Abstract
This paper describes a probabilistic case detection system (CDS) that uses a Bayesian network model of medical diagnosis and natural language processing to compute the posterior probability of influenza and influenza-like illness from emergency department dictated notes and laboratory results. The diagnostic accuracy of CDS for these conditions, as measured by the area under the ROC curve, was 0.97, and the overall accuracy for NLP employed in CDS was 0.91.
Keywords: case detection, disease surveillance, influenza, electronic medical records
1. Introduction
In disease surveillance, the objective of case detection is to notice the existence of a single individual with a disease. We say that this individual is a case of the disease. The importance of case detection is that detection of an outbreak typically depends on detection of individual cases.1
In current practice, cases are detected in four ways: by clinicians, laboratories, screening programs and computers. Some of these methods of case detection employ case definitions. A case definition is a written statement of findings that are both necessary and sufficient to classify an individual as having a disease or syndrome. More commonly, however, the determination of whether an individual has a disease (or syndrome) is left to the expert judgment of a clinician.
Clinicians detect cases as a by-product of routine medical and veterinary care. The strength of case detection by clinicians is that sick individuals seek medical care. Further, clinicians are experts at diagnosing illness, which is fundamental to case detection. However not every sick individual sees a clinician. Also, clinicians may not correctly diagnose every individual they see. Clinicians may forget to report cases or fail to report cases in the time frame required by law.2, 3 Even when a clinician reports a case, the reporting may occur relatively late in the disease process. With some exceptions (e.g., suspected meningococcal meningitis, suspected measles, suspected anthrax), clinicians report cases only after they are certain (or almost certain) about the diagnosis.
A variant of clinician detection is the sentinel clinician approach.4–9, 10 A sentinel clinician reports the number of individuals he or she sees who match a case definition, e.g., for Influenza-Like Illness (ILI). The strength of sentinel clinician case detection is its relative completeness of reporting. Its limitations include that some cases may not be reported and those cases that are reported may be delayed due to its being a manual process.11
Another variant of clinician detection is drop-in surveillance. Drop-in surveillance refers to the practice of asking physicians in emergency rooms to complete a form for each patient seen during the period surrounding a special event.12–19 The clinicians record whether the patient meets the case definition for one or more syndromes of interest. The strength of drop-in surveillance (and sentinel clinician surveillance) is that it detects sick individuals on the day that they first present for medical care. A limitation is that it is labor intensive.
Laboratories detect cases also as a by-product of their routine operation. Laboratories often become aware of cases of notifiable diseases either before or at the same time as the clinician who ordered the test. The strength of laboratories-as-case-detectors is that they are process oriented; therefore, they may report cases more reliably than busy clinicians. A weakness is that there is not a definitive diagnostic test for every disease, and there may not be a test with 100% sensitivity for a disease. Additionally, a laboratory cannot detect a case unless a sick individual sees a clinician, who must suspect the disease and order a definitive test. Lag times for the completion of laboratory work can be substantial.
Screening programs detect cases by interviewing and testing people during a known outbreak to identify additional cases (or carriers of the disease). Screening is most often used for contagious diseases in which it is important to find infected individuals to prevent further infections.
Finally, computers detect cases by applying case definitions or other algorithmic approaches to routinely collect clinical data. The earliest use of automatic case detection was for hospital infections,20–26 followed by notifiable conditions27–30 and syndromes31–43.
The case definitions used in case detection may be represented either using Boolean logical statements or probabilistic statements. Boolean approaches include the clinical findings that are both necessary and sufficient to classify a case, such as a case of ILI. The statements include AND and OR operations. On the other hand, a probabilistic case definition states evidence that supports or refutes a diagnosis using conditional probabilities and provides probability thresholds above which the diagnosis is considered either confirmed, likely, or suspected, e.g., a confirmed case of a Disease might be defined as P(Disease|Data) > 0.99.
In this paper, we describe and evaluate an automated case detection system (CDS) that uses Bayesian network models of diagnosis to represent case definitions. It first derives the likelihood of a disease given the symptoms, signs, and findings (Data) for a patient, namely, P(Data|Disease). It then combines such a likelihood with a prior probability distribution of disease, P(Disease), to derive the posterior probability of disease given the Data, P(Disease|Data).
