Abstract
Post-transcriptional control of gene expression is crucial for the control of cellular differentiation. Erythroid precursor cells loose their organelles in a timely controlled manner during terminal maturation to functional erythrocytes. Extrusion of the nucleus precedes the release of young reticulocytes into the blood stream. The degradation of mitochondria is initiated by reticulocyte 15-lipoxygenase (r15-LOX) in mature reticulocytes. At that terminal stage the release of r15-LOX mRNA from its translational silenced state induces the synthesis of r15-LOX. Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a key regulator of r15-LOX mRNA translation. HnRNP K that binds to the differentiation control element (DICE) in the 3′ untranslated region (UTR) inhibits r15-LOX mRNA translation initiation. During erythroid cell maturation, activation of r15-LOX mRNA translation is mediated by post-translational modifications of hnRNP K and a decrease of the hnRNP K level. To further elucidate its function in the post-transcriptional control of gene expression, we investigated hnRNP K degradation employing an inducible erythroid cell system that recapitulates both nuclear extrusion and the timely controlled degradation of mitochondria, mediated by the activation of r15-LOX synthesis. Interestingly, we detected a specific N-terminal cleavage intermediate of hnRNP K lacking DICE-binding activity that appeared during erythroid differentiation and puromycin-induced apoptosis. Employing mass spectrometry and enzymatic analyses, we identified Caspase-3 as the enzyme that cleaves hnRNP K specifically. In vitro studies revealed that cleavage by Caspase-3 at amino acids (aa) D334-G335 removes the C-terminal hnRNP K homology (KH) domain 3 that confers binding of hnRNP K to the DICE. Our data suggest that the processing of hnRNP K by Caspase-3 provides a save-lock mechanism for its timely release from the r15-LOX mRNA silencing complex and activation of r15-LOX mRNA synthesis in erythroid cell differentiation.
Keywords: hnRNP K, caspase-3 activity, K562 cells, erythroid differentiation, post-transcriptional control, mRNA translation regulation
The regulation of mRNA translation is a fundamental branch of post-transcriptional mechanisms, which control gene expression in development and differentiation.1 Translation of specific mRNAs is under the surveillance of trans acting factors that interact with cis elements located predominantly in their UTRs.2 In differentiating erythroid cells hnRNP K regulates translation of specific mRNAs. Post-translational modifications of hnRNP K have been shown to modulate its capacity in regulatory complex formation.3, 4, 5, 6 Erythroid precursor cells undergo nuclear extrusion and mitochondria degradation in reticulocytes at the terminal step of erythrocyte formation. Mitochondria degradation is initiated by r15-LOX expressed only in mature reticulocytes. HnRNP K silences r15-LOX mRNA translation in premature reticulocytes.7, 8 In late erythroid maturation, translation inhibition is abolished by phosphorylation of Y458 in KH domain 3 that mediates binding to the DICE in the r15-LOX mRNA 3′UTR.4, 5 Additionally, a decreasing hnRNP K level contributes to the release of the silencing complex.9 Although the function of site-specific phosphorylation in r15-LOX mRNA translation regulation has been elucidated, there is no information about the mechanism of hnRNP K degradation in erythroid differentiation. Here we analyze the degradation of hnRNP K during induced erythroid differentiation of K562 cells to get further insight in its function as a regulator of post-transcriptional control of gene expression. We found that the ubiquitin E3 ligase HDM2, which was shown to ubiquitinate hnRNP K in p53-dependent DNA damage repair10 is not expressed in K562 cells (Supplementary Figure S1). Additionally, we show that hnRNP K is not ubiquitinated in K562 cells and proteasome inhibitors fail to stabilize the protein. Caspases not only catalyze site-specific protein cleavage in apoptosis,11 but were also shown to be activated in terminal erythroid differentiation,12, 13, 14 which is not associated with apoptosis.15 Interestingly, a specific N-terminal hnRNP K fragment that migrates at 48 kD accumulates during erythroid differentiation. We purified and analyzed this fragment by mass spectrometry and showed that it is a cleavage product of Caspase-3, which is activated during erythroid differentiation in an apoptosis independent manner. Residues D334–G335 were identified as Caspase-3 cleavage site that separates the DICE-binding KH domain 3 from the N-terminal part, which contains critical protein-protein interaction domains.5, 16, 17 Thus, Caspase-3 mediated cleavage inactivates hnRNP K as a regulator of r15-LOX mRNA translation.
Results
Cleavage of hnRNP K during erythroid differentiation generates an N-terminal fragment that lacks KH domain 3
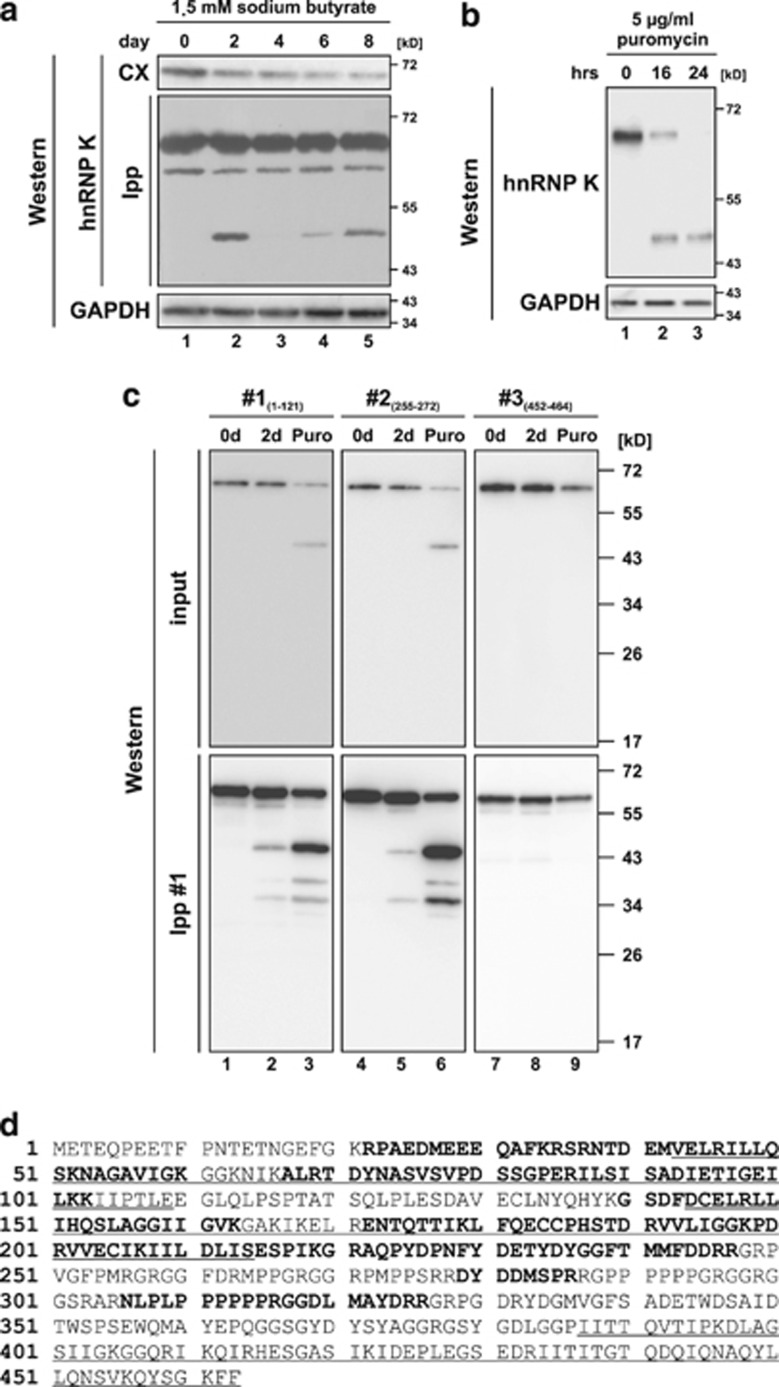
The analysis of proteins involved in r15-LOX mRNA translational control revealed that the level of hnRNP K decreases during erythroid differentiation of K562 cells9 (Figure 1a). Interestingly, a specific fragment migrating at about 48 kD in SDS-PAGE was detected when hnRNP K was enriched by immunoprecipitation (Figure 1a), indicating that a specific protease cleaves the protein. This hnRNP K-derived cleavage product was also detected when K562 cells were treated with puromycin for up to 24 h (Figure 1b). To characterize the fragment initially, hnRNP K was immunoprecipitated from the extracts of non-treated K562 cells, cells induced for erythroid differentiation or cells treated with puromycin. Western blots of the immunoprecipitated protein were incubated with antibodies that recognize the N-terminus (aa 1–121, #1) (Figure 1c, lanes 1–3, Supplementary Figure S2, lanes 4–6), an internal peptide (aa 255–272, #2) (Figure 1c, lanes 4–6) or the C-terminus (aa 452–464, #3) (Figure 1c, lanes 7–9). Antibodies that recognize the N-terminus and the internal peptide, but not the C-terminus detected the immunoprecipitated fragment of about 48 kD and minor smaller fragments (Figure 1c). For further characterization, the cleavage product was purified from puromycin-treated K562 cells (Supplementary Figure S3), in gel digested with trypsin and analyzed by LC-MS/MS. Identified peptides are shown in bold in the hnRNP K sequence (Figure 1d). The C-terminal amino acid identified was R326. No peptides corresponding to KH domain 3 that specifically mediates binding to the DICE in the r15-LOX mRNA5 were detected (Figure 1d).
