidification
Cellular OCR and ECAR data were obtained using a Seahorse XF96 extracellular flux analyzer from Seahorse Bioscience (North Billerica, MA, USA). Experiments were performed according to the manufacturer's instructions. Protein standardization was performed after each experiment, with no noticeable differences in protein concentration and cell type.
Statistical analysis
All values are represented as the mean±S.D. of the indicated number of replicates (n). Significant differences were based on the Student's t-test and the Wilcoxon rank sum test.
Acknowledgments
We thank Pascale Bertrand for helpful discussion and suggestions. We acknowledge the excellent support of the Nice-Sophia Antipolis Functional Genomics Platform and of the ‘Centre Commun de Microscopie Appliquée' from the University of Nice-Sophia-Antipolis. We thank Nicolas Pons and Sandra Schäfer (IPMC) for excellent technical assistance. We thank Thomas Maurin and Eric Lai for providing us with miR-210 sensor construct.This work was supported by CNRS, INSERM, Association pour la Recherche contre le Cancer (ARC, Projet Libre SFI20101201798 to BM), Canceropole PACA, French National Research Agency (ANR), Ligue Nationale Contre le Cancer (JP, NM), the Centre Antoine Lacassagne, European Community (MICROENVIMET, FP7-HEALTH-F2-2008-201279 to PB, BM and METOXIA to JP, NM). SG is a recipient of a post-doctoral fellowship from the ‘Ville de Nice'.
Glossary
- CAIX
carbonic anhydrase IX
- DSB
double-strand break
- ETC
electron transport chain
- HKII
hexokinase II
- HIF-1
hypoxia-inducible factor 1
- HRE
hypoxia responsive element
- miRNA
microRNAs
- NDUFA4
NADH dehydrogenase (ubiquinone) 1α subcomplex, 4
- NSCLC
non-small cell lung carcinoma
- SDHD
succinate dehydrogenase complex, subunit D
- UTR
untranslated region
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by D Aberdam
Supplementary Material
References
- Gray LH, Conger AD, Ebert M, Hornsey S, Scott OC. The concentration of oxygen dissolved in tissues at the time of irradiation as a factor in radiotherapy. Br J Radiol. 1953;26:638–648. doi: 10.1259/0007-1285-26-312-638. [DOI] [PubMed] [Google Scholar]
- Palcic B, Skarsgard LD. Reduced oxygen enhancement ratio at low doses of ionizing radiation. Radiat Res. 1984;100:328–339. [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H. How can we overcome tumor hypoxia in radiation therapy. J Radiat Res. 2011;52:545–556. doi: 10.1269/jrr.11056. [DOI] [PubMed] [Google Scholar]
- Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, et al. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- Unruh A, Ressel A, Mohamed HG, Johnson RS, Nadrowitz R, Richter E, et al. The hypoxia-inducible factor-1 alpha is a negative factor for tumor therapy. Oncogene. 2003;22:3213–3220. doi: 10.1038/sj.onc.1206385. [DOI] [PubMed] [Google Scholar]
- Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–441. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Zhang X, Kon T, Wang H, Li F, Huang Q, Rabbani ZN, et al. Enhancement of hypoxia-induced tumor cell death in vitro and radiation therapy in vivo by use of small interfering RNA targeted to hypoxia-inducible factor-1alpha. Cancer Res. 2004;64:8139–8142. doi: 10.1158/0008-5472.CAN-03-2301. [DOI] [PubMed] [Google Scholar]
- Harada H, Itasaka S, Zhu Y, Zeng L, Xie X, Morinibu A, et al. Treatment regimen determines whether an HIF-1 inhibitor enhances or inhibits the effect of radiation therapy. Br J Cancer. 2009;100:747–757. doi: 10.1038/sj.bjc.6604939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero T, Grosso S, Robbe-Sermesant K, Lebrigand K, Henaoui IS, Puissegur MP, et al. ‘Seed-Milarity' confers to hsa-miR-210 and hsa-miR-147b similar functional activity. PLoS One. 2012;7:e44919. doi: 10.1371/journal.pone.0044919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- White NM, Fatoohi E, Metias M, Jung K, Stephan C, Yousef GM. Metastamirs: a stepping stone towards improved cancer management. Nat Rev Clin Oncol. 2011;8:75–84. doi: 10.1038/nrclinonc.2010.173. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortholan C, Puissegur MP, Ilie M, Barbry P, Mari B, Hofman P. MicroRNAs and lung cancer: new oncogenes and tumor suppressors, new prognostic factors and potential therapeutic targets. Curr Med Chem. 2009;16:1047–1061. doi: 10.2174/092986709787581833. [DOI] [PubMed] [Google Scholar]
- Arora H, Qureshi R, Jin S, Park AK, Park WY. miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1. Exp Mol Med. 2011;43:298–304. doi: 10.3858/emm.2011.43.5.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JS, Kim JJ, Byun JY, Kim IA. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys. 2010;76:5–8. doi: 10.1016/j.ijrobp.2009.08.028. [DOI] [PubMed] [Google Scholar]
