Abstract
Ras association (RalGDS/AF-6) domain family member RASSF5 is a non-enzymatic RAS effector super family protein, known to be involved in cell growth regulation. Expression of RASSF5 is found to be extinguished by promoter hypermethylation in different human cancers, and its ectopic expression suppresses cell proliferation and tumorigenicity. Interestingly, this role in tumorigenesis has been confounded by the fact that regulation at molecular level remains unclear and many transformed cells actually display elevated RASSF5 expression. Here, we demonstrate that E3 ubiquitin ligase Itch is a unique binding partner of RASSF5. Itch can interact with PPxY motif in RASSF5 both in vivo and in vitro through its WW domains. Importantly, the overexpression of Itch induces RASSF5 degradation by poly-ubiquitination via 26S proteasome pathway. In addition, our results indicate that the elevated levels of RASSF5 found in tumor cells due to acetylation, which restricts its binding to Itch and results in a more stable inert protein. Inhibition of RASSF5 acetylation permits its interaction with Itch and provokes proteasomal degradation. These data suggest that apart from promoter methylation, hyperacetylation could also be downregulating RASSF5 function in different human cancer. Finally, results from functional assays suggest that the overexpression of wild type, not the ligase activity defective Itch negatively regulate RASSF5-mediated G1 phase transition of cell cycle as well as apoptosis, suggesting that Itch alone is sufficient to alter RASSF5 function. Collectively, the present investigation identifies a HECT class E3 ubiquitin ligase Itch as a unique negative regulator of RASSF5, and suggests the possibility that acetylation as a potential therapeutic target for human cancer.
Keywords: RASSF5, Itch, ubiquitination, acetylation, cell cycle
Ras association (RalGDS/AF-6) domain family member RASSF5/NORE1 is the founding member of RAS effector super family protein that inhibits tumor growth by promoting G1/S arrest.1, 2, 3, 4, 5 RASSF5 is a proapoptotic factor of RAS and induces apoptosis via p53.2 RASSF5 binds with components (mammalian sterile 20-like kinase, MST1/2 and yes associated protein 1, YAP1) of hippo pathway, and regulates cell proliferation and apoptosis.6, 7, 8 Mice deficient for RASSF5 were resistant to TNF-α-induced apoptosis.4 Furthermore, targeted ablation of RASSF5 resulted in spontaneous immortalization of mouse embryo fibroblasts (MEFs) and the RASSF5-deficient MEFs were transformed by GTP-bound form of K-Ras.4 In addition, loss of RASSF5 expression by promoter hypermethylation has been linked to tumor formation.9, 10 Given its important role in tumor, studies of RASSF5 may provide new insights in our understanding on the mechanism that leads to tumorigenesis.
Although, RASSF5 shares high homology with other RASSF family members, a unique proline-rich (PPxY) motif is present only in RASSF5.11 WW domains are known to mediate protein–protein interactions through the recognition of PPxY motifs.12, 13, 14 Components of the hippo pathway like MST1/2, large associated tumor suppressor 1 (LATS1) and YAP1 are shown to modulate cell growth and apoptotic function through PPxY–WW domain interaction.15, 16, 17, 18, 19 In addition, recent reports suggest that E3 ubiquitin ligase Itch WW domain interacts with p73, p63 and LATS1 through PPxY motifs, and regulates their functions.16, 20, 21 Protein ubiquitination is a fundamental mechanism for regulating steady state levels and functions of many proteins involved in controlling cell proliferation and apoptosis by targeting them to proteasomal-mediated degradation pathway.15, 16, 17, 18, 19, 20, 21 These results prompted us to understand whether WW domain containing E3 ligase regulates RASSF5 function.
Recent studies suggest that post-translational modifications regulate the functions of proteins that are involved in cell cycle regulation, differentiation, oxidative stress response and transcription.3, 22, 23, 24, 25, 26, 27, 28 Interestingly, acetylation of proteins resulted in both the activation and deregulation of protein function.24, 26, 27, 29, 30 In addition, altered expression of deacetylases resulted in tumorigenesis.24, 27 Furthermore, acetylation of proteins modulate ubiquitination resulted in the alteration of protein function suggest that deacetylation may promote ubiquitination.27 Together, these data suggest that acetylation and ubiquitination have a critical role in regulating protein stability and function.
In the present study, we have provided evidence that E3 ubiquitin ligase Itch interacts with RASSF5, and promotes degradation via proteasome pathway. In addition, the acetylated form of RASSF5 fails to interact with Itch in transformed cells. These finding suggests that Itch mediates the degradation of RASSF5 by proteasome-mediated pathway in acetylation-dependent manner.
Results
Ubiquitin ligase Itch interacts with RASSF5
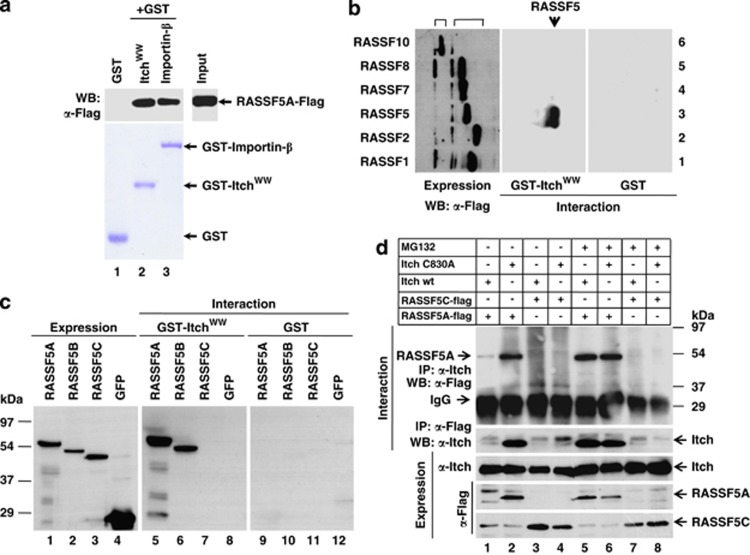
Detailed analysis of the amino acid sequences of RASSF super family proteins (RASSF1 to RASSF10) suggests the presence of PPxY motif only in RASSF5 (Supplementary Figure S1A). It is well established that PPxY motif is responsible for the interaction with WW domain-containing proteins.15, 16, 17, 18, 19 Recent reports suggest that WW domain-containing E3 ligase Itch utilizes PPxY motif to target substrates for ubiquitin-dependent degradation.16, 20, 21 To confirm whether RASSF5 interacts with Itch, we examined glutathione S-transferase (GST)-Itch WW domains (amino acids 251–529) for their ability to associate with RASSF5-Flag in GST pull-down experiments. Interestingly, RASSF5A-Flag from human embryonic kidney 293T (HEK293T) cell lysates specifically interacts with the GST-Itch WW domain to the same extent as the positive control GST-Importin-β (Figure 1a). Despite the presence of PPxY motif only in RASSF5, we tested whether Itch interacts with other RASSF members. Results in figure 1b demonstrate that Itch interacts specifically with RASSF5, not with other RASSF family members. These data suggest that Itch is a specific interacting partner for RASSF5.