Figure 1.
hnRNP K is degraded via an N-terminal fragment of 48 kD during induced erythroid differentiation and puromycin-induced apoptosis of K562 cells. (a) Detection of hnRNP K in cytoplasmic extract (CX) and immunoprecipitates (immunoprecipitation with hnRNP K antibody #1, Ipp) from K562 cells induced for erythroid differentiation with sodium butyrate for up to 8 days in western blot assays. GAPDH was detected as loading control. (b) Detection of hnRNP K and GAPDH in lysates of K562 cells, which were non-treated or treated with puromycin for 16 or 24 h, respectively. (c) Immunoprecipitation of hnRNP K with antibody #1 from cytoplasmic extracts of non-treated K562 cells (lanes 1, 4 and 7), cells induced with sodium butyrate for 2 days (Lanes 2, 5 and 8) or cells treated with puromycin for 16 h (lanes 3, 6 and 9). Western blot analysis of the input and immunoprecipitates with antibodies #1 (directed against aa 1–121), #2 (directed against aa 255–272) and #3 (directed against aa 452–464). (d) Amino acid sequence of human hnRNP K. The three KH domains are underlined. Peptides identified in the mass spectrometry analysis after trypsin digestion of the purified N-terminal hnRNP K fragment are shown in bold
Caspase-3 is activated during erythroid differentiation of K562 cells
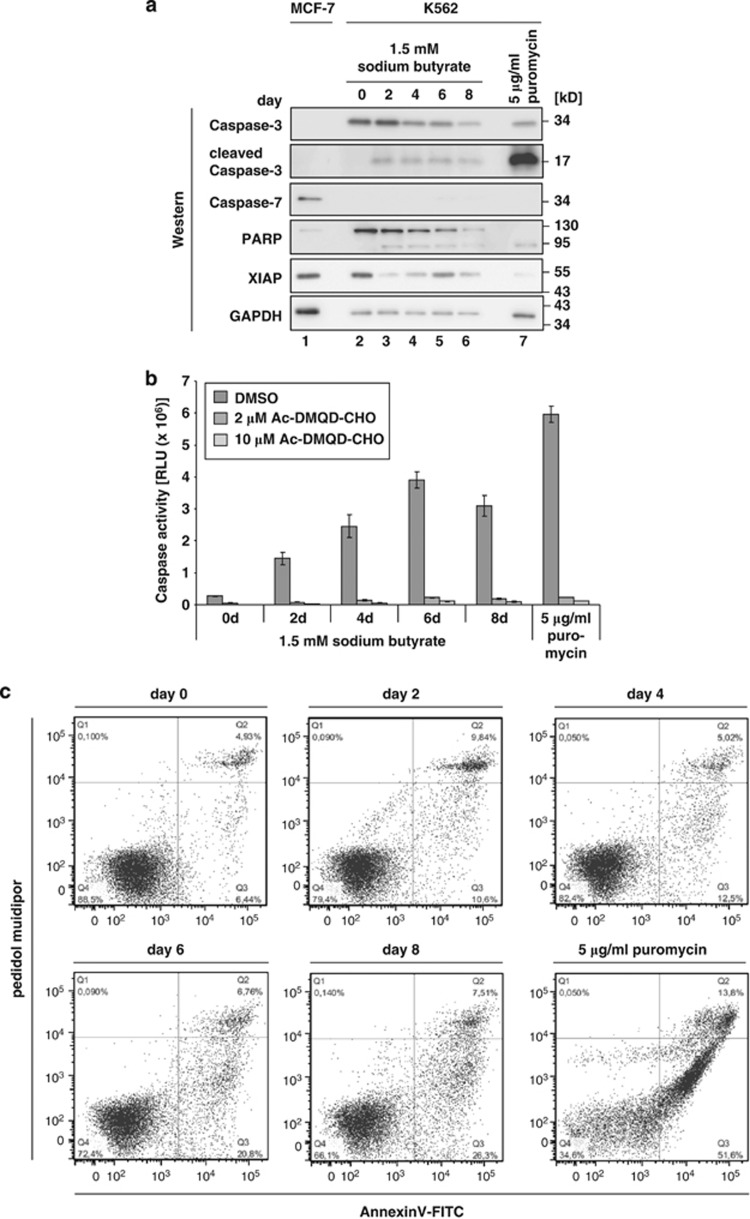
Caspases are not only activated in apoptosis,11 but as well in development and differentiation.18 It was shown that Caspase-3 is activated in erythroid cell differentiation12, 13, 14 and during differentiation of lens fiber cells, neuronal stem cells and skeletal muscle cells (reviewed in Lamkanfi et al18). Therefore, we analyzed whether Caspase-3 becomes activated during sodium-butyrate induced erythroid differentiation of K562 cells. Activation of Caspase-3 was monitored in western blot assays by detection of the p34 precursor cleavage fragment p17.19 When K562 cells were treated with puromycin for 16 h Caspase-3 was strongly activated (Figure 2a, lane 7). During erythroid maturation activation of Caspase-3 was also detectable, but to a lower level (Figure 2a, lanes 2–6). To quantify Caspase-3 activation a luminometric assay was used, which is sensitive to the activity of Caspase-3 and Caspase-7 (Figure 2b). A contribution of Caspase-7 to the read-out of this assay could be excluded, because Caspase-7 that is readily detectable in MCF-7 cells (Figure 2a, lane 1) was undetectable in K562 cells (Figure 2a, lanes 2–7). Caspase-3 activity increases up to 14-fold during induced erythroid differentiation and is enhanced to 22-fold upon puromycin treatment (Figure 2b). The Caspase-3 inhibitor Ac-DMQD-CHO blocks its activity efficiently (Figure 2b). Additionally, during erythroid differentiation cleavage of poly-ADP-ribose polymerase 1 (PARP) that is a substrate of both, Caspase-3 and Caspase-720 was observed (Figure 2a, lanes 2–6). To exclude that sodium butyrate induces apoptosis in K562 cells, AnnexinV-FITC and propidium iodide staining was performed (Figure 2c). A strong increase in the amount of apoptotic cells (AnnexinV-FITC positive) could be observed when cells were treated with puromycin, whereas induction of apoptosis did not occur during erythroid differentiation (Figure 2c). Caspase-3 activity can be regulated by X-chromosome linked inhibitor of apoptosis protein (XIAP), which was shown to bind and inhibit activated Caspase-3 directly.21 Noteworthy XIAP expression is high in non-induced K562 cells, but expression drops strongly during erythroid differentiation (Figure 2a, lanes 2–6), indicating that XIAP is involved in regulation of Caspase-3 activity during erythroid differentiation.