- Salim H, Akbar NS, Zong D, Vaculova AH, Lewensohn R, Moshfegh A, et al. miRNA-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38MAPK, apoptosis and senescence. Br J Cancer. 2012;107:1361–1373. doi: 10.1038/bjc.2012.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One. 2010;5:e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YC, Banerjee J, Choi SY, Sen CK. miR-210: the master hypoxamir. Microcirculation. 2012;19:215–223. doi: 10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman MZ, Buart S, Romero P, Ketari S, Janji B, Mari B, et al. Hypoxia-inducible miR-210 regulates the susceptibility of tumor cells to lysis by cytotoxic T cells. Cancer Res. 2012;72:4629–4641. doi: 10.1158/0008-5472.CAN-12-1383. [DOI] [PubMed] [Google Scholar]
- Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Brigand K, Robbe-Sermesant K, Mari B, Barbry P. MiRonTop: mining microRNAs targets across large scale gene expression studies. Bioinformatics. 2010;26:3131–3132. doi: 10.1093/bioinformatics/btq589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, et al. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ, Souza AL, Clish CB, Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol. 2011;31:2696–2706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan F, Bilton RL, Laferriere J, Trottier E, Roux D, Pouyssegur J, et al. Activation of HIF-1alpha in exponentially growing cells via hypoxic stimulation is independent of the Akt/mTOR pathway. J Cell Physiol. 2009;218:167–174. doi: 10.1002/jcp.21584. [DOI] [PubMed] [Google Scholar]
- Dayan F, Roux D, Brahimi-Horn MC, Pouyssegur J, Mazure NM. The oxygen sensor factor-inhibiting hypoxia-inducible factor-1 controls expression of distinct genes through the bifunctional transcriptional character of hypoxia-inducible factor-1alpha. Cancer Res. 2006;66:3688–3698. doi: 10.1158/0008-5472.CAN-05-4564. [DOI] [PubMed] [Google Scholar]
- Bilton R, Mazure N, Trottier E, Hattab M, Dery MA, Richard DE, et al. Arrest-defective-1 protein, an acetyltransferase, does not alter stability of hypoxia-inducible factor (HIF)-1alpha and is not induced by hypoxia or HIF. J Biol Chem. 2005;280:31132–31140. doi: 10.1074/jbc.M504482200. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Dayan F, Durivault J, Ilc K, Pecou E, Pouyssegur J, et al. The asparaginyl hydroxylase factor-inhibiting HIF is essential for tumor growth through suppression of the p53-p21 axis. Oncogene. 2012;31:2989–3001. doi: 10.1038/onc.2011.471. [DOI] [PubMed] [Google Scholar]
- Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, et al. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci USA. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, et al. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59:928–942. doi: 10.1016/j.ijrobp.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Sun T, Cao J, Liu F, Tian Y, Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res. 2012;318:944–954. doi: 10.1016/j.yexcr.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Quero L, Dubois L, Lieuwes NG, Hennequin C, Lambin P. miR-210 as a marker of chronic hypoxia, but not a therapeutic target in prostate cancer. Radiother Oncol. 2011;101:203–208. doi: 10.1016/j.radonc.2011.05.063. [DOI] [PubMed] [Google Scholar]
- Groussard C, Morel I, Chevanne M, Monnier M, Cillard J, Delamarche A. Free radical scavenging and antioxidant effects of lactate ion: an in vitro study. J Appl Physiol. 2000;89:169–175. doi: 10.1152/jappl.2000.89.1.169. [DOI] [PubMed] [Google Scholar]
- Sattler UG, Meyer SS, Quennet V, Hoerner C, Knoerzer H, Fabian C, et al. Glycolytic metabolism and tumour response to fractionated irradiation. Radiother Oncol. 2010;94:102–109. doi: 10.1016/j.radonc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Sattler UG, Mueller-Klieser W. The anti-oxidant capacity of tumour glycolysis. Int J Radiat Biol. 2009;85:963–971. doi: 10.3109/09553000903258889. [DOI] [PubMed] [Google Scholar]
- Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- Schelter F, Halbgewachs B, Baumler P, Neu C, Gorlach A, Schrotzlmair F, et al. Tissue inhibitor of metalloproteinases-1-induced scattered liver metastasis is mediated by hypoxia-inducible factor-1alpha. Clin Exp Metastasis. 2011;28:91–99. doi: 10.1007/s10585-010-9360-x. [DOI] [PubMed] [Google Scholar]
- Cui H, Grosso S, Schelter F, Mari B, Kruger A. On the pro-metastatic stress response to cancer therapies: evidence for a positive co-operation between TIMP-1, HIF-1alpha, and miR-210. Front Pharmacol. 2012;3:134. doi: 10.3389/fphar.2012.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stuhler K, Meyer HE, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2009;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satzger I, Meier A, Alter M, Kapp A, Gutzmer R. Parameters predicting prognosis in melanoma sentinel nodes. J Clin Oncol. 2011;29:3588–3590. doi: 10.1200/JCO.2011.37.1112. [DOI] [PubMed] [Google Scholar]
- Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, et al. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- Greither T, Wurl P, Grochola L, Bond G, Bache M, Kappler M, et al. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int J Cancer. 2012;130:1230–1235. doi: 10.1002/ijc.26109. [DOI] [PubMed] [Google Scholar]
- Hong L, Yang J, Han Y, Lu Q, Cao J, Syed L. High expression of miR-210 predicts poor survival in patients with breast cancer: a meta-analysis. Gene. 2012;507:135–138. doi: 10.1016/j.gene.2012.07.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.