Figure 1.
E3 ubiquitin ligase is a unique interacting partner for RASSF5. (a) GST pull-down analysis of interaction between RASSF5 and Itch WW domain in vitro. HEK293T cell lysates containing RASSF5A-Flag was incubated with equal amount of either GST or GST-Itch WW domain or GST-Importin-β and the bound proteins were resolved in SDS-12% PAGE followed by western blotting using anti-Flag antibodies. GST-Importin -β was used as positive control. (b) HEK293T cell lysates containing equal amounts of Flag-tagged RAS effectors (classical Ras effectors: RASSF1, RASSF2 and RASSF5; N-terminal Ras association domain containing Ras effectors: RASSF7, RASSF8 and RASSF10) were pull-down with either GST or GST-Itch WW domain followed by western blotting using anti-Flag antibodies. Results indicate that Itch specifically interacts only with RASSF5. (c) HEK293T cell lysates containing equal amounts of alternate spliced forms of GFP-tagged RASSF5 (RASSF5A, RASSF5B and RASSF5C (no PPxY motif)) were incubated with either GST or GST-Itch WW domain, and the bound proteins were resolved in SDS-12%PAGE followed by western blotting using anti-GFP antibodies. (d) RASSF5A-Flag or RASSF5C-Flag was transiently co-transfected with either Itchwt or ItchC830A mutant (ligase activity defective) in HEK293T cells and treated with proteasome inhibitor, MG132. The expression of all the indicated proteins (top) was determined by western blot analysis using anti-Flag or anti-Itch antibodies. Cell lysates containing equal amounts of indicated proteins (top) were used for coimmunoprecipitation with anti-Flag or anti-Itch antibodies followed by western blot analysis using anti-Itch or anti-Flag antibodies as indicated
RASSF5 interacts with Itch both in vitro and in vivo
Comparative sequence analysis of all three alternatively spliced forms of RASSF5 indicates Itch interaction motif, PPxY is present only in RASSF5A and RASSF5B, not in RASSF5C (Supplementary Figure S1B). Results in Figure 1c indicate that WW domain of Itch interacts with RASSF5A–green fluorescent protein (GFP) and RASSF5B–GFP, but not with RASSF5C–GFP or GFP. GST was used as negative control (Figure 1c). These data suggest that PPxY motif in RASSF5 is critical to interact with Itch. To verify this interaction in vivo, full-length Itch expression plasmid was co-transfected with RASSF5A-Flag and RASSF5C-Flag expression vectors in HEK293T cells. Interestingly, RASSF5C was efficiently expressed in the presence of Itch, but not RASSF5A (Figure 1d). These data lead to the hypothesis that Itch and RASSF5A interaction through PPxY motif resulted in the degradation of RASSF5A. To confirm this possibility, RASSF5A and RASSF5C were coexpressed with the catalytically inactive Itch mutant, ItchC830A, and their expression checked. Figure 1d suggests that the amount of RASSF5A protein was similar to that of RASSF5C in ItchC830A coexpressed cells. These data suggest that the catalytic activity of Itch is required for RASSF5A degradation. We next checked the expression pattern of both RASSF5A and RASSF5C in the presence of proteasome inhibitor, MG132 with or without Itchwt expression. Results in Figure 1d indicate that MG132 rescues RASSF5A expression despite the overexpression of Itchwt, while the level of RASSF5C expression was not altered. Together, these data provided evidence that Itch may mediate the degradation of RASSF5A via poly-ubiquitination/proteasome-mediated pathway.
We next performed coimmunoprecipitation experiments from HEK293T cell lysates containing RASSF5A-Flag or RASSF5C-Flag coexpressed with Itchwt or ItchC830A in presence or absence of MG132. Results in Figure 1d indicate that Itch physically interacts with RASSF5A in vivo, and interestingly, interaction of ItchC830A with RASSF5A suggests that the catalytic activity of Itch is not essential for its association. In addition, while a comparable levels of RASSF5A associates with Itchwt or ItchC830A in presence of MG132, no association with RASSF5C detected even in the presence of MG132 (Figure 1d). Furthermore, reciprocal coimmunoprecipitation suggests that RASSF5A was detected in Itch immunoprecipitates (Figure 1d). The observed faint signal for RASSF5A when coexpressed with Itchwt may be due to Itch-mediated degradation. These data provided evidence that Itch physically interacts with RASSF5A in vivo and mediates RASSF5A degradation.
It is well-known that Itch encodes four WW domains,17 and we performed series of GST pull-down assays to find out whether any one or all of Itch WW domains are involved in RASSF5A interaction. Towards this end, several GST-Itch WW domain mutants were generated (Supplementary Figure S2A) and results in Supplementary Figure S2B indicate that Itch mutants W291, 323A; W291, 403A and W291, 443A failed to interact with RASSF5A. Interestingly, exchange of individual tryptophan residues within WW domains did not alter Itch interaction, but the exchange of W291 in combination with W323 or W403 or W443 completely abrogated Itch interaction with RASSF5A (Supplementary Figure S2B). These data suggest that Trp 291 has a critical role in Itch interaction with RASSF5. Taken together, our results demonstrate that Itch interacts with RASSF5 both in vitro and in vivo, and this interaction is critical for RASSF5 turn-over in cells.
Itch-dependent degradation of RASSF5
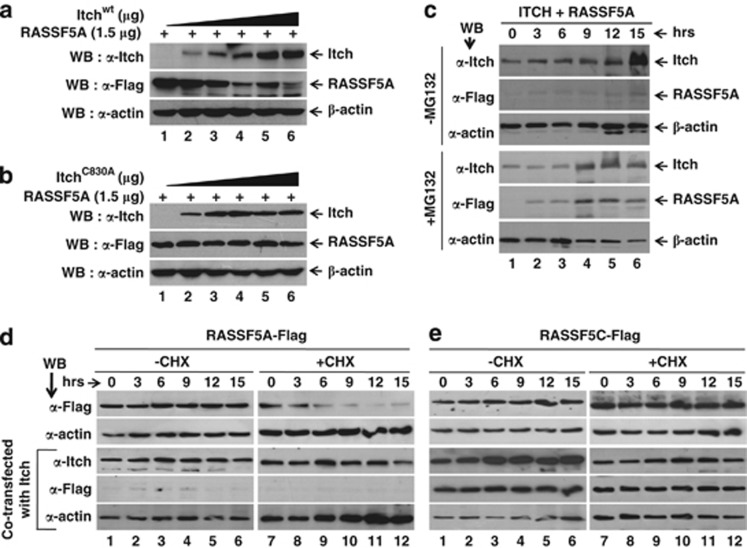
Itch is known to target multiple tumor suppressor proteins for ubiquitination and degradation.17, 20, 21 We next tested whether RASSF5A is an ubiquitin substrate for Itch. Results in figure 2a indicate that the overexpression of Itch significantly down-modulates RASSF5A in a dose-dependent manner. In contrast, coexpression of increasing amounts of ItchC830A has no effect on RASSF5A levels (Figure 2b) despite a robust interaction (Figure 1d). These data suggest that catalytic activity of Itch is important for RASSF5 degradation. In addition, the downregulation of RASSF5A in the presence of Itchwt was blocked by the treatment of MG132 in a time-dependent manner (Figure 2c). To further confirm the specificity of RASSF5A downregulation by Itch at protein not at mRNA levels, we determined whether Itch can degrade RASSF5C in cycloheximide (CHX) chase experiment. Interestingly, overexpression of Itchwt but not ItchC830A efficiently promotes RASSF5A degradation with and without CHX treatment (Figure 2d and Supplementary Figure S3A) in a time-dependent manner. In contrast, RASSF5C level remains stable in presence of CHX (Figure 2e). However, the levels of RASSF5A–GFP mRNA were unchanged with or without CHX treatment upon Itch overexpression (Supplementary Figure S3B). Furthermore, analysis of RASSF5A–GFP and RASSF5C–GFP immunofluorescence clearly indicates that Itch downmodulates RASSF5A, but not RASSF5C in the absence of MG132 in vivo (Supplementary Figure S4). Interestingly, RASSF5A signal was observed in the nucleus when overexpressed with Itchwt in the presence of MG132, and similar signal was observed when RASSF5A–GFP coexpressed with ItchC830A (Supplementary Figure S4). Collectively, these data strongly suggest that the destabilizing effect of Itch on RASSF5A is specific at the protein level in proteasome-dependent manner.