Figure 2.
Caspase-3 is activated independent of apoptosis induction during erythroid differentiation of K562 cells. (a) The expression of Caspase-3, Caspase-7, PARP, XIAP and GAPDH in MCF-7 cells (lane 1) and K562 cells induced for erythroid differentiation for up to 8 days with sodium butyrate (lanes 2–6) or treated with puromycin for 16 h (lane 7) was analyzed in western blots with specific antibodies. (b) Caspase-Glo 3/7 assay of K562 cells induced for erythroid differentiation for up to 8 days or treated with puromycin for 16 h. The Caspase-3 inhibitor Ac-DMQD-CHO was added at the final concentration indicated. (c) Flow cytometric analysis of an AnnexinV-FITC and propidium iodide staining of K562 cells induced for erythroid differentiation for up to 8 days or treated with puromycin for 16 h
Caspase-3 activity is required for hnRNP K cleavage
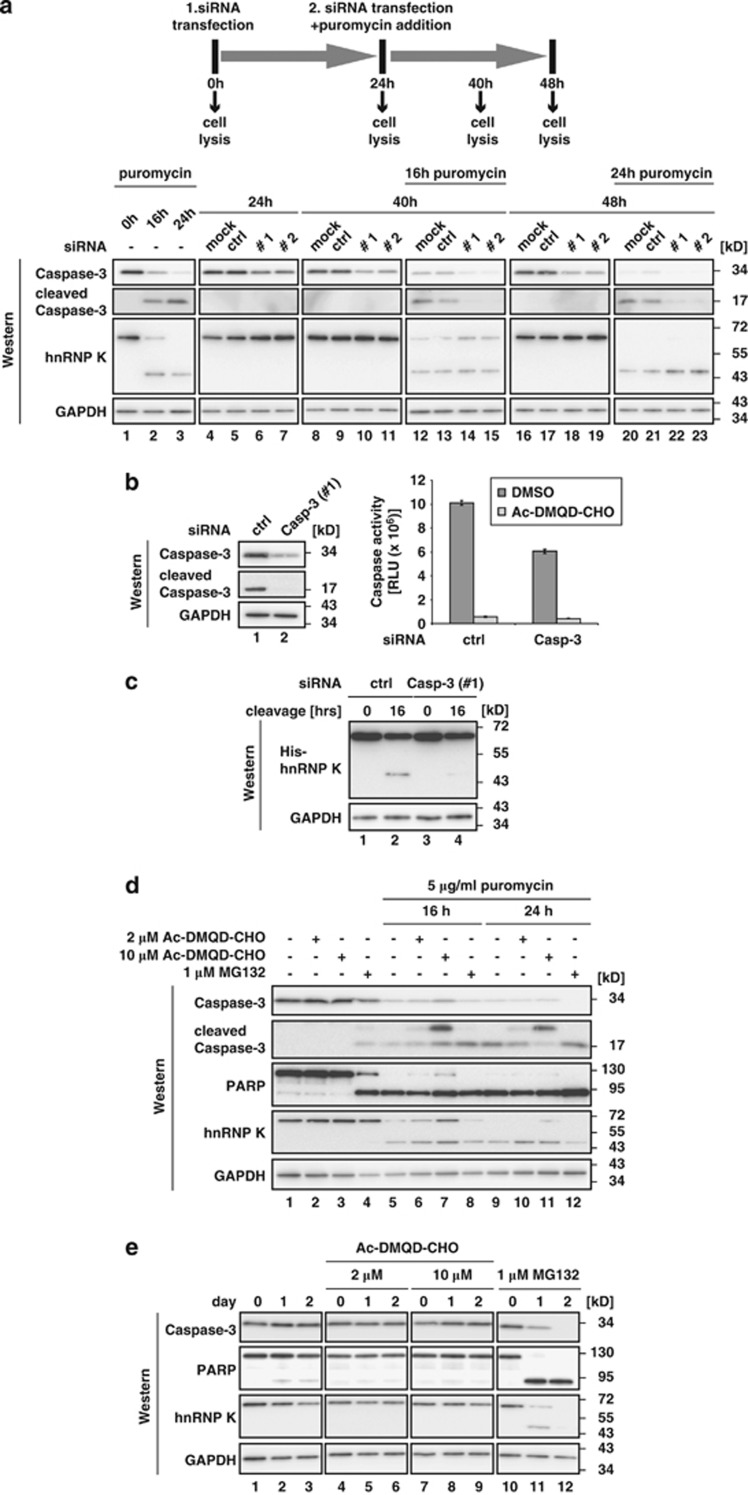
To evaluate a potential role of Caspase-3 in hnRNP K degradation, Caspase-3 expression was reduced by RNA interference (RNAi) in combination with puromycin treatment (Figure 3a). Puromycin treatment of K562 cells for 16 and 24 h resulted in Caspase-3 activation and hnRNP K cleavage (Figure 3a, lower panel, lanes 1–3 and Figure 1b). K562 cells were either mock transfected, transfected with a non-specific control siRNA (ctrl) or two individual siRNAs directed against Caspase-3 (Casp-3 #1, Casp-3 #2) (Figure 3a, lower panel, lanes 4–23). Twenty-four hours post transfection cells were exposed to puromycin (Figure 3a, upper panel and lower panel, lanes 12–15 and 20–23). Knockdown of Caspase-3, results in stabilization of full-length hnRNP K after 16 h of puromycin treatment (Figure 3a, lower panel, lanes 14 and 15). Additionally, a stabilization of the hnRNP K-derived cleavage product was detected after 24-h puromycin treatment, when full-length hnRNP K was below detection level (Figure 3a, lower panel, lanes 22 and 23). To recapitulate cleavage of hnRNP K by Caspase-3 in vitro, cytoplasmic extract of ctrl or Casp-3 #1 siRNA transfected K562 cells was prepared after 16 h puromycin treatment (Figure 3b, left panel). In Caspase-3 knockdown extract, the enzymatic activity was reduced to about 60% compared with extract prepared from ctrl siRNA transfected cells (Figure 3b, right panel), consistent with the reduction of Caspase-3 (Figure 3b, left panel). His-hnRNP K was incubated in these extracts to analyze hnRNP K cleavage with an antibody directed against the N-terminal His-tag. Although the hnRNP K cleavage product could be detected when the reaction was performed in cytoplasmic extract from ctrl siRNA transfected cells (Figure 3c, lane 2), cleavage was strongly reduced in extract prepared from Caspase-3 knockdown cells (Figure 3c, lane 4). The combination of RNAi against Caspase-3 with the induction of erythroid differentiation did not result in a detectable stabilization of hnRNP K (data not shown), probably because a smaller fraction of Caspase-3 is activated in erythroid differentiation compared with puromycin treatment (compare Figure 2a, lanes 2–7 and Figure 3a lower panel, lanes 1–3). When K562 cells were treated with puromycin for 16 or 24 h in the presence of the Caspase-3 inhibitor Ac-DMQD-CHO (Figure 3d), both hnRNP K and the N-terminal fragment were stabilized (Figure 3d, lanes 5–7 and 9–11) and the p20 Caspase-3 cleavage intermediate could be detected.22 In contrast, the proteasome inhibitor MG132 did not stabilize hnRNP K (Figure 3d, lanes 8 and 12). As a control we monitored PARP cleavage, which was also slightly reduced after 16 h in the presence of 10 μℳ Ac-DMQD-CHO (Figure 3d, lanes 5–7). During erythroid maturation, Ac-DMQD-CHO mediated Caspase-3 inhibition results in both, hnRNP K and PARP stabilization (Figure 3e, compare lanes 1–3 and lanes 4–9). The 48 kD hnRNP K cleavage product that could be enriched by immunoprecipitation on day 2 of erythroid maturation (see Figure 1a) can not be visualized in Figure 3e, lanes 1–3, due to the detection limit of the antibody (see Figure 1c). In contrast, treatment with the proteasome inhibitor MG132 that induces apoptosis23 resulted in cleavage of both PARP and hnRNP K (Figure 3e, lanes 10–12).
