Abstract
N-methyl-𝒟-aspartate receptors (NMDAR) overactivation is linked to neurodegeneration. The current prevailing theory suggests that synaptic and extrasynaptic NMDAR (syn- and ex-NMDAR) impose counteracting effects on cell fate, and neuronal cell death is mainly mediated by the activation of ex-NMDAR. However, several lines of evidence implicate the limitation of this theory. Here, we demonstrate that activation of NMDAR bi-directionally regulated cell fate through stimulating pro-survival or pro-death signaling. While low-dose NMDA preferentially activated syn-NMDAR and stimulated the extracellular signal-regulated kinase ½–cAMP responsive element-binding protein–brain-derived neurotrophic factor pro-survival signaling, higher doses progressively activated increasing amount of ex-NMDAR along with syn-NMDAR and triggered cell death program. Interestingly, the activation of syn- or ex-NMDAR alone did not cause measurable cell death. Consistently, activation of syn- or ex-NMDAR alone stimulated pro-survival but not pro-death signaling. Next, we found that memantine, which was previously identified as an ex-NMDAR blocker, inhibited intracellular signaling mediated by syn- or ex-NMDAR. Simultaneous blockade of syn- and ex-NMDAR by memantine dose-dependently attenuated NMDAR-mediated death. Moreover, long- but not short-term treatment with high-dose NMDA or oxygen–glucose deprivation triggered cell death and suppressed pro-survival signaling. These data implicate that activation of syn- or ex-NMDAR alone is not neurotoxic. The degree of excitotoxicity depends on the magnitude and duration of syn- and ex-NMDAR coactivation. Finally, genome-wide examination demonstrated that the activation of syn- and ex-NMDAR lead to significant overlapping rather than counteracting transcriptional responses.
Keywords: intracellular signaling, microarray, NMDA receptor.
N-methyl-𝒟-aspartate receptors (NMDARs) mediate excitatory neural transmission. In addition to their physiological functions, mounting evidence has suggested their involvement in excitotoxicity. Cell death triggered by seizure or ischemic stroke is attributed to NMDAR overactivation.1, 2 Abnormal NMDAR activity is also associated with neurodegenerative disorders, such as Alzheimer's, Huntington's, and Parkinson's disease.3 Thus, it is concluded that, while appropriate NMDAR activation is essential for neuronal survival and physiological functions, excessive activation contributes to pathological changes including cell death.4
Because NMDARs are found at both synaptic and extrasynaptic locations,5, 6 it is attempting to determine whether the two NMDAR populations have differential roles in neuronal survival and death under physiological and pathological conditions.7 An influential study showed that activation of synaptic NMDAR (syn-NMDAR) with bicuculline stimulated pro-survival signaling, such cAMP responsive element-binding protein (CREB) activation, in cultured neurons.8 A subsequent bath application of NMDAR agonist (i.e., glutamate) shut off CREB signaling and triggered neuronal death. It was concluded that, because bath incubation with glutamate activates global NMDAR (including both synaptic and extrasynaptic populations), the participation of extrasynaptic NMDAR (ex-NMDAR) converted survival to death. Increased ex-NMDAR activity is also associated with neurodegeneration such as Huntington's disease.9
Although the current prevailing theory postulates that the activation of ex-NMDAR counteracts syn-NMDAR and triggers cell death,7 some studies have suggested the limitation of this theory. First, activation of ex-NMDAR is not only involved in pathological process but also in normal brain functions.10, 11 Second, younger and developing neurons mainly express ex-NMDAR12 and are resistant to NMDA- or glutamate-induced cell death.13, 14 Moreover, the NMDA and glutamate excitotoxicity is absent in retinal ganglion cells,15 which lack syn-NMDAR, and the NMDAR current is mediated by receptors at the extrasynaptic sites.16, 17 Here, we made effort to isolate syn- and ex-NMDAR, and determined their functions in cell fate. Surprisingly, we found that the activation of syn- or ex-NMDAR alone did not cause NMDAR-dependent cell death, but rather activated pro-survival signaling. We further provide new evidence to show that the degree of NMDAR-dependent excitotoxicity is related to the magnitude and duration of syn- and ex-NMDAR coactivation. Moreover, data from microarray analysis showed that activation of syn- and ex-NMDAR had overlapping but not opposing effects on genomic responses.
Results
NMDAR bi-directionally regulates pro-survival signaling and intracellular Ca2+ homeostasis
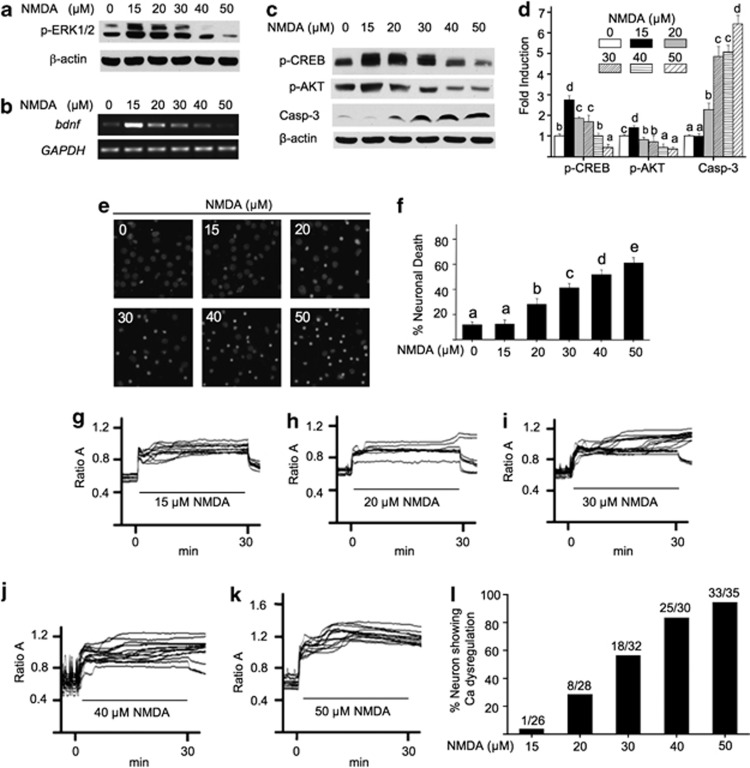
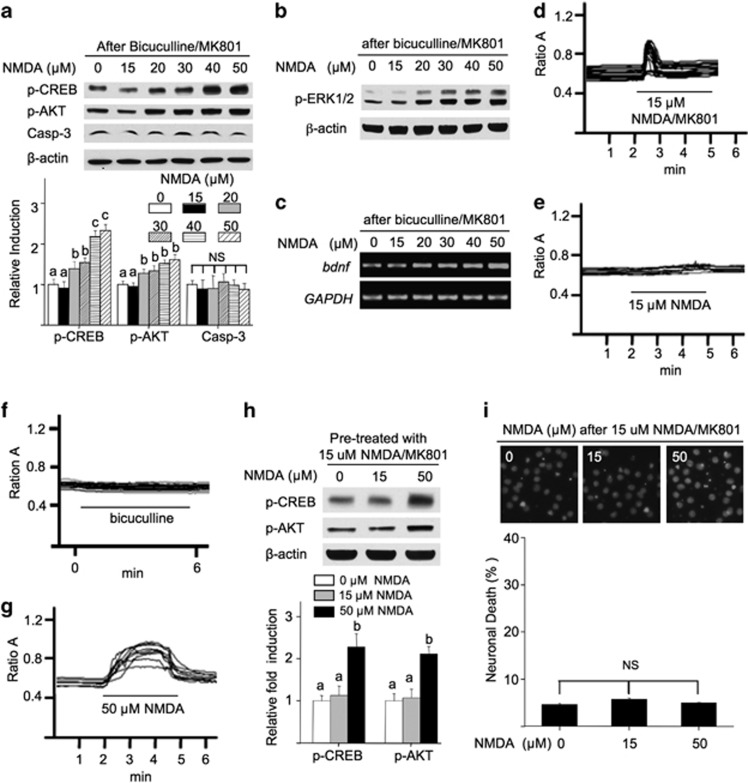
Previous studies show that the activation of NMDAR bi-directionally regulates intracellular responses related to neuronal survival.18 Here, we stimulated neurons with various concentrations of NMDA for 30 min, and examined intracellular responses related to neuronal fate. Consistent with previous studies,18, 19 we found that lower concentration of NMDA (at 15 and 20 μM) upregulated extracellular signal-regulated kinase ½ (ERK1/2) phosphorylation, and higher NMDA deactivated ERK1/2 (Figure 1a). NMDA also bi-directionally regulated the transcription of a pro-survival gene brain-derived neurotrophic factor (Bdnf) (Figure 1b). Moreover, low-dose NMDA activated the pro-survival molecules CREB and AKT; high doses shut them off (Figures 1c and d). In correlation, NMDA triggered the activation of a pro-death molecule caspase-3 (Figures 1c and d), and neuronal death (Figures 1e and f) in a dose-dependent manner. Increasing NMDA concentrations also progressively triggered more lactate dehydrogenase release from damaged neurons (Supplementary Figure 1).