Figure 2.
RASSF5 is downregulated by Itch. Dose-dependent degradation of RASSF5 by Itch. Different amounts of Itchwt (a) or catalytically inactive (ItchC830A) mutant (b) expression plasmids were co-transfected with RASSF5A-Flag in HEK293T cells, treated with CHX for 3 h followed by western blot analysis using anti-Flag antibodies. Results indicate the loss of ligase activity impairs its effect on RASSF5 degradation. (c) Itch mediates the RASSF5 degradation by 26S proteasome pathway. HEK293T cells were co-transfected with RASSF5A-Flag and Itch as described in Materials and Methods. Transfected cells were treated with CHX alone or with proteasome inhibitor MG132. (d and e) CHX chase analysis of RASSF5 degradation upon Itch over expression. HEK293T cells expressing either RASSF5A-Flag (d) or RASSF5C-Flag (e) alone or with Itch were treated with CHX to inhibit de novo protein synthesis. Cell lysates were prepared at indicated time periods and determined RASSF5A and RASSF5C levels by western blot analysis using anti-Flag antibodies
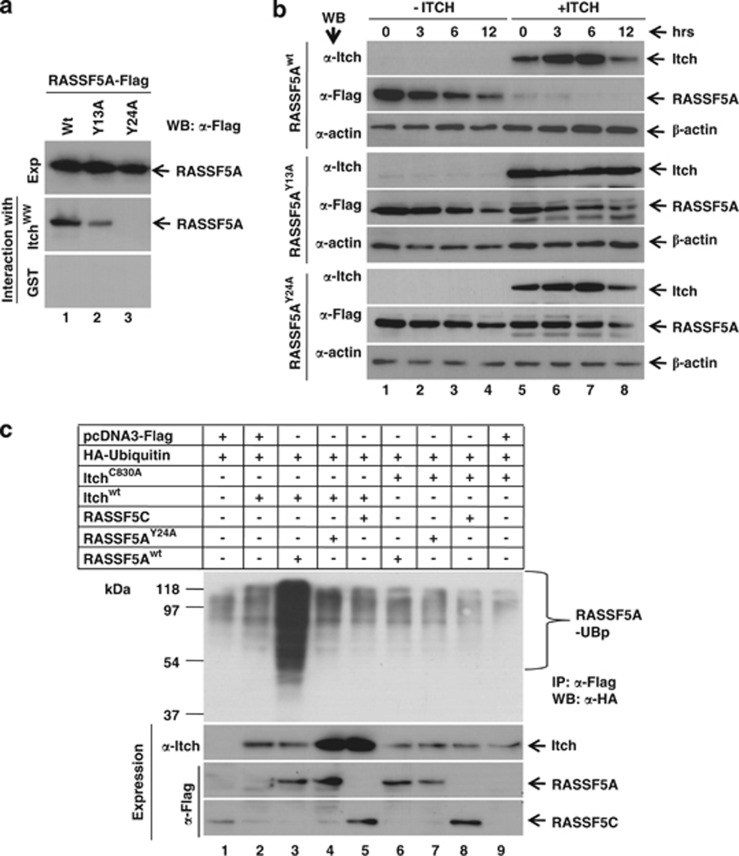
RASSF5A contains 10RPYP14 as well as 21PPRY24 motifs in the amino-terminus (Supplementary Figure S1), and Itch is known to recognize its substrate via PPxY motif (Supplementary Figure S5A). RASSF5AY13A-Flag and RASSF5AY24A-Flag mutants (Supplementary Figure S5B) were generated and GST pull-down assays were performed to determine the contribution of tyrosine residues in RASSF5 interaction with Itch. We observed reduced ability of RASSF5AY13A to interact with Itch compared with RASSF5Awt; however, RASSF5AY24A mutant failed to interact with Itch (Figure 3a). These results suggest that the PPxY motif in RASSF5A is essential for its interaction with Itch. To further understand the role of PPxY motif on Itch-mediated downregulation of RASSF5A, the stability of RASSF5A mutants was determined in the presence of Itch. As expected, while the stability of RASSF5A was susceptible, the steady state level of RASSF5AY24A-Flag was independent of Itch coexpression (Figure 3b). Interestingly, reduced level of RASSF5AY13A-Flag mutant protein degradation was noticed in the presence of Itch coexpression compared with RASSF5Awt (Figure 3b). Collectively, these data provided evidence that PPxY motif mediates the interaction of RASSF5A with Itch and regulate the steady state levels of RASSF5.
Figure 3.