Figure 3.
Caspase-3 functions in cleavage of hnRNP K in K562 cells. (a) Upper panel: schematic representation of the experiment. K562 cells were transfected with a control siRNA (ctrl) or two individual siRNAs directed against Caspase-3 (Casp #1, Casp #2). Twenty-four hours later siRNA transfection was repeated to ensure prolonged knockdown and 5 μg/ml puromycin were added to the cells. Cells were harvested at the indicated time points. Lower panel: western blot analysis of the samples obtained from the experiment described above with antibodies specific for Caspase-3, hnRNP K and GAPDH. (b) Left panel: western blot analysis of cytoplasmic extracts generated from cells transfected with ctrl or Casp-3 #1 siRNA and treated with puromycin for 16 h with Caspase-3 and GAPDH specific antibodies. Right panel: the Caspase-Glo 3/7 assay was performed with extracts characterized at the left. Where indicated Ac-DMQD-CHO was added at a final concentration of 10 μℳ. (c) His-hnRNP K was incubated with cytoplasmic extract of ctrl or Casp-3 (#1) knockdown cells for 16 h at 37 °C. Reactions were stopped by addition of SDS sample buffer and analyzed in western blots with an antibody against the N-terminal His-tag of hnRNP K. GAPDH served as loading control. (d) Puromycin treatment of K562 cells was combined with the inhibition of Caspase-3. Cells were either non-treated or treated for 16 and 24 h with 5 μg/ml puromycin. Additionally, Ac-DMQD-CHO was added at the final concentrations indicated. The proteasome inhibitor MG132 was added as a specificity control. Western blot analysis with antibodies against Caspase-3, PARP, hnRNP K and GAPDH. (e) The inhibition of Caspase-3 and the proteasome as in (d) was combined with the induction of erythroid differentiation by addition of 1.5 mℳ sodium butyrate for up to 2 days. Western blot analysis as in (d)
HnRNP K is cleaved by ectopically expressed Caspase-3 in MCF-7 cells
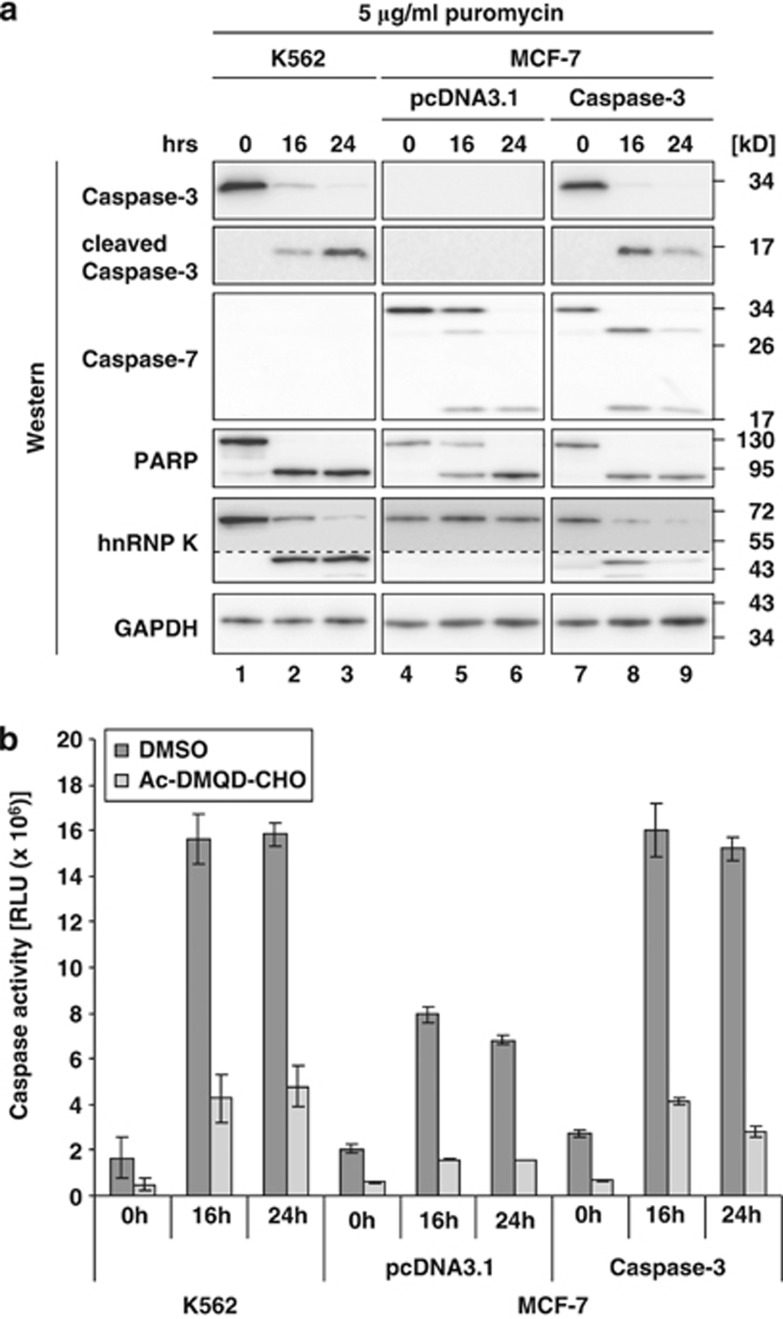
To further verify hnRNP K as a Caspase-3 substrate we employed the breast cancer cell line MCF-7, which in contrast to K562 cells does express Caspase-7, but not Caspase-3 (Figure 4a, lanes 1–6).24 To analyze the effect of Caspase-3 on hnRNP K cleavage, we made use of MCF-7 cells stably transfected with Caspase-3 cDNA (MCF-7(Caspase-3)).24 Caspase-3 was only detected in K562 and MCF-7(Caspase-3) cells, but not pcDNA3.1 transfected MCF-7 cells (MCF-7(pcDNA3.1)) (Figure 4a). Puromycin treatment resulted in the activation of Caspase-3 and -7 (Figure 4a) and induced a strong increase in Caspase activity in extracts of K562 cells and MCF-7(Caspase-3) cells as detected by the enzymatic assay (Figure 4b). The moderate increase of Caspase activity detected in MCF-7(pcDNA3.1) cells (Figure 4b) might result from puromycin-mediated Caspase-7 activation (Figure 4a, lanes 4–6).
Figure 4.