Figure 1.
NMDA bi-directionally regulates intracellular signaling, cell death, and calcium homeostasis. DIV 14 cortical neurons were stimulated with NMDA (at 15, 20, 30, 40, and 50 μM, as indicated). Samples were collected 30 min (a and c) or 60 min (b) after stimulation, and examined for the phosphorylation of ERK1/2 (a), transcription of Bdnf (b), and the phosphorylation of CREB, AKT, and the activated form of caspase-3 (c). The level of p-CREB, p-AKT, and activated cappase-3 was quantified and expressed as average±S.E.M. in (d). For e and f, DIV 14 neurons were subjected to a 30-min treatment with NMDA at different concentrations (in μM as indicated). Immediately after the treatment, neurons were washed with prewarmed media, and maintained in conditioned media plus MK801. Neurons were then fixed 20 h later, and stained with DAPI (e). (f) percentage of cell death induced by NMDA at different concentrations was quantified and expressed as average±S.E.M. Condensed nuclear staining by DAPI was used to determine cell death. For d and f, ANOVA analysis revealed significant effects of NMDA concentration. Different letters indicate distinct SNK groups with a<b<c<d<e for different NMDA treatment. The level of Ca2+i in live neurons was determined by calcium imaging. DIV 14 neurons were stimulated with 15 (g), 20 (h), 30 (i), 40 (j), and 50 μM (k) NMDA as indicated. The representative traces from individual neurons are presented. The percentage of neurons, in which Ca2+i did not return to the baseline level after NMDA withdrawal, was quantified and presented in l. The duration of NMDA stimulation (i.e., 30 min) is indicated by the lines in g–k
The Ca2+ overload and loss of Ca2+ homeostasis triggered by NMDAR overactivation is highly related to neuronal death.20 By measuring intracellular Ca2+ (Ca2+i) in live neurons, we found that NMDA caused elevation of Ca2+i in a dose-dependent manner (Figures 1g–k). When NMDA was withdrawn after the 30-min stimulation, Ca2+i remained elevated and did not return to the baseline level in more neurons treated with higher concentrations of NMDA (Figure 1l). The loss of Ca2+ homeostasis correlated with cell death. The percentage of cell death increased in neurons treated with higher concentration of NMDA (Figures 1e and f). Neurons treated with 15 μM NMDA did not show significant Ca2+ dysregulation (Figure 1g) and cell death (Figures 1e and f). These data demonstrate that increasing amount of NMDA activated more receptors, which contribute to a program switch from survival to cell death.
Activation of syn-NMDAR alone triggers pro-survival signaling but not death
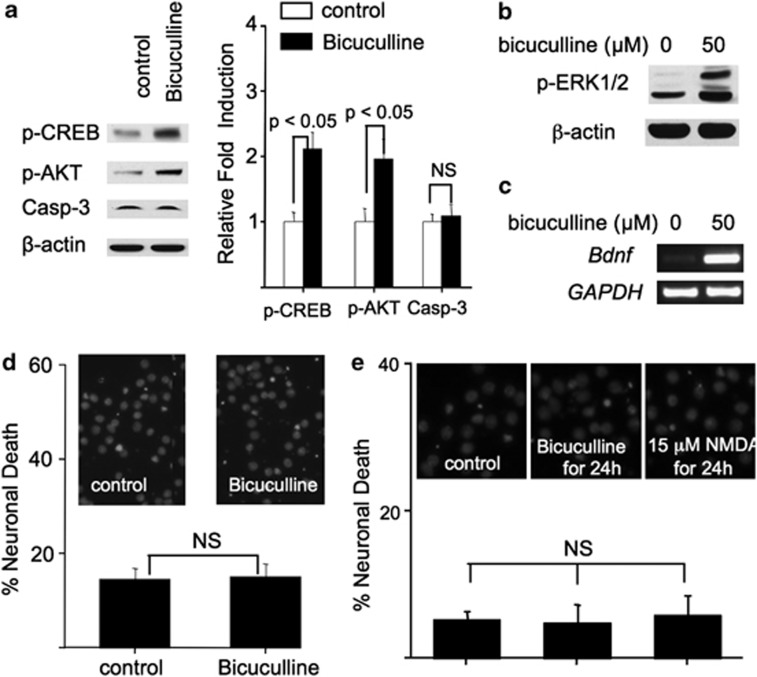
Why does bath incubation with NMDA at different concentrations lead to different neuronal responses? One possibility is that there is a threshold of NMDAR activation, surpassing which will trigger the pro-death program.21 The other possibility is that low-dose NMDA mainly activates syn-NMDAR; high-dose NMDA activates more receptors including ex-NMDAR, which was suggested as the major cause for cell death by previous studies.7 Here, we demonstrate that bicuculline and 15 μM NMDA had similar effects. It is known that bicuculline, a GABAA receptor antagonist, activates syn-NMDAR by blocking synaptic inhibition and thereby leading to glutamate release from presynaptic terminals.22 First, bicuculline treatment activated CREB and AKT but not caspase-3 (Figure 2a). Bicuculline also upregulated ERK1/2 phosphorylation (Figure 2b) and Bdnf transcription (Figure 2c). Second, a 30-min treatment with bicuculline did not cause any detectable cell death (Figure 2d). A long-term 24-h treatment with bicuculline or 15 μM NMDA also failed to induce death (Figure 2e).
Figure 2.
Activation of syn-NMDARs stimulates pro-survival signaling. DIV 14 cortical neurons were treated with bicuculline for 30 min (a and b) or 60 min (c). The levels of p-CREB, p-AKT, and activated caspase-3 were determined by western blot, and shown in a. Quantification (average±S.E.M.) from three independent experiments is shown in the right panel of a. The level of p-ERK1/2 (b) and Bdnf mRNA (c) was determined by western blot and RT-PCR, respectively. (d and e) There was no significant cell death in neurons treated with bicuculline for 30 min (d), or bicuculline or 15 μM NMDA for 24 h (e). NS, not significant
Activation of ex-NMDAR alone does not cause neuronal death but instead activates pro-survival signaling
To determine the function of ex-NMDAR, we used a well-accepted method to block syn-NMDAR by treating neurons with bicuculline and MK801. Because bicuculline only activates syn-NMDAR and MK801 irreversibly blocks the opened channels, cotreatment with bicuculline plus MK801 would block syn-NMDAR. Thus, any subsequent NMDAR-mediated responses would be attributed to ex-NMDAR. Bicuculline and MK801 caused a transient syn-NMDAR-mediated Ca2+ influx (Supplementary Figure 2A). After removing bicuculline/MK801, subsequent application of bicuculline no longer induced any measurable Ca2+ influx (Supplementary Figure 2B). Neurons cotreated with bicuculline/MK801 did not show CREB activation (Supplementary Figure 2C). Following bicuculline/MK801 pretreatment, subsequent bicuculline did not upregulate CREB phosphorylation (p-CREB) (Supplementary Figure 2D). These results demonstrate that cotreatment with bicuculline/MK801 sufficiently blocked syn-NMDAR.