A Bayesian network is a compact representation of a joint probability distribution among the nodes in the network. When a Bayesian network is used to represent the medical diagnosis of a disease, the variables (nodes) include the diagnosis and findings that a physician would use when diagnosing the disease, including significant negative findings that the physician might count against some disease being present. For example, negative lab tests that usually have high sensitivity can help physicians rule out a diagnosis.1, 44, 45Similarly, positive results of tests with high specificity can help rule in a diagnosis.44, 45 The relevance of Bayes rules to medical diagnosis was first introduced theoretically by Ledley and Lusted in 195946 and was used early on in a diagnostic expert system by Homer Warner in 1961.47 Developers of diagnostic expert systems continue to use the same methods as did Warner, as well as more complex Bayesian methods.
Several theoretical advantages of Bayesian case detection over Boolean case detection include: (1) it can use the prior probability of a disease, (2) it can represent the sensitivity and specificity of tests and findings for a disease, (3) it can represent an expert’s knowledge of disease diagnosis in the form of conditional probabilities, (4) it parallels a physician’s diagnosis of reasoning under uncertainty by computing posterior probabilities of diseases, and (5) it assists in decision making when new information becomes available.
The current state-of-the-art automated CDSs are (1) electronic laboratory reporting (ELR) systems that are based on laboratory reports, and (2) syndromic surveillance systems that are based on chief complaints.43, 48 However, the two systems fall into two extremes on diagnostic accuracy and timeliness spectrums. In regards to diagnostic accuracy, electronic lab reporting is at one extreme of generally being very accurate, whereas syndromic surveillance is generally less so. Regarding timeliness, syndromic surveillance can be immediately available at the time of a patient visit, whereas an ELR can be delayed for days from the time a lab was drawn.49
CDS is a component in the probabilistic, decision-theoretic disease surveillance and control system described in an accompanying paper in this issue of the journal.
Bayesian networks have not only been used for case detection but also for outbreak detection during the past decade. As a representative example, Mnatsakanyanet al.50 developed Bayesian information fusion networks that compute the posterior probability of an influenza outbreak by using multiple data sources, such as aggregate counts of emergency department (ED) chief complaints that are indicative of influenza and counts of relevant ICD-9 codes from outpatient clinics. As another example, Cooper et al.51–53 developed the PANDA system and its extensions that derive the posterior probabilities of CDC Category A diseases (including anthrax, plague, tularemia, and viral hemorrhagic fevers) using ED chief complaints and patient demographic information as evidence.
In this paper, we use the diagnoses of influenza and influenza-like-illness as examples, although the approach is general and can be applied to other notifiable conditions or syndromes.
2. Methods
This section describes (1) the Bayesian CDS, and (2) an evaluation of its diagnostic accuracy for the diagnosis of influenza and ILI.
2.1. Bayesian CDS
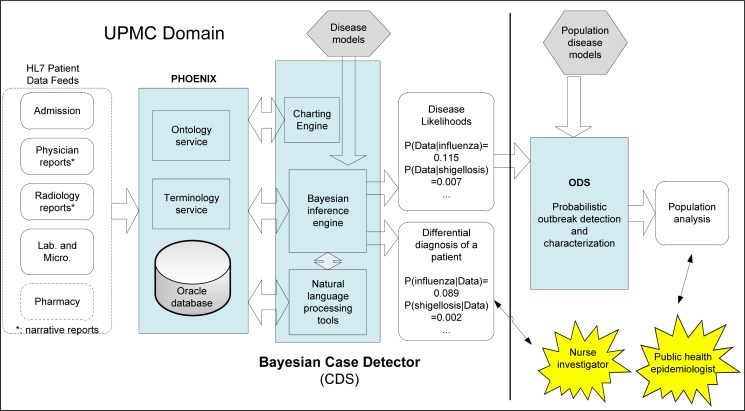
The Bayesian CDS includes (1) a natural language component that process free-text clinical reports and chief complaints, (2) disease models in the form of diagnostic Bayesian networks, (3) a Bayesian inference engine, and (4) a time-series chart reporting engine (Figure 1). The software components, including the inference engine, are implemented in Java.