Ectopically expressed Caspase-3 restores hnRNP K cleavage in MCF-7 cells. (a) K562 cells, MCF-7 cells transfected with pcDNA3.1 (MCF-7(pcDNA3.1)) or with pcDNA3.1-Caspase-3 (MCF-7(Caspase-3)) were treated with puromycin for 16 and 24 h. Western blot analysis with antibodies directed against Caspase-3, Caspase-7, PARP, hnRNP K, GAPDH. The dotted line indicates extended exposure of the hnRNP K cleavage product. (b) Caspase-Glo 3/7 assay of lysates from K562, MCF-7(pcDNA3.1) and MCF-7(Caspase-3) cells treated as in (a). Addition of Ac-DMQD-CHO at a final concentration of 10 μℳ as indicated
The cleavage of PARP and hnRNP K was analyzed in the three cell lines. PARP that is a substrate of several Caspases20 is cleaved in all cell lines upon puromycin treatment (Figure 4a). In contrast, hnRNP K cleavage could be detected in K562 cells and in MCF-7(Caspase-3) cells (Figure 4a, lanes 1–3 and 7–9), but not MCF-7(pcDNA3.1) cells (Figure 4a, lanes 4–6).
Caspase-3 cleaves hnRNP K C-terminal to D334
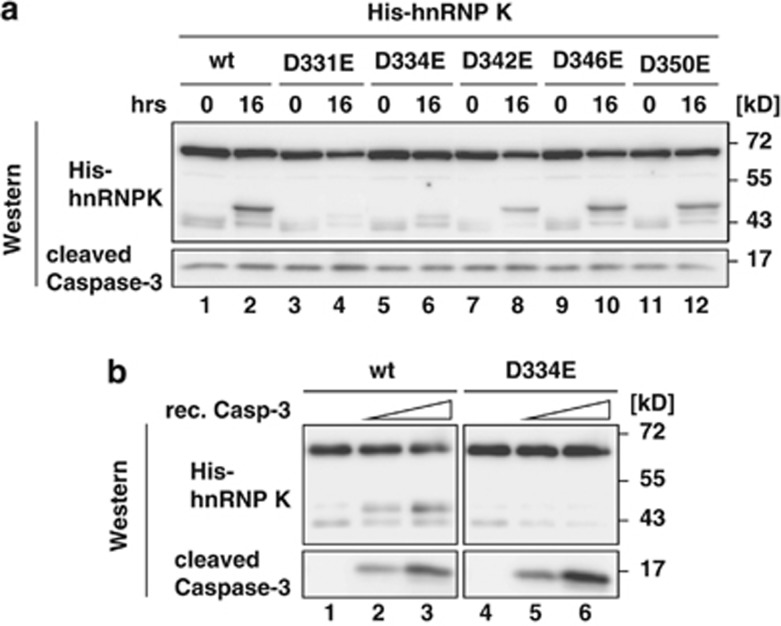
The analysis of the N-terminal hnRNP K cleavage product identified a peptide with R326 as the C-terminal residue (Figure 1d). To characterize the cleavage site of Caspase-3 that strictly cleaves C-terminal to aspartate residues,25 we generated five N-terminal His-tagged hnRNP K variants with aspartate replaced by glutamate C-terminal of R326 (His-hnRNP K D331E, D334E, D342E, D346E and D350E). The recombinant proteins were incubated in cytoplasmic extract from puromycin-treated K562 cells and analyzed in western blots employing a His-tag antibody (Figure 5a). The N-terminal fragment could only be detected when His-hnRNP K wt or variants D342E, D346E and D350E were used (Figure 5, lanes 1, 2 and 7–12). His-hnRNP K D331E and D334E were not cleaved (Figure 5, lanes 3–6), indicating that the protease recognizes a D-x-x-D motif and cleaves C-terminal to D334 of hnRNP K.
Figure 5.
Caspase-3 directly cleaves hnRNP K at D334-G335. (a) His-hnRNP K(wt) or the variants D331E, D334E, D342E, D346E and D350E were incubated 16 h with cytoplasmic extract from K562 cells treated with 5 μg/ml puromycin for 16 h at 37 °C. Reactions were stopped by addition of SDS sample buffer and analyzed in western blots with antibodies against the His-tag and Caspase-3. (b) His-hnRNP K(wt) or His-hnRNP K(D334E) were incubated with different amounts of recombinant Caspase-3 as indicated for 16 h at 37 °C. Analysis as in (a)
The cleavage of hnRNP K detected in MCF-7(Caspase-3) cells (Figure 4a) and the mutational analysis (Figure 5a), strongly indicate that specifically Caspase-3 cleaves hnRNP K. Through the use of recombinant activated Caspase-3 and His-hnRNP K(wt) or His-hnRNP K(D334E) in in vitro cleavage assays we wanted to exclude that another enzyme that is activated by Caspase-3 processing acts on hnRNP K (Figure 5b). The signal of the His-hnRNP K(wt) cleavage product detected in western blots increased when the amount of recombinant Caspase-3 was elevated in the in vitro cleavage reaction (Figure 5b, lanes 1–3), but no cleavage product of His-hnRNP K(D334E) could be detected (Figure 5b, lanes 4–6). The smaller band that appears when His-hnRNP K(wt) or His-hnRNP K(D334E) were used might represent a non-specific E.coli product as it could already be detected when Caspase-3 is not added to the cleavage assay (Figure 5b, lanes 1–6).
Caspase-3 cleavage inactivates hnRNP K as a DICE-binding protein
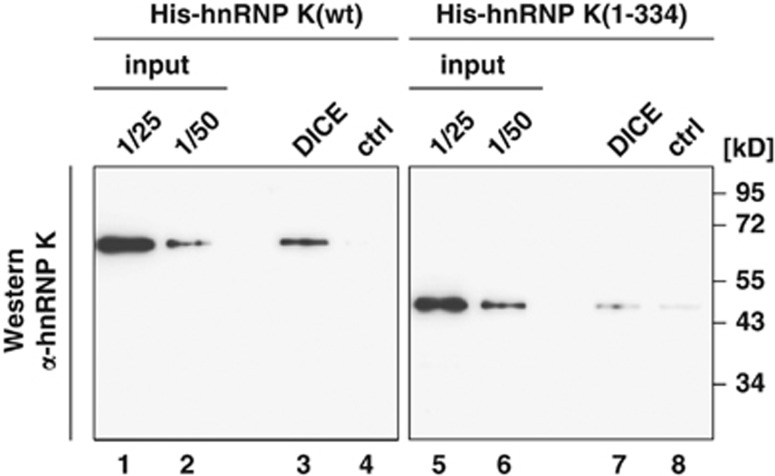
The RNA binding activity of hnRNP K, which is conferred by KH domain 3 that interacts with the 3′UTR DICE of the r15-LOX mRNA5 is critical for its function in translational regulation in erythroid differentiation. To prove that deletion of KH domain 3 by Caspase-3 results in loss of interaction with the target mRNA sequence, in vitro RNA binding assays were performed. His-hnRNP K(wt) and His-hnRNP K(1–334) were incubated with a 5′tobramycin (Tob) aptamer bearing DICE or the β-globin mRNA 5′leader as a control transcript (ctrl) bound to a Tob-matrix (Figure 6). His-hnRNP K(wt) is specifically bound to the DICE RNA, but not to the control RNA (Figure 6, lanes 1–4). In contrast, binding of truncated His-hnRNP K(1–334) to the DICE RNA is strongly reduced and comparable to the non-specific control RNA, due to the loss of the KH domain 3 that mediates RNA binding specificity (Figure 6, lanes 5–8). In vitro cleavage assays in the presence of the DICE or ctrl RNA showed that RNA binding neither affects His-hnRNP K cleavage by recombinant Caspase-3 (Supplementary Figure S4), nor that of endogenous hnRNP K in cytoplasmic extract from non-induced K562 cells by recombinant Caspase-3 (Supplementary Figure S5).
Figure 6.