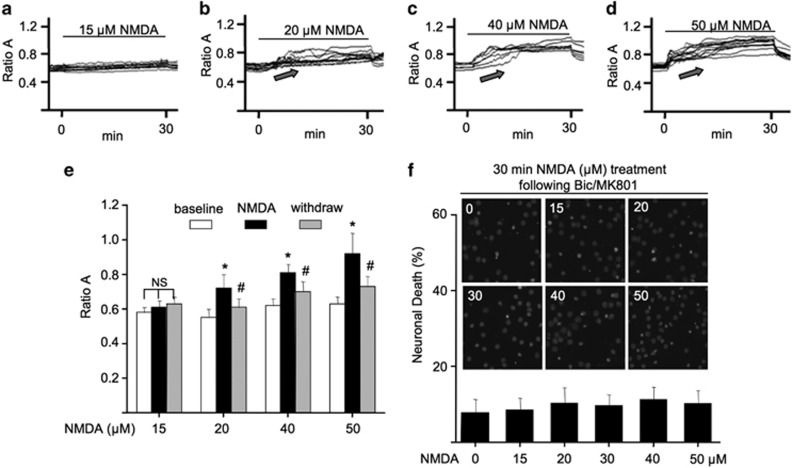
Following bicuculline/MK801 and a subsequent wash, we examined the changes of Ca2+i by applying increasing concentrations of NMDA. We found that NMDA from 15 to 50 μM progressively induced increasing degrees of Ca2+i elevation (Figures 3a–e). This indicates that higher doses of NMDA activated more ex-NMDAR.
Figure 3.
Activation of ex-NMDAR does not cause Ca2+i dysregulation and neuronal cell death. (a–e), DIV 14 cortical neurons were first treated with bicuculline and MK801 for 2 min, and then washed extensively with buffered solution. Calcium-imaging traces show the elevation of Ca2+i by the subsequent application of NMDA at 15 (a), 20 (b), 40 (c), and 50 μM (d) as indicated. The quantification of relative changes (i.e., Ca2+i level at baseline, at the end of NMDA application, and after NMDA withdrawal) is shown in (e). NS, not significant. ‘*' denotes P<0.05 between baseline and NMDA treatment; ‘#' denotes P<0.05 between NMDA and withdrawal. (f) DIV 14 neurons were first pretreated with bicuculline and MK801 for 2 min followed by extensive wash with conditioned medium. Subsequently, neurons were incubated with NMDA at different concentrations (as indicated) for 30 min. Neurons were fixed 20 h after the treatment, and stained with DAPI. Cell death was determined by the appearance of condensed nuclear staining.
We noticed that, after blocking syn-NMDAR, NMDA at 20 to 50 μM no longer caused Ca2+ dysregulation. After NMDA withdrawal, the elevated Ca2+i decreased toward the baseline level (Figures 3b–e). These data suggest that activation of ex-NMDAR alone does not disrupt Ca2+ homeostasis. Consistently, no significant cell death was observed with neurons stimulated with higher NMDA following bicuculline/MK801 pretreatment (Figure 3f). Further, with syn-NMDAR blocked, NMDA or glutamate at as high as 1 mM failed to cause measurable death (Supplementary Figure 3A). Moreover, after blocking syn-NMDARs, 100 μM NMDA or 100 μM glutamate incubation for 24 h failed to induce death (Supplementary Figure 3B). Because previous observations suggest that bicuculline pretreatment has a protective effect on NMDA-induced cell death,23 we wondered whether the presence of bicuculline during the 2-min pretreatment with bicuculline/MK801 has a similar effect. However, we did not observe any protective effect from the 2-min bicuculline pretreatment (Supplementary Figure 4). Together, we show that activation of syn- or ex-NMDAR alone was not sufficient to cause measurable cell death and Ca2+ dysregulation. Activation of syn-NMDAR along with increasing involvement of extrasynaptic receptor, as in the cases when neurons were treated with increasing concentrations of NMDA (Figures 1e and f), was neurotoxic.
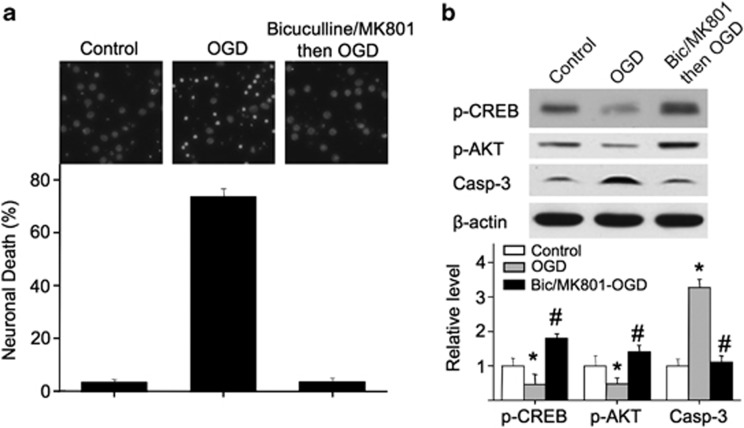
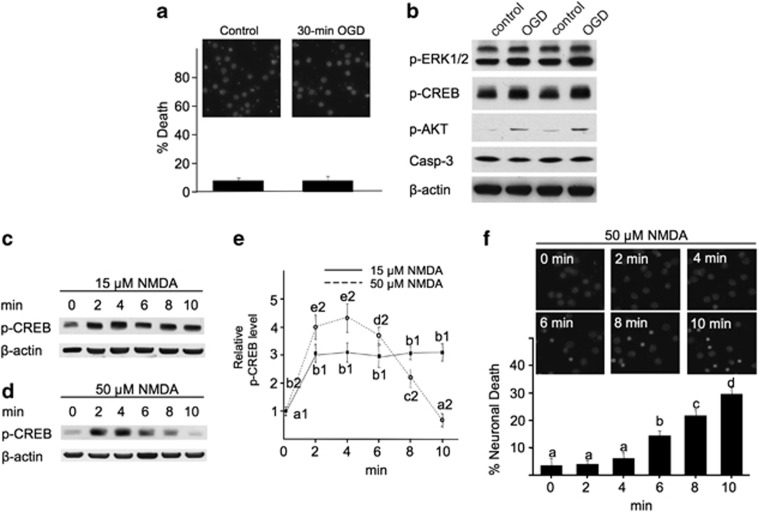
To further determine the function of ex-NMDARs under pathological conditions, we used oxygen–glucose deprivation (OGD) as a cellular ischemic stroke model.24 A 75-min OGD induced 73.6% cell death (Figure 4a). After blocking syn-NMDARs, OGD did not cause significant death (Figure 4a). Consistently, OGD caused a reduction in p-CREB and p-AKT along with an elevation of caspase-3 activity (Figure 4b). After blocking syn-NMDAR, OGD upregulated p-CREB and p-AKT without elevating caspase-3 (Figure 4b). We are aware of the possible NMDAR-independent mechanisms in OGD. Under our experimental setup, 2 h of OGD caused severe cell death with or without bicuculline/MK801 pretreatment (Supplementary Figure 5). When the duration of the OGD reduced to 75 min, there is a clear functional involvement of NMDAR. Our data show that this ‘NMDAR-involved phase' is not sensitive to OGD when synaptic receptors are blocked.