Figure 1:
CDS and its relationship to other components in a probabilistic, decision-theoretic system for disease surveillance and control. CDS currently operates on clinical data from the UPMC Healthcare System. The blue boxes represent software components and hexagons represent models.
CDS sits between clinical data and ODS, an outbreak detection and characterization system. A component called Phoenix, described in an accompanying paper, receives data from an electronic medical record (EMR) system via HL-7 messaging, converts any proprietary codes to LOINC and SNOMED codes, stores the data, and processes requests from CDS. In general, CDS passes the likelihoods P(Dataj | Diseasei) to ODS for each modeled disease i and for each patient j in the monitoring period. For example, for a given patient, CDS would send the probability P(Data | influenza) to ODS, where Data denotes the symptoms, signs, and other findings of that patient. An accompanying paper in this issue describes ODS in more detail. In addition, CDS can output the posterior probabilities of modeled diseases for end users, as shown in Figure 1.
Our design criteria for CDS included computational efficiency sufficient to keep up with the volume of new patient data in a large healthcare system, and portability.54 CDS uses the computationally efficient junction tree algorithm55, 56 for Bayesian inference, which is also used in popular commercial Bayesian inference engines such as Hugin® and Netica®.
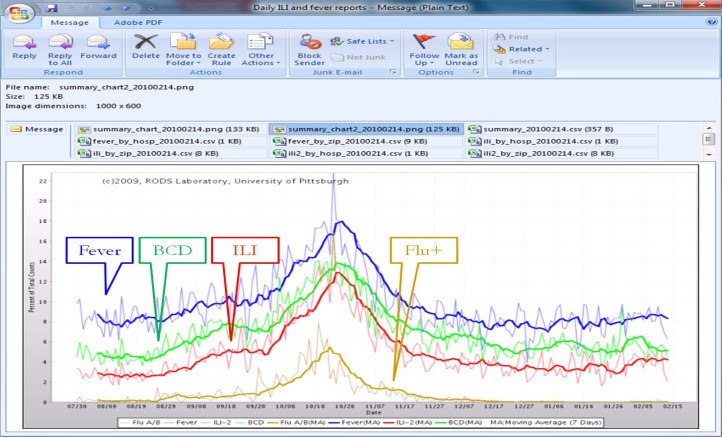
We have operated CDS since 2009.57 It generates daily reports of influenza and ILI and sends them to the Allegheny County Health Department (ACHD) by way of email (Figure 2). The daily report includes a graph of the daily counts of expected influenza cases, which is derived as . It also includes in the graph a daily time-series plot of Boolean-based ILI cases, influenza test orders, and influenza positive cases. Note the Boolean ILI counts in the daily chart are based on the Boolean case definition (Fever) AND (Cough OR Sore Throat), where the symptoms or findings are extracted by NLP.
Figure 2:
Influenza and ILI summary chart for February 15, 2010 (showing data from Aug. 1, 2009 to Feb. 14, 2010) in a daily email report to the Allegheny County Health Department. It comprises daily fever counts (from NLP), accumulated influenza posterior probability counts from Bayesian CDS, ILI counts (from NLP), and influenza (flu) test positive counts.
Public health officials in the ACHD have indicated that they find CDS to be useful. The charts shown in Figure 2 illustrate three areas of impact on practice at ACHD.57 First, CDS provided ACHD with daily updates instead of weekly reports from sentinel physicians. Second, ACHD could provide the charts to local media on a regular basis.58 Finally, ACHD reduced staff time since they no longer had to manually compile ILI reports from sentinel ILI reports (2 days of work for each weekly report).
2.1.1. Disease models
One of the core components in CDS is a knowledge base that contains disease models represented as Bayesian diagnostic networks. A disease model can include symptoms, signs, diagnosis, radiology findings, and laboratory test results (which we refer to as all-data), or it may use selected data, such as laboratory results, in which case we refer to the network as lab-only. CDS has one Bayesian diagnostic network (disease model) per disease.