E3 ubiquitin ligase Itch promotes poly-ubiquitination and degradation of RASSF5. (a) HEK293T cell lysates containing wild type or indicated mutants of RASSF5A-Flag (Y13A and Y24A) were incubated with either GST or GST-Itch WW domain and the bound proteins were resolved in SDS-12%PAGE followed by western blotting using anti-Flag antibodies. (b) Interaction of RASSF5A via PPxY motif with Itch promotes RASSF5 degradation. Wild type and the indicated mutants of RASSF5A-Flag were coexpressed with Itch in HEK293T cells and treated with CHX for different time periods. The expression levels of RASSF5A mutants and Itch were determined by western blot analysis using anti-Flag and anti-Itch antibodies respectively. β-Actin was used as internal control. (c) HA-ubiquitin was transiently coexpressed with indicated combination of wild type or mutants of RASSF5 (RASSF5Awt, RASSF5AY24A and RASSF5C) and wild type or catalytic activity deficient Itch (ItchC830A) in HEK293T cells. Expression of all the indicated Itch and RASSF5 variants were determined by western blotting using anti-Itch or anti-Flag antibodies, respectively. Poly-ubiquitinated forms of RASSF5 was detected by immunoprecipitation with anti-Flag antibodies, followed by the detection of ubiquitin using anti-HA antibody
Itch degrades RASSF5 via poly-ubiquitination
We next carried out an in vivo ubiquitination assay to determine whether Itch can catalyze ubiquitination of RASSF5A in cells. Results in Figure 3c indicate that high-molecular weight poly-ubiquitinated forms of RASSF5A were observed only when the Itchwt but not the ItchC830A coexpressed with RASSF5A. In contrast, RASSF5C or RASSF5AY24A are not decorated by poly-ubiquitin chains when both are coexpressed with Itch (Figure 3c). These results suggest that interaction as well as catalytic activity of Itch is essential for poly-ubiquitination of RASSF5A. Interestingly, RASSF5 level was stabilized in presence of MG132 suggests that Itch mediates RASSF5 degradation via 26S proteasome pathway (Figure 1d). Together, these results provide evidence for the first time that Itch promotes downregulation of RASSF5A by catalyzing its poly-ubiquitination followed by 26S proteasome-mediated proteolysis.
Itch failed to degrade endogenous RASSF5 in cancer cell lines
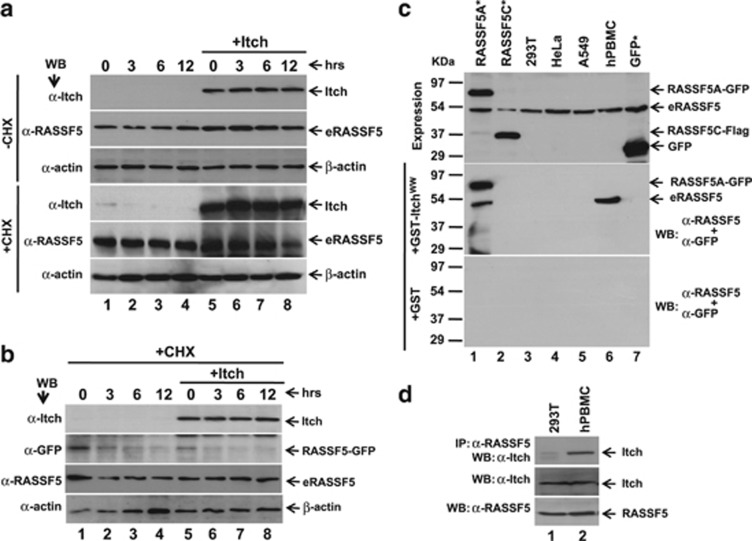
We next determined the steady state levels of endogenous RASSF5 (eRASSF5) in HEK293T cells. Results in Figure 4a indicate that the stability of eRASSF5 was maintained despite Itch overexpression even after the de novo protein synthesis was inhibited by CHX. Conversely, we observed the destabilization of RASSF5 in human primary peripheral blood mononuclear cell (hPBMCs) treated with CHX (Supplementary Figure S6). These data suggest that eRASSF5 in HEK293T cells is resistant to Itch-mediated degradation. To further confirm this, RASSF5–GFP was coexpressed with Itch, and determined the stability of both the ectopically expressed as well as the eRASSF5 in the presence of CHX under the same experimental condition. As shown in Figure 4b, Itch is able to degrade the ectopically expressed but not the eRASSF5. Interestingly, RASSF5–GFP degraded within 3 h after CHX treatment without Itch overexpression (Figure 4b). Taken together, these data suggest the possibility that RASSF5 in cancer cells may undergo some post-translational modification(s) that may inhibit its degradation.
Figure 4.
Itch failed to mediate the degradation of eRASSF5 in transformed cells. (a) Itch expression plasmid was transiently transfected in HEK293T cells and treated with CHX for various time periods. eRASSF5 upon Itch overexpression were determined by western blot analysis using anti-RASSF5 antibody. (b) RASSF5A–GFP was expressed alone or together with Itch in HEK293T cells and treated with CHX. At indicated time intervals, the cell lysates were prepared and checked the levels of ectopically expressed RASSF5–GFP and the eRASSF5 by western blotting using anti-GFP and anti-RASSF5 antibodies, respectively. (c) RASSF5 from human primary cells but not from transformed cells interacts with Itch in vitro. Indicated cell lysates (HEK293T, HeLa, A549 and hPBMC) containing equal amounts of RASSF5 (eRASSF5 as well as GFP-tagged RASSF5A and RASSF5C) were incubated with either GST or GST-Itch WW domain, and the bound proteins were detected by western blot analysis using anti-RASSF5 (eRASSF5) and anti-GFP (ectopically expressed RASSF5–GFP) antibodies. (d) RASSF5 from human primary cells but not from transformed cells interacts with Itch in vivo. HEK293T and hPBMC lysates containing equal amount of eRASSF5 and Itch were subjected to coimmunoprecipitation with anti-RASSF5 antibody followed by western blotting with anti-Itch antibody
To understand further, we checked whether the eRASSF5 in transformed cell lines interacts with Itch. Towards this end, the levels of eRASSF5 were determined in various transformed cell lines as well as in primary cells. In addition, RASSF5A–GFP and RASSF5C–GFP were expressed in HEK293T cells as positive and negative controls for Itch interaction. Results in Figure 4c indicate that equal levels of eRASSF5 expression in both transformed cells and primary hPBMCs. Surprisingly, eRASSF5 from hPBMC but not from transformed cells interact with Itch under the same experimental conditions (Figure 4c). Interestingly, Itch interaction with eRASSF5 was observed when it coexpressed with RASSF5A–GFP, (Figure 4c) suggesting the possibility of homotypic interaction between eRASSF5 and the RASSF5A–GFP. To confirm further, we performed coimmunoprecipitation to determine eRASSF5 interaction with Itch in vivo. RASSF5 and Itch complexes were coimmunoprecipitated with anti-RASSF5 antibody from the lysates of hPBMC and HEK293T cells followed by western blot using anti-Itch antibody. Results in Figure 4d indicate that eRASSF5 from hPBMC but not from HEK293T cells interact with Itch despite equal expression of Itch and RASSF5. Together, these data suggest that RASSF5 in transformed cells may be post-translationally modified or mutation within RASSF5 ORF that may prevent protein–protein interaction. This was intruiging as it may explain the persistence of RASSF5 in tumor where the expression is not silenced by promoter methylation.