Figure 4.
Blocking synaptic receptors results in resistance to OGD. DIV 14 neurons were first treated with bicuculline plus MK801 for 2 min, washed extensively, and subjected to a 75-min OGD. (a) Cell death was determined 20 h later by DAPI staining. (b) Neuronal extracts were harvested immediately after OGD, and analyzed for p-CREB, p-AKT, and activated caspase-3 by western blot. ‘*' denotes P<0.05 when compared with the control; ‘#' denotes P<0.05 when compared with the OGD group
To seek direct evidence on why extrasynaptic activation failed to induce death, we stimulated neurons with different concentrations of NMDA after blocking the synaptic receptors. In neurons pretreated with bicuculline/MK801, NMDA at 15 μM did not cause significant phosphorylation of CREB, AKT (Figure 5a), and ERK1/2 (Figure 5b), as well as Bdnf transcription (Figure 5c). This is consistent with the result in Figure 3a, which shows that 15 μM NMDA failed to significantly activate ex-NMDAR and trigger Ca2+ influx. Activation of ex-NMDAR by higher concentrations of NMDA (at 20, 30, 40 or 50 μM) stimulated these survival-related signaling pathways (Figures 5a–c), without activating caspase-3 (Figure 5a) and lactate dehydrogenase release (Supplementary Figure 6). These results provide molecular mechanisms supporting that activation of ex-NMDAR alone is not sufficient to cause death.
Figure 5.
Activation of extrasynaptic receptor stimulates pro-survival signaling. DIV 14 neurons were first pretreated with bicuculline and MK801 for 2 min (a, b, and c) or 15 μM NMDA and MK801 for 3 min (d–i), washed, and treated with different doses of NMDA as indicated for 30 min (a, b, h, and i) or 60 min (c). Immediately after NMDA treatment, samples were collected, and analyzed for the level of p-CREB (a and h), p-AKT (a and h), activated caspase-3 (a), p-ERK1/2 (b), and Bdnf transcription (c). Neuronal cell death was analyzed 20 h after the NMDA treatment by DAPI staining (i). (d–g) Calcium imaging shows changes of Ca2+i in neurons treated with 15 μM NMDA along with MK801 for 3 min (d). Following a 5-min wash, 15 μM NMDA (e), or bicuculline (f), or 50 μM NMDA (g) was applied (as indicated by the line for the duration of application). Calcium-imaging traces from individual neurons are shown. Statistical difference (P<0.05) was revealed by ANOVA for the data in a and h. Different letters indicate distinct SNK groups with a<b<c for different measurements in a, and a<b in h. NS, not significant
Because we observed that bicuculline and 15 μM NMDA had similar effects on cell fate and intracellular signaling, we next blocked receptors that are sensitive to 15 μM NMDA. Neurons were first treated with 15 μM NMDA and MK801 (Figure 5d). Subsequent application of 15 μM NMDA (Figure 5e) failed to cause detectable elevation of Ca2+i, indicating that receptors sensitive to 15 μM NMDA were effectively blocked. Although subsequent application of bicuculline failed to trigger calcium influx (Figure 5f), subsequent application of 50 μM MNDA did (Figure 5g). These data demonstrated that, after pretreatment with 15 μM NMDA and MK801, there was no available syn-NMDAR, and the subsequent application of 50 μM NMDA mainly activated ex-NMDAR. Furthermore, following 15 μM NMDA/MK801, subsequent application of 50 μM (but not 15 μM) NMDA caused upregulation of p-CREB and p-AKT (Figure 5h) without triggering significant cell death (Figure 5i) and lactate dehydrogenase release (Supplementary Figure 7). Together, we suggest that activation of either syn- or ex-NMDAR alone is not sufficient to promote cell death. Switching intracellular program from pro-survival to pro-death may require certain degree of coactivation of NMDAR at both sub-cellular locations.
Memantine blocks intracellular signaling mediated by either syn- or ex-NMDAR
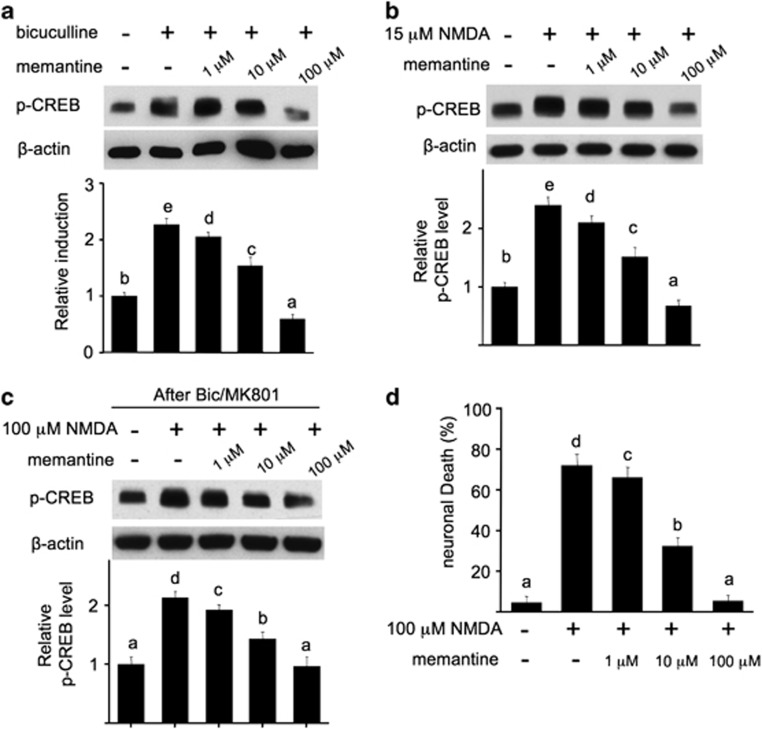
Memantine, a non-competitive low affinity NMDAR antagonist, has been used for the treatment of moderate-to-severe Alzheimer's disease.25 Some electrophysiological examinations implicated that memantine preferentially affects ex-NMDAR (Xia et al.26 but also see Wroge et al.27), although how memantine affects intracellular signaling related to survival and death remains unclear. To test its effects on syn-NMDAR-mediated signaling, we treated neurons with bicuculline in the presence of different concentrations of memantine. Memantine at 1 μM marginally suppressed bicuculline-stimulated p-CREB (Figure 6a). The phosphorylation of CREB was significantly and fully blocked by 10 and 100 μM memantine, respectively. Similarly, memantine also blocked 15 μM NMDA-induced p-CREB elevation in a concentration-dependent manner (Figure 6b). Next, we found that memantine at 1 and 10 μM marginally and partially blocked p-CREB triggered by ex-NMDAR, respectively (Figure 6c). Memantine at 100 μM completely blocked p-CREB stimulated by ex-NMDAR activation (Figure 6c). These data implicate that memantine non-selectively blocks both synaptic and extrasynaptic receptors.
Figure 6.