CDS uses an existing Bayesian network design tool named GeNIe59 as a front end graphical user interface for disease model editing. GeNIe, which was developed at the University of Pittsburgh, can be downloaded from the Web60 for free. GeNIe can convert proprietary Bayesian network file formats used by Hugin® and Netica® (two of the most popular commercial Bayesian inference engines) into an XML file that can be then fed into CDS, allowing CDS users to import networks already developed by other groups. Note that the GeNIe tool is only needed when a user wishes to revise or create a Bayesian network. Figure 3 shows the GeNIe graphical user interface that allows an physician expert in clinical infectious disease to construct the influenza diagnostic model shown in right panel.
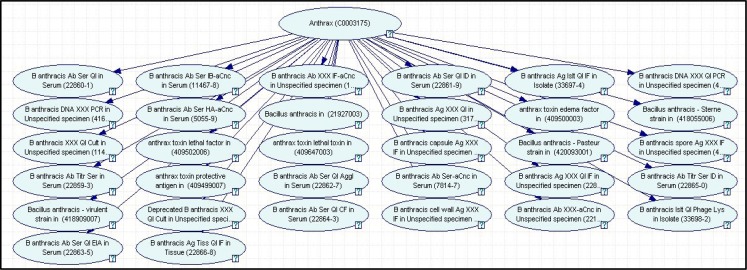
Figure 3:
Anthrax lab-only diagnostic network. A Bayesian network model for detection of anthrax cases using only laboratory results.
For portability, CDS disease models use standard terminology. For variables in a disease model representing symptoms and signs, we use concept unique identifiers (CUIs) from the UMLS. Both NLP tools in CDS--the well-known Medical Language Extraction and Encoding system (MedLEE)61, and a locally developed Topaz, use CUIs to represent extracted symptoms and findings; note that we used Topaz results in this paper. For laboratory tests, we use Logical Observation Identifiers Names and Codes (LOINC).
2.1.1.1. Lab-only Diagnostic Bayesian Network
Figure 3 shows a lab-only diagnostic model for B. anthracis. The laboratory tests in this model come from the Reportable Condition Mapping Tables (RCMT).62 This disease model comprises 33 nodes that represent 32 lab tests for B. anthracis. The names and results of the tests are represented using the Logical Observation Identifiers Names and Codes (LOINC) and Systematized Nomenclature of Medicine (SNOMED) coding systems. The parent node, labeled Anthrax, denotes whether the diagnosis of anthrax equals True or False. The 32 child nodes denote, for each laboratory test, whether the result was positive, negative, or unknown (because it has not been obtained). The structure of this particular model indicates that we are assuming that the tests are independent, given the diagnosis. Any dependencies among tests can be modeled in hidden nodes or by the inclusion of direct arcs among the nodes that denote tests.
We can apply this network to report cases in a manner similar to current electronic laboratory reporting systems. The conditional probability distributions in the network represent the sensitivity and specificity of each laboratory test for the disease anthrax. Let R denote the results of a set of laboratory tests for a given individual. We can perform inference on the network to derive P(anthrax | R). If that probability is above a threshold Tanthrax, then the case is reported. If the specificities of the tests are assumed to be 1, then any positive test result will lead to a probability of anthrax of 1, which will result in the reporting of the case if Tanthrax≤ 1. More generally, however, the sensitivities and specificities of the tests will not be 1, and in turn the probability of anthrax given test results will not be 0 or 1. Thus, in general, there is a need for case reporting that is based on probabilistic modeling and inference.
2.1.1.2. All-data Diagnostic Bayesian Network for Influenza
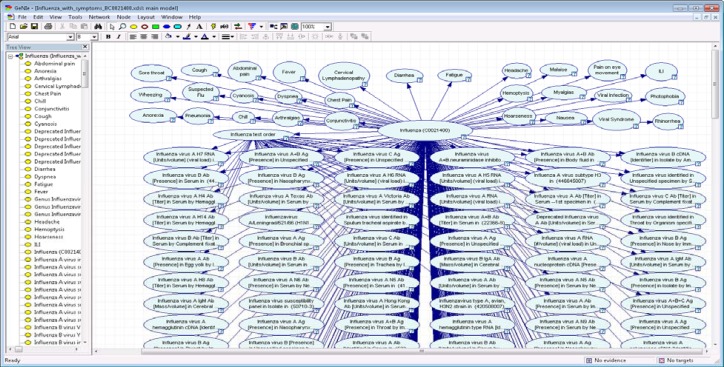
We developed an all-data influenza/ILI diagnostic Bayesian network that comprises flu symptoms, findings, and lab tests defined in the RCMT (Figure 4). The symptom and sign nodes and their corresponding conditional probabilities were initially built by author JD, who is board-certified in infectious diseases. The network comprises a total of 368 nodes including 29 symptom nodes, 337 lab test nodes, one test-order node, and one disease node (influenza), which can take the values “true” or “false”. The 337 lab nodes are those tests defined as reporting conditions in RCMT.63 Note that an NLP algorithm extracts symptoms and signs from free-text clinical reports, and they are used to set the values of the finding nodes.