Acetylation regulates the steady state level of RASSF5 in transformed cells
Recent reports suggest that acetylation regulates the stabilization as well as destabilization of proteins in a context-dependent manner.24, 27 This prompted us to test the status of RASSF5 acetylation in transformed and primary cells. Results in Supplementary Figure S7A indicate that eRASSF5 from HEK293T cells is more acetylated compared with hPBMCs despite equal amount of RASSF5 expression. Surprisingly, no acetylation was observed for the ectopically expressed RASSF5A–GFP (Supplementary Figure S7A). To confirm the specificity of RASSF5 acetylation, RASSF5 was immuno-depleted with anti-RASSF5 antibodies, and acetylated forms of RASSF5 was determined. Results in Supplementary Figure S7A reveal that acetylated RASSF5 was disappeared from immuno-depleted cell lysates. These results provide evidence that RASSF5 is acetylated both in transformed as well as in primary cells, but level of RASSF5 acetylation is elevated upon transformation.
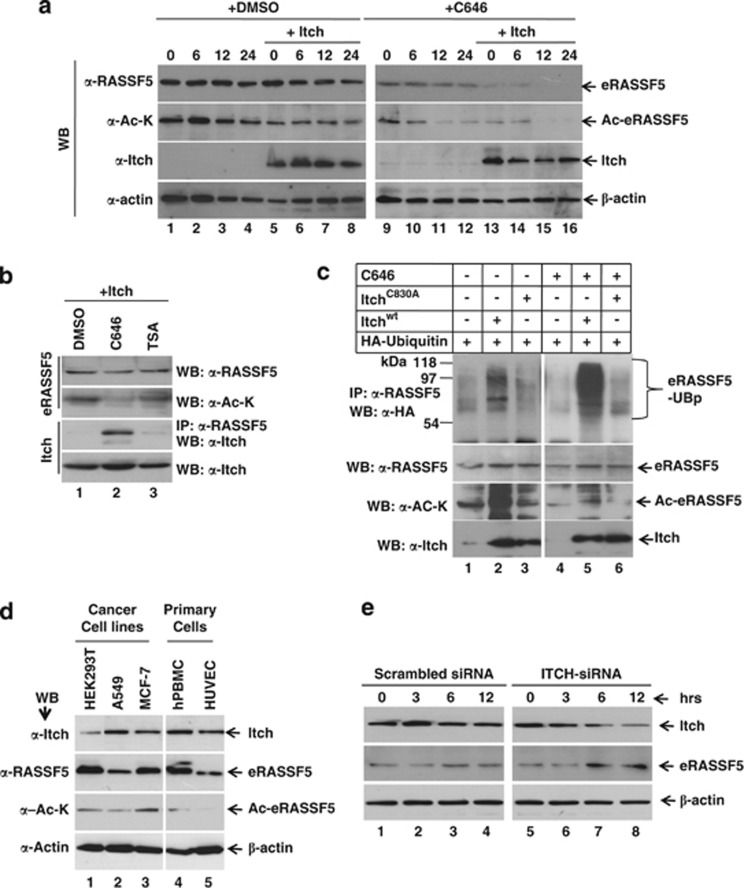
We next determined whether acetylation modulates the stability of RASSF5 in transformed cell lines. As shown in Supplementary Figure S7B, acetylation inhibitors (anacardic acid (AA) and C646) reduced RASSF5 acetylation while, in contrast, increased levels of RASSF5 acetylation was observed in trichostatin A (TSA; inhibitor of deacetylation)-treated cells. As same level of inhibition of RASSF5 acetylation was observed by both AA and C646, we used C646 for further experiments. Results in Figure 5a indicate that inhibition of acetylation by C646 significantly promotes the destabilization of eRASSF5 in a time-dependent manner in contrast to DMSO-treated cells. To confirm whether deacetylated form of RASSF5 can interact with Itch, HEK293T cells were transfected with Itch expression plasmids and treated with C646 or TSA. Interestingly, reduced RASSF5 acetylation promoted interaction with Itch, whereas increased acetylation attenuated its association with Itch (Figure 5b). We next determined whether the observed destabilization of nonacetylated RASSF5 is due to poly-ubiquitination. Results in Figure 5c suggest that Itchwt but not ItchC830A, induces poly-ubiquitination of eRASSF5 in C646-treated cells, suggesting that acetylation negatively modulates RASSF5 interaction with Itch. To understand further, we correlated acetylation status as well as the levels of RASSF5 expression in various human primary as well as cancer cell lines. Higher levels of RASSF5 acetylation were observed in cancer cell lines compared with the indicated primary cells (Figure 5d) despite comparable levels of RASSF5 and Itch expression. To further confirm the specificity of RASSF5 degradation by Itch, human umbilical vein endothelial cells (HUVEC) were transfected with Itch siRNA, and the levels of both Itch and RASSF5 were determined. Results indicate that the RASSF5 level was stabilized when Itch was downregulated by specific siRNA compared with scrambled siRNA (Figure 5e). These results suggest that RASSF5 is degraded by Itch-mediated ubiquitination. Collectively, these data provide evidence that acetylation in transformed cell lines stabilizes RASSF5 by preventing its interaction with Itch.
Figure 5.
Acetylation prevents RASSF5 interaction with Itch in transformed cell lines. (a) Acetylation stabilizes RASSF5 in transformed cell lines. HEK293T cells were transfected with Itch expression plasmids and treated with CHX in the presence or absence of acetylation inhibitor C646. Acetylation status as well as steady state level of RASSF5 were determined by western blot analysis using anti-acetyl-lysine or anti-RASSF5 antibodies, respectively. β-Actin was used as internal control. (b) Acetylation prevents RASSF5 interaction with Itch. Itch was overexpressed in HEK293T cells and treated with inhibitors of acetylation (C646) or deacetylation (TSA). Cell lysates containing equal amounts of eRASSF5 and Itch were subjected to coimmunoprecipitation with anti-RASSF5 antibodies followed by western blot using anti-Itch antibodies. Acetylation status of RASSF5 was determined using anti-acetyl-lysine antibodies. (c) Inhibition of acetylation promotes ubiquitin-mediated degradation of RASSF5. HA-ubiquitin was coexpressed with wild type or ligase activity defective Itch in HEK293T cells and treated with C646. Poly-ubiquitinated eRASSF5 was detected by immunoprecipitation with anti-RASSF5 antibodies, followed by detection of ubiquitin using anti-HA antibody. (d) Expression and acetylation status of RASSF5 in different transformed as well as primary cells. Endogenous level of RASSF5 and Itch expression were determined by western blot analysis using anti-RASSF5 and Itch antibodies, respectively. RASSF5 acetylation was determined using acetylated-lysine antibodies. (e) Downregulation of endogenous Itch stabilize RASSF5. HUVEC cells were transfected with indicated siRNA and cells were collected in different time periods after CHX treatment. The expression levels of RASSF5 and Itch were determined by western blot analysis using anti-RASSF5 and Itch antibodies, respectively
Itch negatively regulates RASSF5 function
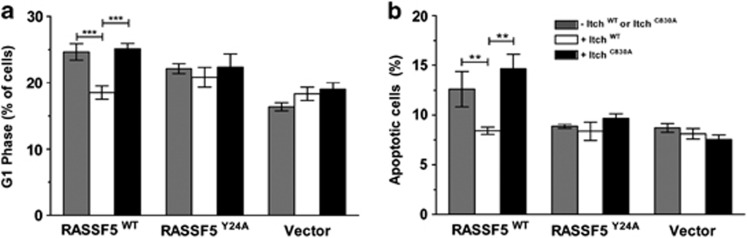
To understand the physiological role of RASSF5 downregulation by Itch during cell cycle progression, RASSF5A was coexpressed with either Itchwt or ItchC830A in HEK293T cells. As shown in Figure 6A, transient overexpression of Itchwt but not ItchC830A significantly suppresses RASSF5A-mediated G1 arrest. However, the profile of S and G2/M phases of the cell cycle was not significantly altered when RASSF5A coexpressed with Itch (Supplementary Figures S8A, B and C). Furthermore, C646-promoted G1 arrest was downregulated by Itch overexpression (Supplementary Figures S9A and B). The patterns of G1 arrest inversely correlated with the acetylation status of RASSF5 (Figure 5a), suggesting that acetylation modulates RASSF5 function during cell cycle. Interestingly, RASSF5-mediated apoptosis was impaired by Itchwt but not ItchC830A (Figure 6b). The specificity of regulation was further confirmed by the fact that Itch failed to alter the G1 cell cycle pattern as well as apoptosis when it is overexpressed with RASSF5AY24A (defective for Itch interaction)(Figures 6a and b). These data indicate that the association of RASSF5 with Itch, or additional PPxY-dependent protein interaction, may be required to elicit apoptosis (Figure 6b). This later fact is a likely reason behind the inconsistency between RASSF5A expression in tumor cells and its role as a tumor suppressor, as high levels may indicate enhance protein acetylation and inactivation of the ability to promote apoptosis of tumor cells.