Memantine blocks intracellular CREB signaling mediated by either syn- or ex-NMDAR. DIV 14 neurons were stimulated with bicuculline (a), or 15 μM NMDA (b), or 100 μM NMDA following a 2-min pretreatment with bicuculline/MK801 (c). Memantine at different concentrations was coapplied with the stimulants. Thirty minutes after the stimulation, samples were harvested, and the level of p-CREB was determined by western blot. (d) Memantine blocks NMDAR-mediated cell death in a dose-dependent manner. Different doses of memantine were coapplied with 100 μM NMDA for 30 min. Twenty hours after the removal of NMDA, cell death was determined by DAPI staining. Statistical difference (P<0.05) was revealed by ANOVA. Different letters indicate distinct SNK groups with a<b<c<d<e for different measurements
We next investigated the effects of simultaneous blockage of synaptic and extrasynaptic receptors (i.e., by memantine) on NMDAR-dependent excitotoxicity. Memantine at 1 μM slightly reduced NMDA-induced neuronal death (72.1±5.6% versus 66.2±5.1%, P<0.05). Memantine at 10 μM greatly attenuated cell death (32.4±4.2%), and 100 μM memantine reduced cell death to the control level (5.4±2.9 versus 4.5±3.2% in control) (Figure 6d). These results along with the data in Figure 1 implicate that the degree of NMDA-induced death depends on the magnitude of coactivation of NMDAR at both sub-cellular locations.
Transient coactivation of syn- and ex-NMDAR does not cause cell death
It has been established that OGD triggers massive release of glutamate and induces death through NMDAR overactivation. However, such insult-induced damages may depend on the duration of OGD. Although 75-min OGD caused significant death (Figure 4), a short-term OGD for 30 min did not (Figure 7a). Intriguingly, a 30-min OGD upregulated the activity of ERK1/2, CREB, and AKT without activating caspase-3 (Figure 7b). These data support the importance of the ‘short therapeutic time window' to treat ischemic stoke, and also suggest that short-term coactivation of syn- and ex-NMDAR is not toxic. Next, we examined how short-term incubation with NMDA affected death. We used 15 μM NMDA to preferentially activate syn-NMDA, and 50 μM NMDA to coactivate both syn- and ex-NMDAR. As shown in Figures 7c and e, a 2- to 10-min stimulation with 15 μM NMDA caused significant p-CREB upregulation. However, while a short (2- to 4-min) stimulation with 50 μM NMDA upregulated p-CREB, a longer stimulation (more than 6 min) failed to activate CREB (Figures 7d and e). As shown in Figure 1c, a 30-min incubation with 50 μM NMDA significantly deactivated CREB. Consistently, incubation with 50 μM NMDA for up to 4 min did not cause significant death, but longer incubation did (Figure 7f). By extending the duration of NMDA insult to 60 min, 20 μM but not 15 μM NMDA started to deactivate CREB (Supplementary Figure 8). While stimulation with 20 μM NMDA for 30 min caused mild cytotoxicity (Figure 1f), a 24-h stimulation resulted in severe cell death (Supplementary Figure 9). These data indicate that the NMDAR-mediated cell death depends on both the magnitude and duration of synaptic and extrasynaptic receptor coactivation.
Figure 7.
Short-term insults with OGD or high-dose NMDA do not cause cell death, but rather stimulate the pro-survival signaling. DIV 14 neurons were subjected to a 30-min OGD (a and b), or treated with 15 (c and e) or 50 μM NMDA (d and e) for 2, 4, 6, 8, and 10 min. Samples were collected immediately after the treatment, and examined for the level of p-ERK1/2, p-CREB, p-AKT, and activated caspase-3 (b), or p-CREB (c and d). Quantification of p-CREB after NMDA treatment is shown in (e). ANOVA analysis revealed significant difference (P<0.05) between different groups. The letters a1 and b1 (a1<b1) are different SNK groups for the 15-μM NMDA treatment. The letters a2–e2 (a2<b2<c2<d2<e2) are different SNK groups for the 50-μM NMDA treatment. (a and f) Neurons were washed with culture medium after OGD or NMDA treatment. The neurons were stained with DAPI 20 h later, and cells with condensed nuclear were counted to determine cell death. One-way ANOVA and the post-hoc SNK analysis revealed significant difference (P<0.05) among different groups with a<b<c<d
Activation of syn- and ex-NMDAR leads to overlapping genomic responses
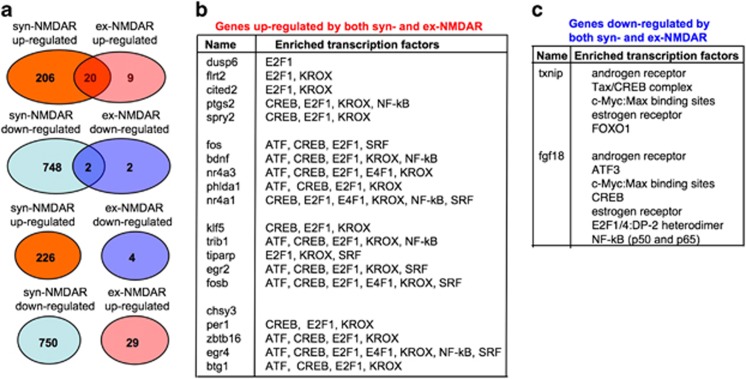
Although previous studies suggested that syn- and ex-NMDAR causes opposing effects, our data showed that they both stimulated pro-survival signaling and turned on the transcription of a CREB target gene Bdnf. We further determined how genome-wide expression profiles were regulated by NMDAR at different locations. By using false discovery rate of 25% as the threshold, we identified upregulated and downregulated genes from the Affymetrix GeneChip (Affymetrix, Santa Clara, CA, USA) Rat Gene 1.0 ST Arrays. Activation of syn-NMDAR up-regulated the expression of 226 genes, and downregulated 750 genes (Supplementary Table 1). Following ex-NMDAR activation, 29 genes were upregulated and four genes were downregulated (Supplementary Table 2). Interestingly, activation of syn- and ex-NMDAR triggered overlapping rather than opposing genomic responses (Figure 8a). Over 60% of the genes upregulated by ex-NMDAR were also upregulated by syn-NMDAR (Figures 8a and b). Two genes were downregulated by both syn- and ex-NMDAR (Figure 8c). The microarray data also demonstrated that Bdnf expression was upregulated by both syn- and ex-NMDAR (Figure 8b). TRANSFACT analysis of the coactivated genes revealed that both syn- and ex-NMDAR might have stimulated the transcription factors such as CREB, ATF, KROX, E2F, and SRF (Figure 8b).
Figure 8.
Activation of syn- and ex-NMDAR leads to overlapping but not opposing genomic responses. The genome-wide expression changes after the activation of syn- or ex-NMDAR were examined by using the Affymetrix GeneChip Rat Gene 1.0 ST Arrays. (a) Venn diagram showing the number of overlapping genes upregulated or downregulated by both syn- and ex-NMDAR. The upregulated genes mediated by syn-NMDAR do not overlap with the downregulated genes by ex-NMDAR; the downregulated genes mediated by syn-NMDAR do not overlap with the upregulated genes by ex-NMDAR. (b) List of the overlapped upregulated genes and their putative transcription factors. (c) List of the overlapped downregulated genes and their putative transcription factors
Discussion
Since the discovery of NMDAR involvement in neurodegeneration, numerous molecular determinants of cell death have been identified by stimulating neurons with NMDAR agonists. Intriguingly, neurons often do not show the same response to different concentrations of the agonists. Chandler et al.18 showed that NMDA at 10 or 20 μM upregulated ERK1/2 activity, but 100 μM NMDA caused down-regulation. Consistent with the bi-directional regulation of the pro-survival molecule ERK1/2, we demonstrated that high but not low-dose NMDA triggered cell death. Although most studies presume that bath NMDA or glutamate treatment causes death through ex-NMDAR activation, we provide new evidence demonstrating that the degree of NMDAR-mediated excitotoxicity depends on the magnitude and duration of syn- and ex-NMDAR coactivation.