Figure 4:
Influenza all-data diagnostic network. A Bayesian network model for diagnosing Influenza. The network utilizes data from free text clinical reports, orders for laboratory tests and the results of laboratory tests.
2.1.2. Parameter Estimation
Each model we built has two sets of parameters: expert assigned conditional probability tables (CPTs) and machine learning estimated CPTs. We have access to a large corpus of EMRs through the UPMC health System. We implemented a variation of the well-known Expectation Maximization-Maximum-A-Posteriori (EM-MAP) algorithm56 for learning network parameters from data. The EM-MAP is implemented in Java. The algorithm is able to learn network parameters by combining the data with prior knowledge (e.g., from our infectious disease experts and the literature), while being tolerant of missing values in the data.
2.1.3. Natural Language Processing
We developed an NLP application called Topaz that determines the presence, missing, or absence (negation) of 51 findings (e.g., signs, symptoms, and diagnoses) that are expected in influenza and shigellosis cases, or that are significant negative findings. Note that CDS will not assign any value for a variable in a disease model when the variable identified by Topaz has a value missing. Topaz comprises three modules. Module 1 looks for relevant clinical conditions and annotates all instances of those conditions in the report. Module 2 determines which annotations are negated, historical, hypothetical, or non-patient. Module 3 integrates the information from the annotations in the first two steps to assign values of present, absent (negated), or missing to each clinical condition for each patient.
2.1.4. User Interface/Data Viewer
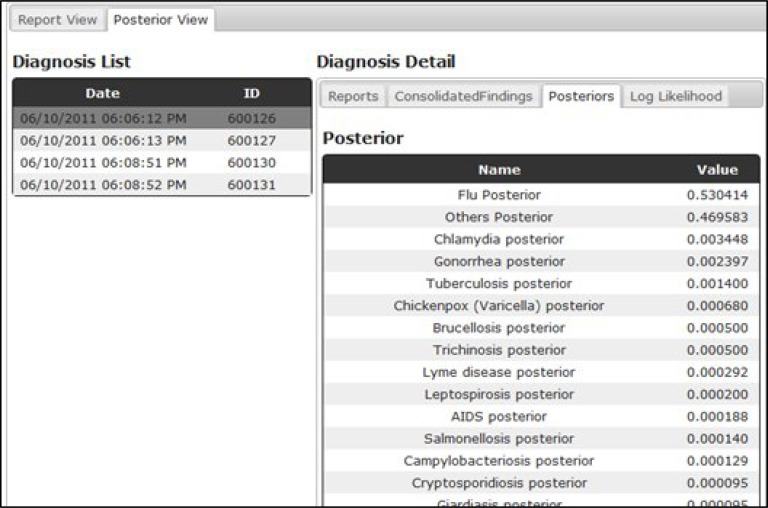
Figure 5 is a screen capture of a data viewer, which gives a patient care-episode view of the data for internal development purposes and serves as a prototype for a health department end-user interface. It displays all data associated to a patient’s visit, including extracted symptoms and signs from free text reports, lab findings and CDS output (posterior probabilities).
Figure 5.
Case review web page. A web page that allows users to review disease posterior probabilities (CDS output) and patient data including lab reports, free text reports. The posterior probabilities are displayed in a descending order with the highest disease probability on the top.
2.1.5. Event Driven Process
An event driven process is a software process that defines how a system reacts to an event.64 We define an event as data that triggers the execution of CDS, such as a laboratory test report or an ED report for a patient’s visit. When an event is available to CDS, CDS computes the posterior probabilities of the patient.