Figure 6.
Itch negatively regulates RASSF5 function. Wild type not the catalytically defective Itch suppress the RASSF5-mediated G1 phase of the cell cycle arrest (a) and apoptosis (b). HEK293T cells were transiently co-transfected with wild type or indicated mutant of RASSF5 alone or in combination with either Itchwt or ItchC830A, and the cell cycle profile as well as apoptosis were analyzed using Flowcytometry as described in Materials and Methods. All numbers are mean and S.D. of three independent experiments. (**P<0.001) Statistically significant difference
Discussion
Ras effector RASSF5 (NORE1) is found to be downregulated in many tumors, but little is known about the molecular mechanism of its function during cell proliferation. Detailed analysis of RASSF5 amino acid sequences and the data from the in vitro and in vivo protein–protein interaction experiments identified E3 ubiquitin ligase Itch as a unique interacting partner. Results from the present study indicate that Itch promotes the poly-ubiquitination and degradation of RASSF5 by 26S proteasome-mediated pathway. In addition, RASSF5 is acetylated in transformed cells and its interaction with Itch is dependent on the status of acetylation. Furthermore, by destabilizing RASSF5, Itch showed a strong inhibitory effect on RASSF5-mediated G1 phase of cell cycle arrest and apoptosis.
To the best of our knowledge, Itch is the first negative regulator of RASSF5, and the downregulation is dependent upon the status of acetylation in transformed cells. Recent reports demonstrate that Itch downregulates LATS1, an essential component of hippo pathway, have an important role in the regulation of various biological processes such as cancer development, organ size control, differentiation, apoptosis and cell growth.8, 16, 17, 18, 31, 32 On the other hand, RASSF5 interacts with MST1/2, which are essential for the activation of LATS1/2 in the hippo pathway to regulate the fundamental cellular processes.22, 25 Together, these data suggest that an interplay between RASSF5, Itch and hippo pathway may be essential to regulate the cell growth and apoptosis. In this investigation, we demonstrated that RASSF5–Itch interaction is mediated by WW domain of Itch with PPxY motif in RASSF5. Itch directly recognizes many of its substrates such as p73, p63, and LATS1 through PPxY motifs.16, 20, 21 It is possible that Itch may compete with other WW domain containing ubiquitin ligases to regulate RASSF5 half-life. On the other hand, recent study suggests that PPxY motifs interact with the transcriptional activator YAP1 and modulates many cellular processes.15, 19, 33, 34 Together, these results suggest that the PPxY motif not only interacts with ubiquitin ligase but also with transcriptional regulators. It is possible to hypothesize that RASSF5 may interact with the transcriptional regulators in addition to Itch to modulate downstream signaling pathway to control cell proliferation. It is important to further examine how RASSF5 co-ordinate with ubiquitin ligases and other downstream targets to regulate various signaling pathways.
It is interesting to note that, in addition to the repression of RASSF5 expression by promoter hypermethylation,9, 10 yet another unknown mechanism may downregulates RASSF5 function in cancer cells. Our data suggest that acetylation of RASSF5 prevents its interaction with Itch and resistant to Itch-mediated degradation in transformed cells. Recent reports suggest that acetylation increases the stability of transcription factor such as FOXO3 and attenuate the transcriptional activity by impairing its DNA binding ability.24, 27 In addition, acetyl transferases like p300 was found to be overexpressed in tumors and resulted in poor survival rate of patients with prostate cancer, colon carcinoma, breast cancer and non-small cell lung cancers.35, 36 On the other hand, downregulation of deacetylases like SIRT1 activity resulted in tumorigenesis.27 Interestingly, inhibition of eRASSF5 acetylation in HEK293T cells by acetyl transferase inhibitor, C646 promotes its binding with Itch, and resulted in RASSF5 degradation. As lysine residues in proteins undergo both acetylation and ubiquitination,27 the data from the present study speculate the hypothesis that acetylated form of RASSF5 may not be recognized by ubiquitin ligase Itch, which leads to the stabilization of RASSF5 in tumors. Furthermore, the acetylated form of RASSF5 may be a non-functional protein in tumor cells, suggesting the possibility that acetylation may have an important role in negative regulation of RASSF5 function during tumorigenesis. This is in agreement with the recent reports that demonstrate that acetylation attenuates the transcriptional activity of FOXO1 and FOXO3.24, 27 Together, these observations lead to the hypothesis that acetylation may act as both positive and negative regulator of protein function in a context-dependent manner. Therefore, further experiments are needed to examine whether RASSF5 is a transcriptional regulator, and how acetylation regulates its function during tumorigenesis.
In conclusion, the present investigation provides evidence that E3 ubiquitin ligase Itch negatively regulates the Ras effector RASSF5 in acetylation-dependent manner. Further characterization of their functional interaction as well as the role of acetylation on RASSF5 function in clinical cancer patients will provide new insights in diagnostics and treatment of human cancers.
Materials and Methods
Plasmids
Generation of wild type and mutants (Y13A and Y24A) of RASSF5 expression plasmids has been described previously.3, 11 The plasmid encoding wild type and variants of Itch WW domains were generated by Quick-change site-directed mutagenesis according to the manufacturer's instruction (Stratagene, La Jolla, CA, USA), and were expressed as GST fusion proteins. Itch (wild type as well as ligase activity defective) expression plasmid was kindly provided by Dr. Pweson37 and Dr. Melino.21 The primers used for PCR amplification to create RASSF5 and Itch fusion constructs were listed in Supplementary Table 1. All mutant constructs were sequenced to verify the integrity of each clone.