One novel and surprising finding of this study is that we found no obvious causal effects of ex-NMDAR activation on death. This is in contrast to the prevailing hypothesis, emphasizing that ex-NMDAR contributes to excitotoxicity by shutting off pro-survival signaling.7, 8, 28 By carefully controlling NMDAR activation at different locations, we showed that blocking syn-NMDAR ablated cell death after high-dose NMDA or OGD. Supportively, after blocking syn-NMDAR, the activation of the available ex-NMDAR stimulated the ERK1/2-CREB-Bdnf pro-survival signaling. This is consistent with a recent study, which demonstrated that 50 μM glutamate failed to induce death after bicuculline/MK801 pretreatment.27 A previous study also showed that partial reduction of syn-NMDAR attenuates OGD-induced death.29 Further, Gao et al.30 showed that ERK1/2 activity was not suppressed by ex-NMDAR activation. The fact that immature developing neurons or retinal ganglion cells, which mainly express ex-NMDAR, 1216, 17 are resistant to NMDA- or glutamate-induced cell death13, 14, 15 is in line with our conclusion.
An important support for the deleterious role of ex-NMDAR is from the pro-survival effect of memantine, which has been found to preferentially block ex-NMDAR.26 However, this is based on the assumption that bath application of 100 μM NMDA only activates ex-NMDAR. Recently, Wroge and colleagues27 showed that memantine significantly attenuates current mediated by both syn- and ex-NMDAR. We demonstrate that memantine at both low (e.g., 1 or 10 μM) and high doses (e.g., 100 μM) blocks CREB activation mediated by either syn- or ex-NMDAR. Its effects on syn-NMDAR is consistent with previous reports, showing that memantine partially inhibits the induction of LTP and learning, which depend on syn-NMDAR function.31 Because simultaneous blocking syn- and ex-NMDAR by memantine dose-dependently suppressed excitotoxicity, we conclude that the magnitude of coactivation of these receptors determines the degree of cell death.
Although we demonstrated that syn-NMDAR activation did not cause death, we cannot exclude the detrimental effects of syn-NMDAR overactivation under some extreme conditions. For example, blocking the removal of synaptically released glutamate by TBOA (a glutamate transporter inhibitor) exaggerates hypoxic excitotoxic death.27 In cocultures that contain 95% neuron and 5% glia, promoting glutamate uptake reversal causes significant cell death via syn-NMDAR activation.32
Another important finding of this study is that NMDAR-mediated cell death also depends on the duration of syn- and ex-NMDAR coactivation. A short-term OGD or a brief exposure to the excitotoxic dose of NMDA was not effective to cause death. Rather, short-term coactivation of syn- and ex-NMDAR triggered pro-survival responses. One possibility is that these brief insults do not lead to full coactivation of syn- and ex-NMDAR. Figures 3b–d show that the rise in Ca2+i triggered by ex-NMDAR activation was gradual, and not as rapid as that triggered by global NMDAR activation (Figures 1g–k). A significant component of such gradual elevation occurred at 5–10 min, following application of 20, 40, and 50 μM NMDA (as indicated by the gray arrows in Figures 3b–d). Gradual Ca2+i elevation persisted for up to 30 min after the application of 50 μM NMDA (Figure 3d). This implicates that ex-NMDARs may be slow-responding receptors, whose full activation may need prolonged ligand exposure. Therefore, when both synaptic and extrasynaptic receptors are available, OGD or bath incubation with high NMDA (e.g., 50 μM) may first rapidly activate synaptic receptors along with a small population of ex-NMDAR, and then gradually activate increasing number of ex-NMDARs. Such feature of ex-NMDAR may help neurons to escape from death during sub-damaging and short-term insults.
Although there were attempts to determine how the whole-genome programming responds to NMDAR activation, this is the first microarray study that isolates the role of syn- and ex-NMDAR. The study by Hong and colleagues33 examined limited number of genes (i.e., 1152) by using a custom microarray. There is minimal overlap of genomic changes when comparing their study with ours, possibly because they mainly examined late responses at up to 24 h after NMDA stimulation. The other study by Zhang et al.34 compared early genomic changes triggered by bicuculline and 20 μM glutamate, which only shut off CREB without causing acute necrosis. By using a different neuron type (i.e., hippocampal neurons), Zhang et al.34 found opposing effects of bicuculline and 20 μM glutamate on transcriptional responses. However, the study also revealed that significant amount of changes triggered by 20 μM glutamate (53/106 upregulated genes and 10/51 downregulated genes) overlapped with those triggered by bicuculline. Here, we clearly demonstrate that activation of syn- and ex-NMDAR did not lead to opposing genomic responses. The overlap in expression changes was significant, consistent with that syn- and ex-NMDAR both activated CREB signaling and certain CREB target genes.
We found that syn-NMDAR triggered changes in significantly more genes than ex-NMDAR. This is consistent with that, comparing with broader brain functions mediated by syn-NMDAR, ex-NMDAR has limited roles. Certain physiological functions of ex-NMDAR are implicated in enhancing synaptic signaling and temporal coordination of synaptic firing.10, 11 Ex-NMDAR may also serve as reserves for the activity-dependent lateral incorporation to synapses.35, 36 The different genomic responses triggered by syn- and ex-NMDAR may be attributed to difference in receptor density and regulatory property. For example, syn- and ex-NMDAR are differentially regulated by calcium and tyrosine phosphorylation.12 Differential interaction with the scaffolding network and receptor subunit composition along with different splicing variants may lead to distinct linkage to different intracellular cascades.37, 38 The cellular and molecular determinants require further investigation.
Materials and Methods
Primary neuronal culture
Primary cortical neurons were obtained from postnatal day 0 Sprague Dawley rats as previously described.39 The Institutional Animal Care and Use Committee at Michigan State University has approved the procedures used in this study. Cultures were maintained in Neurobasal A, 1 × B27 supplement (Invitrogen, Carlsbad, CA, USA), 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 0.5 mM glutamine. One-third of the medium was replenished every 3 days during the culturing. Neurons at DIV (days in vitro) 14 were used for experiments.
Neuronal stimulation
All stimulations were done after a 30-min pretreatment with 20 μM CNQX (Sigma, St Louis, MO, USA) and 5 μM nifedipine (Sigma), which block the non-NMDA-type glutamate receptors and ℒ-type voltage-gated calcium channels, respectively. NMDAR coagonist glycine (2 μM) was coapplied with stimulants. To achieve global activation of NMDAR, neurons were stimulated with NMDA at different concentrations (as indicated). To stimulate syn-NMDAR, 50 μM bicuculline was applied. To stimulate ex-NMDAR, neurons were first treated with 50 μM bicuculline and 10 μM MK801 for 2 min, washed three times with prewarmed conditioned media, and then incubated in conditioned media with 50 μM NMDA (or at other concentrations as indicated). All drugs were applied directly to the medium, and the neurons were maintained at 37 °C with 5% CO2 during all treatments.