Since a patient’s visit may have multiple events (such as chief complaint, ED reports, laboratory test reports) that are available at different points in time, a disease’s posterior probability may change over time. For example, a lab report followed by a free text discharge report could raise the influenza posterior probability from 0.5 to very close to 1 when the lab report states an influenza test is positive. Note that a free-text ED report could be available a few hours after the patient visit whereas a lab report could take days.
To obtain an accurate patient diagnosis, when an event becomes available, CDS retrieves all patient events across different types up to the current time of the patient visit by using a data linkage key. In particular, CDS uses the visit number as the data linkage key.
2.2. Evaluation of Bayesian CDS
We evaluated Bayesian CDS in two ways: 1) case detection performance for one illness (i.e., influenza) from processing one data type, namely ED reports, and 2) NLP (Topaz) performance for extracting findings from ED reports.
2.2.1.1. Diagnostic Bayesian Network Study
For the study of case detection performance, we evaluated two influenza (all-data) Bayesian networks: 1) an expert influenza network constructed by a board-certified infectious disease domain expert, who assessed both the structure and parameters and the Bayesian network, and 2) an EM-MAP trained influenza network. Note that both networks share the same structure but different parameters.
2.2.1.1.1. Training and Testing Data for Case Detection Evaluation
In this study, we used ED reports from UPMC Heath System to measure the CDS performance for influenza case detection. All the ED reports used for evaluation were de-identified by an honest broker using the De-ID tool.65 The training data comprised 182 influenza cases and 47,062 non-influenza cases. The test data consisted of 58 influenza positive cases and 522 non-influenza cases. All cases were selected randomly from EMRs in the UPMC HS.
We considered a patient to have influenza if: 1) a polymerase chain reaction (PCR) test was positive, and 2) the linked ED reports had the keywords of flu, influenza, or H1N1 in the Impression section or Diagnosis section.
We considered a patient to not have influenza if: 1) no flu tests were ordered, and 2) the ED visits were during July 1, 2010 through August 31, 2010 for the training data, and during July 1, 2011 through July 31, 2011 for the test data.
2.2.1.1.2. Evaluation Metrics
The evaluation metrics used in this study include: ROC curves, area under a ROC curve (AUROC), probability of data given each of two diagnostic Bayesian networks as stated in the above paragraph, and the average speed for processing one case.
2.2.1.2. Topaz (NLP) Evaluation
We randomly selected 201 ED reports with flu PCR tests positive. The gold standard for evaluating Topaz was experts’ annotation. Three board certified physicians annotated the ED reports for a set of 51 signs, symptoms, and other findings that are expected in influenza and shigellosis cases. To ensure reliability, all the three annotators first went through training sessions; when the measured kappa value was above 0.8, they started annotating the 201 ED reports.
The evaluation metrics used in this study include kappa values, accuracy, and recall and precision.
3. Results
This section provides the evaluation results.
3.1. Diagnostic Bayesian Networks
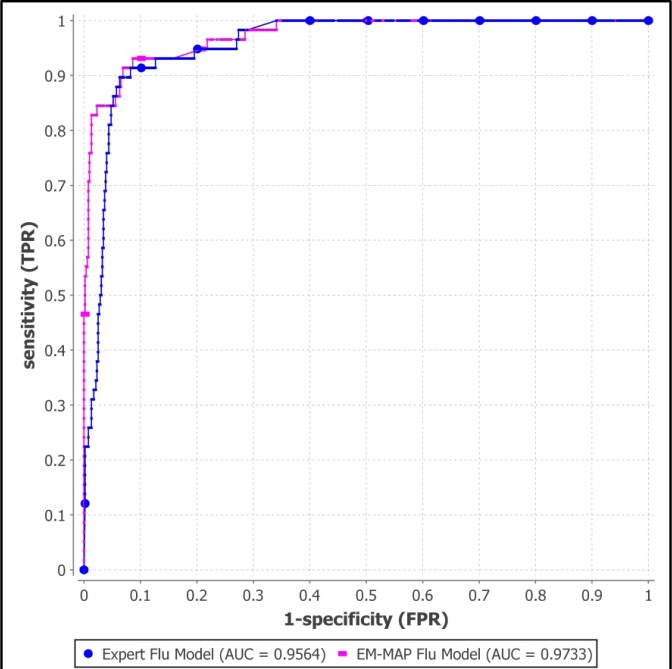
Figure 6 shows the two ROC curves for the expert model and the EM-MAP trained model for the total 580 test cases. The expert model has AUROC 0.956 (95% CI: 0.936–0.977) and the EM-MAP model has AUROC 0.973 (95% CI: 0.955–0.992).