Chemicals
Cycloheximide (Sigma-Aldrich, St Louis, MO, USA) was dissolved in water and used at 25 μg/ml. MG132 (Calbiochem, Billerica, MA, USA) was dissolved in DMSO and used at a final concentration of 20 μM. Anacardic (Calbiochem, USA) acid and C646 (Calbiochem, USA) was dissolved in DMSO and used at a final concentration of 30 μM. Trichostatin-A (Calbiochem, USA) was used at a final concentration of 6 μM.
Cell culture, transfection, coimmunoprecipitation and western blotting
A549, HeLa and HEK293T cells were maintained in Dulbecco's modified Eagle's Medium as described elsewhere.3, 11 HUVEC were isolated from umbilical cords by digestion with collagenase as described elsewhere.38 HEK293T cells were transfected with RASSF5 or Itch expression plasmids using Lipofectin (Invitrogen Life Technologies, Grand Island, NY, USA) as described elsewhere.3, 11 To check the expression of RASSF5 or Itch, transfected cells were lysed in 1X cell lysis buffer (25 mM Tris-HCl, pH7.4, 150 mM KCl, 1 mM Na2EDTA, 1 mM EGTA, 1% TritonX100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 0.4 mM PMSF, 1 mM Na-fluoride, 1 mM Na-orthovanadate and 1 μg/ml each of aprotinin, leupeptin and pepstatin) after 32-h post-transfection, and separated on a SDS-12%PAGE. For coimmunoprecipitation, equal amounts of cell lysates were incubated with anti-RASSF5 (Abcam, Cambridge, UK) or anti-Itch antibodies (BD Transduction Laboratories, San Jose, CA, USA), and the bound protein complexes were eluted and resolved in SDS-12%PAGE. Separated proteins were transferred to a Hybond-P membrane (GE HealthCare, Stockholm, Sweden) and probed with appropriate antibodies (anti-RASSF5 or anti-Itch or anti-GFP antibodies). Anti-rabbit acetylated-lysine antibody (Cell Signaling, Danvers, USA) was used to determine the acetylation status of RASSF5. Protein bound antibodies were probed with horseradish peroxidase conjugated specific secondary antibodies and developed using the enhanced chemiluminescence-plus detection system (GE HealthCare).
Fluorescence microscopy
HeLa cells grown on chamber culture slides (BD Biosciences, Franklin Lake, NY, USA) were transfected with expression plasmids containing RASSF5–GFP or Itch using Lipofectin as described elsewhere.3, 11 After 32 h of transfection, cells were fixed with 3% paraformaldehyde for 20 min and incubated with anti-Itch antibodies. Alexa Flour 594 were used as secondary antibodies and then mounted in mounting medium (Vector Laboratories, Burlingame, CA, USA) containing 4,6-diamidino-2-phenylindole to stain nuclei. RASSF5–GFP signals were visualized directly. Samples were viewed with LSM710 laser scanning confocal microscopy (Carl Zeiss, Jena, Germany). Image acquisition was carried out using Zen 2009 Image software (Carl Zeiss) and Adobe Photoshop CS3 was used for image processing.
Expression and purification of GST-Itch WW domain fusion proteins
Wild type and different variants of Itch WW domain containing expression vectors were transformed into Escherichia coli BL21DE-3, single colony was grown at 37 °C in kanamycin-containing medium, and the protein expression was induced for 4 h at 37 °C with 1 mM IPTG. Cells were harvested and cell lysates were mixed with Glutathione–Sepharose beads (GE Healthcare). Bound proteins were eluted in elution buffer (10 mM reduced glutathione in 50 mM Tris-Cl, pH7.4) and the integrity of fusion proteins were analyzed by SDS-12%PAGE followed by staining with Coomassie blue.
Half-life determination
HEK293T cells were transfected with RASSF5A, RASSF5C and Itch expression plasmids. Twenty-four hours after transfection cell were treated with 25 μg/ml of CHX for different time periods. Cells were collected at different time periods and equal amount of total cellular protein was loaded on SDS-12%PAGE followed by western blot analysis.
GST pull-down assay
HEK293T cells were transfected with RASSF5A-Flag (wild type or mutants) expression constructs as described above. Cell lysates containing equal amounts of RASSF5 were incubated with glutathione–sepharose beads prebound with Itch WW domain (wild type or mutants). After 4 h, glutathione–sepharose beads containing proteins were washed and the eluted proteins were separated by SDS-12% PAGE followed by western blot analysis using anti-Flag or anti-RASSF5 antibodies.
In vivo ubiquitination assay
HEK293T cells were transiently transfected with plasmid expressing HA-ubiquitin and RASS5A-Flag or RASSF5C-Flag alone or together with Itch (wild type or ligase defective mutant, ItchC830A). After 24 h of transfection, cells were treated with 20 μM MG132 for 12 h before being harvested. Cell extracts were subjected to immunoprecipitation using anti-Flag antibodies (Sigma) and ubiquitin conjugates were detected by western blot analysis using anti-HA antibodies (Santa Cruz, Santacruz, CA, USA).
RNA interference
HUVEC cells were transiently transfected with control siRNA or Itch siRNA by using Oligofectamine (Invitrogen Life Technologies) according to the manufacturer's instructions. The sequence of Itch specific siRNAs (VBC Biotech, Vienna, Austria) are as follows: Itch-1: 5′-GGUGACAAAGAGCCAACAGAGdTdT-3′ and scrambled siRNA 5′-ACAGACUUCGGAGUACCUGdTdT-3′. After 36 h of post-transfection, cells were collected and the levels of eRASSF5 and Itch, and the acetylation status of eRASSF5 were determined by western blot analysis using respective antibodies as described above.
Cell cycle and apoptosis assay
HEK293T cells were transfected with RASSF5 alone or in combination with Itch (wild type or ligase activity defective mutant) using lipofectamine. For cell cycle analysis, cells were treated with hypertonic buffer (0.1% sodium citrate, 0.1% NP40, 45 μg/ml propidium iodide, 50 μg/ml ribonuclease A pH 7.4) for 10 min at room temperature after 36 h of post-transfection. For apoptosis assay, cells were collected after 36 h of post-transfection and incubated with buffer containing 5 μl FITC-Annexin V, 5 μl Propidium Iodide (BD Pharmingen, Sandiego, CA, USA), 0.1 M Hepes/NaOH (pH 7.4), 1.4 M NaCl, 25 mM CaCl2 for 15 min at room temperature. All the samples were analyzed by flow cytometry (FACSCanto II, BD Biosciences) and the data were processed using FACS Diva software.
Acknowledgments
We thank Dr. T Pawson, Samuel Lunenfeld Research Institute, Canada and Dr. G Melino, University of Leicester, UK for providing wild-type and C830A Itch expression plasmids. We thank Dr. Eric O'Neill, University of Oxford, UK for critical reading of the manuscript. This work was supported by grants from the Department of Science and Technology (VI-D&P/242/07-08/TDT), Govt. of India, the Department of Atomic Energy (2008/21/08-BRNS/2790), Govt. of India to SM.