Caspase-3 cleavage abolishes DICE-binding activity of hnRNP K. Tobramycin RNA affinity purification was performed employing the DICE or the β-globin mRNA 5′leader as control transcript (ctrl) and recombinant His-hnRNP K(wt) or His-hnRNP K(1-334). Western blot analysis with hnRNP K antibody #1
Discussion
In this study, we show that hnRNP K is a Caspase-3 substrate, which is cleaved during erythroid differentiation and puromycin-induced apoptosis of K562 cells. It was known before that the level of hnRNP K decreases during erythroid differentiation9 and employing hnRNP K immunoprecipitation we detected a specific cleavage product of about 48 kD (Figure 1a). Previously it was shown that hnRNP K10 and also other RNA binding proteins as HuR are degraded by the ubiquitin proteasome system26 in response to genotoxic stress. In DNA damage response, the function of hnRNP K in p53 activation has been reported to be modulated by sumoylation, which enables the protein to function as p53 cofactor27 or prevents HDM2-dependent ubiquitination and proteasomal degradation of hnRNP K.28 For YB-1 an ubiquitin and ATP independent cleavage by the 20S proteasome in response to DNA damage was shown,29 which generates a stable N-terminal fragment accumulating in the nucleus of stressed cells. Therefore we investigated a potential contribution of the ubiquitin proteasome system to hnRNP K degradation. Treatment of K562 cells with the proteasome inhibitor MG132 does not result in a stabilization of hnRNP K (Figures 3d and e), but even induced cleavage, presumably due to apoptosis induction.23 Additionally, we performed ubiquitin pull-down assays, but could not detect ubiquitination of hnRNP K during erythroid differentiation. In contrast, ubiquitination of Tak130 and XIAP31 was detectable (Supplementary Figure S6). The simultaneous knockdown of the two ubiquitin activating enzymes Uba132 and Uba633, 34 combined with either puromycin treatment or erythroid differentiation did not result in a stabilization of hnRNP K (Supplementary Figures S7 and S8), indicating that the ubiquitin proteasome system does not initiate hnRNP K degradation. To identify the so far unknown protease we characterized the hnRNP K-derived fragment. Our analysis indicated that the remaining stable N-terminal fragment lacks KH domain 3 (Figures 1c and d) that mediates binding to the DICE.5
Several reports described an activation of Caspase-3 during erythroid differentiation.12, 13, 14 Caspase-3 activation was also detectable during sodium-butyrate induced erythroid differentiation of K562 cells (Figures 2a and b). Cell viability is not influenced by sodium-butyrate treatment9 and an induction of apoptosis could be excluded by AnnexinV staining (Figure 2c).
The knockdown of Caspase-3 in puromycin-treated K562 cells showed that cleavage of hnRNP K is impaired in vivo and in vitro (Figures 3a–c). The combination of erythroid differentiation with Caspase-3 knockdown had no effect on hnRNP K degradation (not shown). This may be due to the fact that during erythroid differentiation a smaller fraction of Caspase-3 is activated than in puromycin-treated cells (Figure 2a). The inhibition of Caspase-3 with a specific peptide based inhibitor resulted in a stabilization of hnRNP K in K562 cells (Figures 3d–e), indicating that Caspase-3 has a function in hnRNP K degradation.
To further elucidate hnRNP K cleavage, we made use of Caspase-3 deficient MCF-7 cells and MCF-7 cells stably transfected with Caspase-3 cDNA24 (Figure 4). Analysis of puromycin-induced apoptosis in these cells revealed that hnRNP K is only cleaved in MCF-7(Caspase-3) cells but not MCF-7(pcDNA3.1) cells, (Figure 4a) strongly indicating that hnRNP K is a Caspase-3 substrate.
Mutational analysis implied that Caspase-3 cleaves hnRNP K C-terminal to D334 (Figure 5a) and recognizes the favored D-x-x-D-G motif.35 The minor cleavage products detected in (Figure 1c) might result from cleavage at position D282–M283. However, D334 seems to represent the major cleavage site of Caspase-3 in hnRNP K, because G335 is strongly favored in P1′ position compared with M283.35 The smaller fragments were only detectable after enrichment by hnRNP K immunoprecipitation (Figure 1c). In vitro cleavage reactions employing recombinant activated Caspase-3 excluded cleavage by an unknown protease, which is activated by Caspase-3 (Figure 5b). A comparison of the RNA binding capacity of His-hnRNP K(wt) and the N-terminal 48 kD fragment His-hnRNP K(1–334) revealed that the 48 kD fragment lost its specific DICE-binding activity (Figure 6). Binding to RNA does not influence hnRNP K cleavage in vitro. The DICE does not affect the Caspase-3 catalyzed cleavage of His-hnRNP K(wt) (Supplementary Figure S4) or of endogenous hnRNP K in cytoplasmic K562 cell extract (Supplementary Figure S5).
The identification of the specific 48 kD hnRNP K fragment and mutational analysis of putative Caspase recognition sites prove that Caspase-3 mediated cleavage separates the C-terminal KH domain 3 of hnRNP K that confers binding specificity to the r15-LOX mRNA DICE5 from the N-terminal part, which bears protein-interaction sites like the kinase binding domain.16, 17 Site-specific cleavage of hnRNP K represents a second molecular switch besides c-Src dependent Y458 phosphorylation, which modulates mRNA interaction specifically.4, 5 Both, phosphorylation of Y458 in KH domain 3 of hnRNP K that abrogates DICE-binding5 and the separation from the N-terminus, which confers protein-protein interactions, contribute to the release of r15-LOX mRNA from the translational repressed state.
Analysis of XIAP expression (Figure 2a) suggests that the activity of Caspase-3 is regulated by XIAP during erythroid differentiation.
Activation of Caspase-3 is well studied in apoptosis where it results in cleavage of multiple proteins.11 Several reports also suggest a role for Caspase-3 activation in proliferation and differentiation processes in erythroid cells,12 lens fiber cells,36 neuronal cells,37 skeletal muscle cells38 and in monocyte to macrophage differentiation.39, 40 There Caspase-3 catalyzes the cleavage of a more restricted and distinct set of proteins.18 Common characteristics of erythropoiesis and the differentiation of lens fiber cells are the exclusion of the nucleus and degradation of other organelles such as mitochondria. R15-LOX initiates the degradation of mitochondria in mature reticulocytes and during lens fiber cell differentiation.41 The restricted expression of r15-LOX to late differentiation stages and activation of Caspase-3 suggests that r15-LOX expression is regulated by similar mechanisms in these differentiation processes.
Recently, it was reported that Granzyme M cleaves hnRNP K at multiple sites in human cytomegalovirus infection.42 The fragment of hnRNP K that is generated by Granzyme M in infected cells has a similar molecular weight as the Caspase-3 cleavage product in erythroid differentiation, but cleavage at amino acids D331 or D334 was not detected, instead M359 was mapped as cleavage site.42
In future studies, it would be interesting to analyze whether cleavage and inactivation of the translational regulator hnRNP K by Caspase-3 is restricted to the hematopoietic lineage or represents a more general mechanism in development and differentiation.