Figure 6.
ROC curves for two influenza Bayesian networks. The blue line (with dots) represents the influenza model with parameters assigned by a domain expert and the pink line (with dashes) represents the influenza model with parameters learned by EM-MAP algorithm.
We measured the computational speed for computing the posterior probabilities and EM-MAP training. The average run time for computing influenza posterior probability is 15 milliseconds per case. The speed performance was measured on a desktop computer with Intel® Core™ 2 Quad CPU Q9550, 2.83 GHz and 4GB RAM.
3.2. Topaz
Table 1 summarizes the performance of Topaz. The kappa value between the gold standard and Topaz was 0.79. The overall accuracy including absent (negated), present, and missing findings was 0.91 and the accuracy for only absent and present was 0.77.
Table 1.
Topaz performance
| Recall | Precision | |
|---|---|---|
| Absent (negated) | 0.82 (1022/1249) | 0.84 (1022/1220) |
| Present | 0.73 (1109/1526) | 0.84 (1109/1319) |
| Missing | 0.96 (7205/7476) | 0.93 (7205/7712) |
4. Discussion
The results of the evaluation of the two influenza Bayesian networks (expert model and EM-MAP model) show high diagnostic accuracy. Additionally, augmenting the expert’s conditional probability distributions used in the model with empirical data about the distributions improves the diagnostic accuracy for influenza case detection.
The performance of the Topaz natural language processing algorithm for influenza findings approaches that of medical experts, as indicated by the kappa value 0.79 and overall accuracy of 91%.
A limitation of the evaluation study of Bayesian diagnostic models is as follows. Although we obtained non-influenza cases from patient visits that occurred in the summer and were not associated with an order for an influenza test, it is possible that there are influenza cases in the non-influenza training and testing data. However, any such contamination would be expected to bias the experiment against finding good diagnostic accuracy.
We also note that our current influenza model (Figure 4) should be modified to distinguish between Influenza A and B, which we plan as future work.
Of the four types of case detection discussed in the introduction—clinician, laboratory, screening, and computerized—the principle role of Bayesian CDS is in computerized (automatic) case detection. CDS can be used to augment laboratory, clinician, and screening case detection systems. To assist clinical diagnosis, the differential diagnoses output by CDS can be fed back directly to clinicians, or to other computer systems that provide decision support to clinicians at the point of care—reminding clinicians of diagnoses, notification requirements, vaccination, and history items to obtain or laboratory tests to order.
For laboratory-based case detection, the lab-only approach for Bayesian case detection discussed in this paper is a superset of current ELR approaches, which has the advantage of being able to represent the uncertainty associated with lower sensitivity or specificity tests.
For screening, the ability of the Bayesian CDS to represent a probabilistic case definition could be a significant advantage for emerging diseases that have case definitions that may be evolving or are dependent on constellation of symptoms and signs.
5. Conclusion
We developed an automatic case detection system that uses Bayesian networks as disease models and NLP to extract patient information from free-text clinical reports. The system computes disease probabilities given data from electronic medical records. The system is in use for influenza monitoring in Allegheny County, PA, automatically reporting daily summary charts to public health officials.57 The Bayesian CDS can function as a probabilistic ELR system or an all-data case-detection system. CDS is capable of integrating diagnostic information about a patient with prior probabilities of diseases to compute a probabilistic differential diagnosis that can be used in clinical decision support. The case probabilities derived by CDS can also be used as a key component for a system that detects and characterizes outbreak diseases in the population; a companion paper in this issue discusses a system called ODS that does just that.
Acknowledgments
This research was funded by grant P01-HK000086 from the Centers for Disease Control and Prevention in support of the University of Pittsburgh Center for Advanced Study of Informatics.