Western blot and quantification
After stimulation (30 min or as indicated), samples were collected in 60 μl of SDS sample buffer (10 mM Tris-HCl buffer, pH 6.8, 10% glycerol, 2% sodium dodecylsulfate, 0.01% bromophenol blue and 5% β-mercaptoethanol), and boiled at 100 °C for 10 min. The extracts (from about 8 × 105 cells) were separated by 10% SDS-PAGE, transferred to nitrocellulose membranes, and blocked with 5% non-fat milk in PBS-T (PBS, 0.1% Triton X-100) for 30 min at room temperature. The membranes were then incubated with primary antibodies against phosphorylated ERK1/2 (p-ERK1/2; 1:1000; Cell Signaling Technology, Beverly, MA, USA), phosphorylated CREB (p-CREB at Ser133; 1:1000; Upstate, Lake Placid, NY, USA), phosphorylated AKT (p-AKT at Ser473; 1:1000; Cell Signaling), and cleaved caspase-3 (1:1000, Cell signaling) in PBS-T overnight at 4 °C. After extensive wash and incubation with horseradish peroxidase-conjugated goat anti-rabbit antibodies (1:5000; Pierce, Rockford, IL, USA), the signals were detected by the chemiluminescence method (SuperSignal West Pico, Pierce, Rockford, IL, USA). For the purpose of normalization, the membranes were stripped, and re-probed with antibodies against β-actin (1:10,000; Sigma, St Louis, MO, USA). Several exposure times were used to obtain signals in the linear range. The bands were quantified by using Scion Image Beta 4.0.2 software (Scion Corp. Frederick, MD).
Semi-quantitative RT-PCR
To measure the agonist-stimulated transcription of Bdnf exon 4, control and stimulated neurons were lysed, and total RNA was purified by using the TRIzol method (Invitrogen). The cDNA was synthesized from 0.5 μg total RNA by using the reverse transcription kit (SuperScript, Invitrogen), and subjected to PCR. The primers used for exon 4-containing Bdnf mRNA are: 5′-CTCCGCCATGCAATTTCCAC-3′ (forward), and 5′-GCCTTCATGCAACCGAAGTA-3′ (reverse). These primers produced a 274-bp PCR product. PCR amplification of GAPDH using 5′-TCCATGACAACTTTGGCATTGTGG-3′ and 5′-GTTGCTGTTGAAGTCGCAGGAGAC-3′ was used as the internal control. The cycle number for Bdnf exon 4 and GAPDH was 26 and 20, respectively. The semi-quantitative RT-PCR products were analyzed by agarose gel electrophoresis, and quantified by Scion Image software.
Calcium imaging of cultured neurons
The elevation of Ca2+i triggered by NMDAR activation was determined by calcium imaging in live neurons. After incubation with fura-2 AM (3 μM) for 30 min, cultures were placed in a perfusion chamber mounted on a Nikon Eclipse TE-2000U inverted microscope (Nikon Instruments, Inc. Melville, NY, USA). Cultures were then perfused with buffered solution (147 mM NaCl, 5.3 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, 26.1 mM NaHCO3, 0.5 mM sodium pyruvate, and 30 mM glucose) containing CNQX and nifedipine at 32–34 °C to remove the extracellular dye. The perfusion flow rate was at 1-2 ml/min. Individual neuron was monitored during the stimulation with NMDA or bicuculline along with glycine, CNQX and nifedipine. Illumination was provided by a Sutter DG-5 high-speed wavelength switcher, and fluorescent images were acquired with a Roper Coolsnap Cascade 512B cooled CCD camera (Photometrics, Tucson, AZ, USA). Ca2+i levels were calculated from the ratio of background-corrected fura-2 emission (520 nm) at two excitation wavelengths (340 and 380 nm). Metamorph/Metafluor was used for image acquisition and analysis.
Analysis of NMDAR-mediated cell death
Neurons were stimulated by NMDA (concentrations as indicated) or bicuculline along with glycine, CNQX, and nifedipine for 30 min (or as indicated). Then, the solution was replaced with warm conditioned medium containing 1 μM MK801, which prevented additional neuronal death due to excessive glutamate release from dead neurons. Neurons were fixed with 4% paraformaldehyde 20 h after the 30 min NMDA or bicuculline treatment, and stained with DAPI (1 μg/ml). Images were recorded using an inverted Nikon fluorescence microscope (Nikon Instruments, Inc. Melville, NY, USA). Uniformly stained nuclei were scored as healthy viable neurons, whereas nuclei with condensed staining were used as an indicator for cell death. The number of neurons was counted in six randomly chosen fields per well (about 120–180 neurons, in total), and two wells per condition from three separate experiments were examined.
OGD-induced cell death
OGD-induced cell death was used as a cellular ischemic stroke model as described previously.24 First, culture medium was replaced by glucose-free solution (147 mM NaCl, 5.3 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, 26.1 mM NaHCO3, 0.5 mM sodium pyruvate). Then, the neuronal culture was placed in a hypoxia chamber (Billups-Rothenberg, Inc., Del Mar, CA, USA) supplied with 95% N2 and 5% CO2 at 37 °C. OGD was terminated by removing the culture from the hypoxia chamber, and by replacing the glucose-free buffer with the mixed media (1:1 mixture of fresh and conditioned media with 1 μM MK801). Cell death was determined by DAPI staining 20 h after OGD.
Microarray analysis
The Affymetrix GeneChip Rat Gene 1.0 ST Arrays were used to determine the whole-genome expression changes after the activation of syn- or ex-NMDAR. Total RNA was extracted 2 h after vehicle (control for the syn-NMDAR group, n=3), bicuculline (syn-NMDAR group, n=3), bicuculline plus MK801 (control for the ex-NMDAR group, n=6), and bicuculline plus MK801 followed by 50 μM NMDA (ex-NMDAR group, n=5) treatment, and used for the microarray analysis.
Data analysis
Data (presented as average±S.E.M.) were analyzed by one-way ANOVA and post-hoc Student–Newman–Keuls (SNK) for multiple comparisons. Student's t-test was used to assess significance between two groups.
Acknowledgments
This work was supported by NIH grants (R01MH076906 and R03NS072668) to HW. XZ was supported by a postdoctoral fellowship from American Heart Association (10POST4450000).
Author contributions
XZ and HW designed the research, and analyzed the data. XZ performed the experiments. DH, JL, and EA specifically contributed to the design and analysis related to the genome-wide screening. XZ, EA, and HW wrote the manuscript.