Materials and Methods
Plasmids
pET16b-hnRNP K7 was used to generate amino acid substitutions D331E, D334E, D342E, D346E and D350E by site-directed mutagenesis with the following primers:
D331E fw: GGAGACCTGGAGAGCGTTACGACGGCATGG
D331E rv: CCATGCCGTCGTAACGCTCTCCAGGTCTCC
D334E fw: GAGACCGTTACGAGGGCATGGTTGGTTTCA
D334E rv: TGAAACCAACCATGCCCTCGTAACGGTCTC
D342E fw: GTTTCAGTGCTGAAGAAACTTGGGACTCTG
D342E rv: CAGAGTCCCAAGTTTCTTCAGCACTGAAAC
D346E fw:GATGAAACTTGGGAGTCTGCAATAGATACATGG
D346E rv: CCATGTATCTATTGCAGACTCCCAAGTTTCATC
D350E fw: GACTCTGCAATAGAAACATGGAGCCCATCA
D350E rv: TGATGGGCTCCATGTTTCTATTGCAGAGTC
To generate pET16b-hnRNP K(1-334) the nucleotides that encode amino acids 1–334 of human hnRNP K were amplified from pET16b-hnRNP K7 with the following primers K1–334 BamHI fw: GATAGGAGGATCCGATGGAAACTGAACAGCCAGA, K1–334 BamHI rv: CTATCGGATCCTCTTAGTCGTAACGGTCTCCAGG and cloned into the BamHI site of pET16b. To generate Tob-ctrl, the β-globin 5′leader43 was cloned into XhoI/SpeI of pBSIIKS(+) containing the Tob aptamer in the KpnI site (pKS-Tob). pBSIISK-10R44 was digested with NotI and ligated to an oligo encoding the Tob aptamer: Tob NotI fw: GGCCGCGGCTTAGTATAGCGAGGTTTAGCTACACTCGTGCTGAGCCACTGGGCCAGTGC, Tob NotI rv: GGCCGCACTGGCCCAGTGGCTCAGCACGAGTGTAGCTAAACCTCGCTATACTAAGCCGC.
Cell culture
K562 cells were cultured and induced for erythroid differentiation with 1.5 mℳ sodium butyrate.9 MCF-7 cells stably transfected with pcDNA3.1 or pcDNA3.1-Caspase-3 were described previously.24 K562 (1 × 105/ml) and MCF-7 cells were treated with 5 μg/ml puromycin (Sigma-Aldrich, St Luis, MO, USA) for 16 or 24 h. To purify the hnRNP K-derived fragment, K562 cells (3.5 × 105/ml) were treated for 16 h with puromycin (20 μg/ml). Where indicated, the Caspase-3 specific inhibitor Ac-DMQD-CHO (Enzo Life Sciences, Farmingdale, NY, USA) was added to a final concentration of 2 or 10 μℳ. The proteasome inhibitor MG132 (Biomol, Humburg, Germany) was used at 1 μℳ final concentration.
Cytoplasmic extract preparation
K562 cytoplasmic extracts were generated as described previously.9
Lysate preparation and western blotting
Lysates were prepared and analyzed by western blotting according to A Ostareck-Lederer et al.4
Antibodies
Antibodies were purchased from Santa Cruz (Dallas, TX, USA) (hnRNP K #1, Caspase-3, Caspase-7, His), Abcam (Cambridge, UK) (GAPDH) and Cell signaling (Cell Signaling, Boston, MA, USA) (PARP, XIAP).
The monoclonal hnRNP K antibody #2 was generated against a human hnRNP K peptide (255MRGRGGFDRMPPGRGGRP272),9 polyclonal hnRNP K antibody #3 was described.45
hnRNP K immunoprecipitation
HnRNP K was immunoprecipitated essentially as in IS Naarmann et al.9
HnRNP K fragment purification and mass spectrometry analysis
Purification of the hnRNP K-derived fragment is described in the Supplementary Methods.
The fragment was excised from a colloidal Coomassie stained NuPAGE 4%–12% Bis-Tris gel (Invitrogen, Carlsbad, CA, USA), in gel digested with trypsin, extracted46 and peptides were analyzed in a capillary HPLC coupled electrospray ionization quadrupole time of flight (ESI-Q-TOF, Ultima, Waters, Milford, MA, USA) mass spectrometer as described.47 Peptide assignments and data analysis were performed by Mascot, Matrix Science (London, UK).
Caspase-3/7 activity assay
Caspase-3/7 activity was measured using the Caspase-3/7 Glo Assay (Promega) with either 2 × 104 cells (Figure 2b) or 2 μg lysate (Figures 3b and 4b). Reactions were spiked with DMSO or the Caspase-3 specific inhibitor Ac-DMQD-CHO at a final concentration of 2 μℳ and 10 μℳ. Measurements were done in triplicate. The mean of three biological replicates is shown.
AnnexinV and propidium iodide staining
For AnnexinV and propidium iodide staining the AnnexinV-FITC Apoptosis Detection Kit I (BD Biosciences, San Jose, CA, USA) was employed. Flow cytometric analysis was performed on a FACS Canto II (BD Biosciences), data were analyzed using Flowjo (Tree Star, Ashland, OR, USA).
RNAi
For RNAi K562 cells were transfected with two individual Caspase-3 siRNAs (Casp-3 #1: GCAGCAAACCUCAGGGAAAdTdT, Casp-3 #2: AGUGAAGCAAAUCAGAAACdTdT) or a non-specific control siRNA (ctrl) as described previously.9 Transfection was repeated after 24 h to ensure prolonged Caspase-3 knockdown.
Expression of recombinant hnRNP K
His-hnRNP K and variants were expressed and purified as described,5, 7 except that proteins were dialyzed against 20 mℳ Tris pH 8.0, 300 mℳ potassium chloride, 10 mℳ sodium citrate and 10% sucrose.
In vitro cleavage assay
Five hundred nanograms His-hnRNP K(wt) or variants were incubated with 40-μg cytoplasmic extract from puromycin-treated K562 cells for 16 h, 37 °C in hnRNP K ipp buffer.9 Reactions were stopped by SDS sample buffer addition and analyzed in western blot assays with a His-tag antibody. Reactions containing 0, 1 or 2 μl of activated recombinant Caspase-3 (Enzo Life Sciences) were incubated accordingly. Where indicated, 500 ng in vitro transcribed DICE or control RNA (ctrl)47 were added to the assays.
In vitro transcription
RNAs used for affinity purification and in the in vitro cleavage assay were transcribed with T3 or T7 MEGAscript Kit (Applied Biosystems, Foster City, CA, USA), respectively.
RNA affinity purification
Tob aptamer RNA affinity purification48 was modified: Recombinant His-hnRNP K(wt) or His-hnRNP K(1-334) (40 pmol) were incubated with 2 pmol of in vitro transcribed RNAs as indicated in BB (20 mℳ Tris-HCl, pH 8.0, 1 mℳ CaCl2, 60 mℳ KCl, 3.5 mℳ MgCl2, 0.05% NP-40, 0.2 mℳ DTT) for 10 min at room temperature before immobilization at the Tob-matrix (1.5 h, 4 °C). Beads were washed three times with BB and complexes were eluted with SDS sample buffer and subjected to western blotting.
Acknowledgments
We thank N Simons for technical assistance, B Moritz for help with the purification of the hnRNP K-derived cleavage product as well as F Heymann and A Liepelt for support with flow cytometry. Stably transfected MCF-7 cells were kindly provided by M Bushell and hnRNP K antibody #3 by K Bomsztyk. This work was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) to AOL and DHO (OS 290/3-2).
Author contributions
Unless otherwise stated experiments were performed by ISNdV. Mass spectrometry analysis was performed by HU. ISNdV, DHO and AOL designed the experiments and wrote the paper.