Glossary
- RASSF
Ras association (RalGDS/AF-6) domain family member
- MEF
mouse embryo fibroblasts
- MST
mammalian sterile 20-like kinase
- YAP1
yes associated protein 1
- LATS1
large associated tumor suppressor 1
- SH2/SH3
Src homology domain 2 and 3
- GST
glutathione S-transferase
- GFP
green fluorescent protein
- PBMC
peripheral blood mononuclear cell
- HEK293T
human embryonic kidney 293T cells
- HUVEC
human umbilical vein endothelial cells
- CHX
cycloheximide
- AA
anacardic acid
- TSA
trichostatin A
- DAPI
4, 6-diamidino-2-phenylindole
- eRASSF5
endogenous RASSF5
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Rufini
Supplementary Material
References
- Bee C, Moshnikova A, Mellor CD, Molloy JE, Koryakina Y, Stieglitz B, et al. Growth and tumor suppressor NORE1A is a regulatory node between Ras signaling and microtubule nucleation. J Biol Chem. 2010;285:16258–16266. doi: 10.1074/jbc.M109.081562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvisi DF, Donninger H, Vos MD, Birrer MJ, Gordon L, Leaner V, et al. NORE1A tumor suppressor candidate modulates p21CIP1 via p53. Cancer Res. 2009;69:4629–4637. doi: 10.1158/0008-5472.CAN-08-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari G, Singhal PK, Suryaraja R, Mahalingam S. Functional interaction of the Ras effector RASSF5 with the tyrosine kinase Lck: critical role in nucleocytoplasmic transport and cell cycle regulation. J Mol Biol. 2010;397:89–109. doi: 10.1016/j.jmb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Park J, Kang SI, Lee SY, Zhang XF, Kim MS, Beers LF, et al. Tumor suppressor ras association domain family 5 (RASSF5/NORE1) mediates death receptor ligand-induced apoptosis. J Biol Chem. 2010;285:35029–35038. doi: 10.1074/jbc.M110.165506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavvas D, Li X, Avruch J, Zhang XF. Identification of Nore1 as a potential Ras effector. J Biol Chem. 1998;273:5439–5442. doi: 10.1074/jbc.273.10.5439. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-hippo pathway—An emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Hwang E, Ryu KS, Pääkkönen K, Güntert P, Cheong HK, Lim DS, et al. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc Natl Acad Sci USA. 2007;104:9236–9241. doi: 10.1073/pnas.0610716104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Djos A, Martinsson T, Kogner P, Caren H. The RASSF gene family members RASSF5, RASSF6 and RASSF7 show frequent DNA methylation in neuroblastoma. Mol Cancer. 2012;11:40. doi: 10.1186/1476-4598-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli J, Kogner P, Lanner F, Natalishvili N, Juhlin C, Kiss N, et al. Assessment of NORE1A as a putative tumor suppressor in human neuroblastoma. Int J Cancer. 2008;123:389–394. doi: 10.1002/ijc.23533. [DOI] [PubMed] [Google Scholar]
- Kumari G, Singhal PK, Rao MR, Mahalingam S. Nuclear transport of Ras-associated tumor suppressor proteins: different transport receptor binding specificities for arginine-rich nuclear targeting signals. J Mol Biol. 2007;367:294–1311. doi: 10.1016/j.jmb.2007.01.026. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Chan DC, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilsley JL, Sudol M, Winder SJ. The WW domain: linking cell signalling to the membrane cytoskeleton. Cell Signal. 2002;14:183–189. doi: 10.1016/s0898-6568(01)00236-4. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- Ho KC, Zhou Z, She YM, Chun A, Cyr TD, Yang X. Itch E3 ubiquitin ligase regulates large tumor suppressor 1 stability. Proc Natl Acad Sci USA. 2011;108:4870–4875. doi: 10.1073/pnas.1101273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah Z, Melino G, Aqeilan RI. Negative regulation of the hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 2011;71:2010–2020. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- Sudol M. Newcomers to the WW domain–mediated network of the hippo tumor suppressor pathway. Genes Cancer. 2011;1:1115–1118. doi: 10.1177/1947601911401911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Reuven N, Shaul Y. A regulatory circuit controlling Itch-mediated p73 degradation by Runx. J Biol Chem. 2008;283:27462–27468. doi: 10.1074/jbc.M803941200. [DOI] [PubMed] [Google Scholar]
- Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama Y, Avruch J, Zhang XF. Nore1 inhibits tumor cell growth independent of Ras or the MST1/2 kinases. Oncogene. 2004;23:3426–3433. doi: 10.1038/sj.onc.1207486. [DOI] [PubMed] [Google Scholar]
- Gao M, Labuda T, Xia Y, Gallagher E, Fang D, Liu YC, et al. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science. 2004;306:271–275. doi: 10.1126/science.1099414. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci USA. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- Spangea S, Wagner T, Heinzel T, Krämer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Wang F, Chan CH, Chen K, Guan X, Lin HK, Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2012;31:1546–1557. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- Yang C, Zhou W, Jeon MS, Demydenko D, Harada Y, Zhou H, et al. Negative regulation of the E3 ubiquitin ligase itch via Fyn-mediated tyrosine phosphorylation. Mol Cell. 2006;21:135–141. doi: 10.1016/j.molcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Dokmanovic M, Clarke C, Marks PA. Histone deacetylase inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- Visser S, Yang X. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle. 2010;9:3892–3903. doi: 10.4161/cc.9.19.13386. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Salah Z, Aqeilan RI. WW domain interactions regulate the hippo tumor suppressor pathway. Cell Death Dis. 2011;2:e172. doi: 10.1038/cddis.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Li Y, Luo RZ, Fu JH, He JH, Zhang LJ, et al. High expression of the transcriptional co-activator p300 predicts poor survival in resectable non-small cell lung cancers. Eur J Surg Oncol. 2012;38:523–530. doi: 10.1016/j.ejso.2012.02.180. [DOI] [PubMed] [Google Scholar]
- Liao ZW, Zhou TC, Tan XJ, Song XL, Liu Y, Shi XY, et al. High expression of p300 is linked to aggressive features and poor prognosis of nasopharyngeal carcinoma. J Transl Med. 2012;10:110. doi: 10.1186/1479-5876-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg G, Matskova L, Chen F, Plant P, Rotin D, Gish G, et al. Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol Cell Biol. 2000;20:8526–8535. doi: 10.1128/mcb.20.22.8526-8535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri H, Muthuramu I, Dhar M, Rathnakumar K, Ram U, Dixit M. Protein tyrosine phosphatase SHP2 mediates chronic insulin-induced endothelial inflammation. Arterioscler Thromb Vasc Biol. 2012;32:1943–1950. doi: 10.1161/ATVBAHA.111.239251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.