Glossary
- Bdnf
brain-derived neurotrophic factor
- CREB
cAMP responsive element-binding protein
- ERK1/2
extracellular signal-regulated kinase ½
- NMDAR
N-methyl-𝒟-aspartate receptor
- OGD
oxygen–glucose deprivation
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by D Bano
Supplementary Material
References
- Choi DW, Koh JY, Peters S. Pharmacology of glutamate neurotoxicity in cortical cell culture: attenuation by NMDA antagonists. J Neurosci. 1988;8:185–196. doi: 10.1523/JNEUROSCI.08-01-00185.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villmann C, Becker CM. On the hypes and falls in neuroprotection: targeting the NMDA receptor. Neuroscientist. 2007;13:594–615. doi: 10.1177/1073858406296259. [DOI] [PubMed] [Google Scholar]
- Benarroch EE. NMDA receptors: recent insights and clinical correlations. Neurology. 2011;76:1750–1757. doi: 10.1212/WNL.0b013e31821b7cc9. [DOI] [PubMed] [Google Scholar]
- Szydlowska K, Tymianski M. Calcium, ischemia and excitotoxicity. Cell Calcium. 2010;47:122–129. doi: 10.1016/j.ceca.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Harris AZ, Pettit DL. Extrasynaptic and synaptic NMDA receptors form stable and uniform pools in rat hippocampal slices. J Physiol. 2007;584:509–519. doi: 10.1113/jphysiol.2007.137679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CG, Miller AJ, Westbrook GL. Synaptic and extrasynaptic NMDA receptor NR2 subunits in cultured hippocampal neurons. J Neurophysiol. 2006;95:1727–1734. doi: 10.1152/jn.00771.2005. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11:682–696. doi: 10.1038/nrn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- Milnerwood AJ, Gladding CM, Pouladi MA, Kaufman AM, Hines RM, Boyd JD, et al. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington's disease mice. Neuron. 2010;65:178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Harris AZ, Pettit DL. Recruiting extrasynaptic NMDA receptors augments synaptic signaling. J Neurophysiol. 2008;99:524–533. doi: 10.1152/jn.01169.2007. [DOI] [PubMed] [Google Scholar]
- Fellin T, Pascual O, Gobbo S, Pozzan T, Haydon PG, Carmignoto G. Neuronal synchrony mediated by astrocytic glutamate through activation of extrasynaptic NMDA receptors. Neuron. 2004;43:729–743. doi: 10.1016/j.neuron.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Li B, Chen N, Luo T, Otsu Y, Murphy TH, Raymond LA. Differential regulation of synaptic and extra-synaptic NMDA receptors. Nat Neurosci. 2002;5:833–834. doi: 10.1038/nn912. [DOI] [PubMed] [Google Scholar]
- Choi DW, Maulucci-Gedde M, Kriegstein AR. Glutamate neurotoxicity in cortical cell culture. J Neurosci. 1987;7:357–368. doi: 10.1523/JNEUROSCI.07-02-00357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LK, Segal M. Early exposure of cultured hippocampal neurons to excitatory amino acids protects from later excitotoxicity. Int J Dev Neurosci. 2010;28:195–205. doi: 10.1016/j.ijdevneu.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Ullian EM, Barkis WB, Chen S, Diamond JS, Barres BA. Invulnerability of retinal ganglion cells to NMDA excitotoxicity. Mol Cell Neurosci. 2004;26:544–557. doi: 10.1016/j.mcn.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Chen S, Diamond JS. Synaptically released glutamate activates extrasynaptic NMDA receptors on cells in the ganglion cell layer of rat retina. J Neurosci. 2002;22:2165–2173. doi: 10.1523/JNEUROSCI.22-06-02165.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Diamond JS. Distinct perisynaptic and synaptic localization of NMDA and AMPA receptors on ganglion cells in rat retina. J Comp Neurol. 2006;498:810–820. doi: 10.1002/cne.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler LJ, Sutton G, Dorairaj NR, Norwood D. N-methyl D-aspartate receptor-mediated bidirectional control of extracellular signal-regulated kinase activity in cortical neuronal cultures. J Biol Chem. 2001;276:2627–2636. doi: 10.1074/jbc.M003390200. [DOI] [PubMed] [Google Scholar]
- Soriano FX, Papadia S, Hofmann F, Hardingham NR, Bading H, Hardingham GE. Preconditioning doses of NMDA promote neuroprotection by enhancing neuronal excitability. J Neurosci. 2006;26:4509–4518. doi: 10.1523/JNEUROSCI.0455-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong ZG, Zhu XM, Chu XP, Minami M, Hey J, Wei WL, et al. Neuroprotection in ischemia: blocking calcium-permeable acid-sensing ion channels. Cell. 2004;118:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Sattler R, Tymianski M. Molecular mechanisms of glutamate receptor-mediated excitotoxic neuronal cell death. Mol Neurobiol. 2001;24:107–129. doi: 10.1385/MN:24:1-3:107. [DOI] [PubMed] [Google Scholar]
- Avoli M, Hwa G, Louvel J, Kurcewicz I, Pumain R, Lacaille JC. Functional and pharmacological properties of GABA-mediated inhibition in the human neocortex. Can J Physiol Pharmacol. 1997;75:526–534. [PubMed] [Google Scholar]
- Bengtson CP, Dick O, Bading H. A quantitative method to assess extrasynaptic NMDA receptor function in the protective effect of synaptic activity against neurotoxicity. BMC Neurosci. 2008;9:11. doi: 10.1186/1471-2202-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MP, Choi DW. Combined oxygen and glucose deprivation in cortical cell culture: calcium-dependent and calcium-independent mechanisms of neuronal injury. J Neurosci. 1993;13:3510–3524. doi: 10.1523/JNEUROSCI.13-08-03510.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeage K. Memantine: a review of its use in moderate to severe Alzheimer's disease. CNS Drugs. 2009;23:881–897. doi: 10.2165/11201020-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Xia P, Chen HS, Zhang D, Lipton SA. Memantine preferentially blocks extrasynaptic over synaptic NMDA receptor currents in hippocampal autapses. J Neurosci. 2010;30:11246–11250. doi: 10.1523/JNEUROSCI.2488-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroge CM, Hogins J, Eisenman L, Mennerick S. Synaptic NMDA receptors mediate hypoxic excitotoxic death. J Neurosci. 2012;32:6732–6742. doi: 10.1523/JNEUROSCI.6371-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveille F, El Gaamouch F, Gouix E, Lecocq M, Lobner D, Nicole O, et al. Neuronal viability is controlled by a functional relation between synaptic and extrasynaptic NMDA receptors. FASEB J. 2008;22:4258–4271. doi: 10.1096/fj.08-107268. [DOI] [PubMed] [Google Scholar]
- Sattler R, Xiong Z, Lu WY, MacDonald JF, Tymianski M. Distinct roles of synaptic and extrasynaptic NMDA receptors in excitotoxicity. J Neurosci. 2000;20:22–33. doi: 10.1523/JNEUROSCI.20-01-00022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Gill MB, Tronson NC, Guedea AL, Guzman YF, Huh KH, et al. Hippocampal NMDA receptor subunits differentially regulate fear memory formation and neuronal signal propagation. Hippocampus. 2010;20:1072–1082. doi: 10.1002/hipo.20705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry AE, Klyubin I, Mc Donald JM, Mably AJ, Farrell MA, Scott M, et al. Alzheimer's disease brain-derived amyloid-beta-mediated inhibition of LTP in vivo is prevented by immunotargeting cellular prion protein. J Neurosci. 2011;31:7259–7263. doi: 10.1523/JNEUROSCI.6500-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouix E, Leveille F, Nicole O, Melon C, Had-Aissouni L, Buisson A. Reverse glial glutamate uptake triggers neuronal cell death through extrasynaptic NMDA receptor activation. Mol Cell Neurosci. 2009;40:463–473. doi: 10.1016/j.mcn.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Hong SJ, Li H, Becker KG, Dawson VL, Dawson TM. Identification and analysis of plasticity-induced late-response genes. Proc Natl Acad Sci USA. 2004;101:2145–2150. doi: 10.1073/pnas.0305170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Steijaert MN, Lau D, Schutz G, Delucinge-Vivier C, Descombes P, et al. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron. 2007;53:549–562. doi: 10.1016/j.neuron.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. Mobile NMDA receptors at hippocampal synapses. Neuron. 2002;34:255–264. doi: 10.1016/s0896-6273(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Groc L, Heine M, Cognet L, Brickley K, Stephenson FA, Lounis B, et al. Differential activity-dependent regulation of the lateral mobilities of AMPA and NMDA receptors. Nat Neurosci. 2004;7:695–696. doi: 10.1038/nn1270. [DOI] [PubMed] [Google Scholar]
- Kohr G. NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res. 2006;326:439–446. doi: 10.1007/s00441-006-0273-6. [DOI] [PubMed] [Google Scholar]
- Gladding CM, Raymond LA. Mechanisms underlying NMDA receptor synaptic/extrasynaptic distribution and function. Mol Cell Neurosci. 2011;48:308–320. doi: 10.1016/j.mcn.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Zhou X, Moon C, Zheng F, Luo Y, Soellner D, Nunez JL, et al. N-methyl-𝒟-aspartate-stimulated ERK1/2 signaling and the transcriptional up-regulation of plasticity-related genes are developmentally regulated following in vitro neuronal maturation. J Neurosci Res. 2009;87:2632–2644. doi: 10.1002/jnr.22103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.