Glossary
- aa
amino acid
- CX
cytoplasmic extract
- DICE
differentiation control element
- hnRNP K
heterogeneous nuclear ribonucleoprotein K
- KH
hnRNP K homology domain
- PARP
poly-ADP-ribose polymerase 1
- r15-LOX
reticulocyte 15-lipoxygenase
- RNAi
RNA interference
- siRNA
small interfering RNA
- Tob
5′ tobramycin
- UTR
untranslated region
- XIAP
X-chromosome linked inhibitor of apoptosis protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschellá
Supplementary Material
References
- de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3' UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Abaza I, Gebauer F. Trading translation with RNA-binding proteins. RNA. 2008;14:404–409. doi: 10.1261/rna.848208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, et al. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, et al. c-Src-mediated phosphorylation of hnrnp K Drives translational activation of specifically silenced mRNAs. Mol Cell Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messias AC, Harnisch C, Ostareck-Lederer A, Sattler M, Ostareck DH. The DICE-binding activity of KH domain 3 of hnRNP K is affected by c-Src-mediated tyrosine phosphorylation. J Mol Biol. 2006;361:470–481. doi: 10.1016/j.jmb.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Rucknagel KP, Schierhorn A, Moritz B, Huttelmaier S, et al. Asymmetric arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src. J Biol Chem. 2006;281:11115–11125. doi: 10.1074/jbc.M513053200. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3'end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: the 3'UTR regulatory complex controls 60s ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Naarmann IS, Harnisch C, Flach N, Kremmer E, Kühn H, Ostareck DH, et al. mRNA silencing in human erythroid cell maturation. J Biol Chem. 2008;283:18461–18472. doi: 10.1074/jbc.M710328200. [DOI] [PubMed] [Google Scholar]
- Moumen A, Masterson P, O'Connor MJ, Jackson SP, hnRNP K. An HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- Zermati Y, Garrido C, Amsellem S, Fishelson S, Bouscary D, Valensi F, et al. Caspase activation is required for terminal erythroid differentiation. J Exp Med. 2001;193:247–254. doi: 10.1084/jem.193.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile GW, Smith DH, Wiedmann M. Caspase-3 has a nonapoptotic function in erythroid maturation. Blood. 2004;103:4310–4316. doi: 10.1182/blood-2003-09-3362. [DOI] [PubMed] [Google Scholar]
- Sztiller-Sikorska M, Jakubowska J, Wozniak M, Stasiak M, Czyz M. A non-apoptotic function of caspase-3 in pharmacologically-induced differentiation of K562 cells. Br J Pharmacol. 2009;157:1451–1462. doi: 10.1111/j.1476-5381.2009.00333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka K, Toné S, Mukaida M, Takano-Ohmuro H. The apoptotic and nonapoptotic nature of the terminal differentiation of erythroid cells. Exp Cell Res. 1998;240:206–217. doi: 10.1006/excr.1997.3927. [DOI] [PubMed] [Google Scholar]
- Adolph D, Flach N, Mueller K, Ostareck DH, Ostareck-Lederer A. Deciphering the cross talk between hnRNP K and c-Src: the c-Src activation domain in hnRNP K is distinct from a second interaction site. Mol Cell Biol. 2007;27:1758–1770. doi: 10.1128/MCB.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsztyk K, Denisenko O, Ostrowski J, hnRNP K. One protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Festjens N, Declercq W, Berghe TV, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14:44–55. doi: 10.1038/sj.cdd.4402047. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Soldani C, Scovassi AI. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: an update. Apoptosis. 2002;7:321–328. doi: 10.1023/a:1016119328968. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Caspases—controlling intracellular signals by protease zymogen activation. Biochim Biophys Acta. 2000;1477:299–306. doi: 10.1016/s0167-4838(99)00281-2. [DOI] [PubMed] [Google Scholar]
- Shinohara K, Tomioka M, Nakano H, Toné S, Ito H, Kawashima S. Apoptosis induction resulting from proteasome inhibition. Biochem J. 1996;317:385–388. doi: 10.1042/bj3170385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänicke RU, Sprengart ML, Wati MR, Porter AG. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J Biol Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- Howard AD, Kostura MJ, Thornberry N, Ding GJ, Limjuco G, Weidner J, et al. IL-1-converting enzyme requires aspartic acid residues for processing of the IL-1 beta precursor at two distinct sites and does not cleave 31-kDa IL-1 alpha. J Immunol. 1991;147:2964–2969. [PubMed] [Google Scholar]
- Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisch F, Pozzi B, Risso G, Munoz MJ, Srebrow A. DNA damage-induced heterogeneous nuclear ribonucleoprotein K SUMOylation regulates p53 transcriptional activation. J Biol Chem. 2012;287:30789–30799. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Lee MH, Park JH, Kang SH, Yoo HM, Ka SH, et al. SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J. 2012;31:4441–4452. doi: 10.1038/emboj.2012.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin AV, Selyutina AA, Skabkin MA, Guryanov SG, Nazimov IV, Richard C, et al. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 2005;24:3602–3612. doi: 10.1038/sj.emboj.7600830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yan J, Mao A-P, Li C, Ran Y, Shu H-B, et al. Tripartite motif 8 (TRIM8) modulates TNFα- and IL-1βtriggered NF-κB activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci USA. 2011;108:19341–19346. doi: 10.1073/pnas.1110946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Heller H, Katz-Etzion R, Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc Natl Acad Sci USA. 1981;78:761–765. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer C, Kassner I, Matentzoglu K, Singh RK, Wollscheid H-P, Scheffner M, et al. UBE1L2, a Novel E1 enzyme specific for ubiquitin. J Biol Chem. 2007;282:23010–23014. doi: 10.1074/jbc.C700111200. [DOI] [PubMed] [Google Scholar]
- Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- Ishizaki Y, Jacobson MD, Raff MCA. Role for caspases in lens fiber differentiation. J Cell Biol. 1998;140:153–158. doi: 10.1083/jcb.140.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando P, Brunette S, Megeney LA. Neural stem cell differentiation is dependent upon endogenous caspase-3 activity. FASEB J. 2005;19:1671–1673. doi: 10.1096/fj.04-2981fje. [DOI] [PubMed] [Google Scholar]
- Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci USA. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordet O, Rébé C, Plenchette S, Zermati Y, Hermine O, Vainchenker W, et al. Specific involvement of caspases in the differentiation of monocytes into macrophages. Blood. 2002;100:4446–4453. doi: 10.1182/blood-2002-06-1778. [DOI] [PubMed] [Google Scholar]
- Cathelin S, Rébé C, Haddaoui L, Simioni N, Verdier F, Ml Fontenay, et al. Identification of proteins cleaved downstream of caspase activation in monocytes undergoing macrophage differentiation. J Biol Chem. 2006;281:17779–17788. doi: 10.1074/jbc.M600537200. [DOI] [PubMed] [Google Scholar]
- van Leyen K, Duvoisin RM, Engelhardt H, Wiedmann M. A function for lipoxygenase in programmed organelle degradation. Nature. 1998;395:392–395. doi: 10.1038/26500. [DOI] [PubMed] [Google Scholar]
- van Domselaar R, de Poot SAH, Remmerswaal EBM, Lai KW, ten Berge IJM, Bovenschen N. Granzyme M targets host cell hnRNP K that is essential for human cytomegalovirus replication. Cell Death Differ. 2013;20:419–429. doi: 10.1038/cdd.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, et al. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Standart N, Thiele BJ. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Seuningen I, Ostrowski J, Bustelo XR, Sleath PR, Bomsztyk K. The K Protein domain that recruits the interleukin 1-responsive K protein kinase lies adjacent to a cluster of c-Src and Vav SH3-binding Sites. J Biol Chem. 1995;270:26976–26985. doi: 10.1074/jbc.270.45.26976. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Naarmann IS, Harnisch C, Müller-Newen G, Urlaub H, Ostareck-Lederer A, Ostareck DH. DDX6 recruits translational silenced human reticulocyte 15-lipoxygenase mRNA to RNP granules. RNA. 2010;16:2189–2204. doi: 10.1261/rna.2211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinlich S, Hüttelmaier S, Schierhorn A, Behrens S-E, Ostareck-Lederer A, Ostareck DH. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3'UTR. RNA. 2009;15:1528–1542. doi: 10.1261/rna.1578409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.