References
- 1.Wagner M, Gresham L, Dato V. Chapter 3 Case detection, outbreak detection, and outbreak characterization. In: Wagner M, Moore A, Aryel R, editors. Handbook of Biosurveillance. New York: Elsevier; 2006. [Google Scholar]
- 2.Ewert DP, Frederick PD, Run GH, Mascola L. The reporting efficiency of measles by hospitals in Los Angeles County, 1986 and 1989. Am J Public Health. 1994 May;84(5):868–9. doi: 10.2105/ajph.84.5.868-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewert DP, Westman S, Frederick PD, Waterman SH. Measles reporting completeness during a community-wide epidemic in inner-city Los Angeles. Public Health Rep. 1995 Mar-Apr;110(2):161–5. [PMC free article] [PubMed] [Google Scholar]
- 4.Schoub B, McAnerney J, Besselaar T. Regional perspectives on influenza surveillance in Africa. Vaccine. 2002;20(Suppl 2):S45–6. doi: 10.1016/s0264-410x(02)00129-9. [DOI] [PubMed] [Google Scholar]
- 5.Snacken R, Bensadon M, Strauss A. The CARE Telematics Network for the surveillance of influenza in Europe. Methods of Information in Medicine. 1995;34:518–22. [PubMed] [Google Scholar]
- 6.Snacken R, Manuguerra JC, Taylor P. European Influenza Surveillance Scheme on the Internet. Methods Inf Med. 1998 Sep;37(3):266–70. [PubMed] [Google Scholar]
- 7.Fleming D, Cohen J. Experience of European Collaboration in Influenza surveillance in the winter 1993–1994. J Public Health Medicine. 1996;18(2):133–42. doi: 10.1093/oxfordjournals.pubmed.a024472. [DOI] [PubMed] [Google Scholar]
- 8.Aymard M, Valette M, Lina B, Thouvenot D, the members of Groupe Régional d’Observation de la Grippe and European Influenza Surveillance Scheme Surveillance and impact of influenza in Europe. Vaccine. 1999;17:S30–S41. doi: 10.1016/s0264-410x(99)00103-6. [DOI] [PubMed] [Google Scholar]
- 9.Manuguerra J, Mosnier A, on behalf of EISS (European Influenza Surveillance Scheme) Surveillance of influenza in Europe from October 1999 to February 2000. Eurosurveillance. 2000;5(6):63–8. [PubMed] [Google Scholar]
- 10.Zambon M. Sentinel Surveillance of influenza in Europe, 1997/1998. Eurosurveillance. 1998;3(3):29–31. doi: 10.2807/esm.03.03.00091-en. [DOI] [PubMed] [Google Scholar]
- 11.Vogt RL, LaRue D, Klaucke DN, Jillson DA. Comparison of an active and passive surveillance system of primary care providers for hepatitis, measles, rubella, and salmonellosis in Vermont. American journal of public health. [Comparative Study] 1983 Jul;73(7):795–7. doi: 10.2105/ajph.73.7.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran GJ, Talan DA. Syndromic surveillance for bioterrorism following the attacks on the World Trade Center-New York City, 2001. Ann Emerg Med. 2003 Mar;41(3):414–8. doi: 10.1067/mem.2003.102. [DOI] [PubMed] [Google Scholar]
- 13.Das D, Weiss D, Mostashari F, Treadwell T, McQuiston J, Hutwagner L, et al. Enhanced drop-in syndromic surveillance in New York City following September 11, 2001. J Urban Health. 2003 Jun;80(2 Suppl 1):i76–88. doi: 10.1007/PL00022318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.County of Los Angeles. Democratic National Convention - Bioterrorism syndromic surveillance. 2000 [cited 2005 July 1]; Available from: http://www.lapublichealth.org/acd/reports/spclrpts/spcrpt00/DemoNatConvtn00.pdf.
- 15.Arizona Department of Health Services Syndromic Disease Surveillance in the Wake of Anthrax Threats and High Profile Public Events. Prevention Bulletin. 2002 Jan-Feb;2002:1–2. [Google Scholar]
- 16.CDC Syndromic surveillance for bioterrorism following the attacks on the World Trade Center--New York City, 2001. MMWR. 2002 Sep 11;51(Special issue):13–5. [PubMed] [Google Scholar]
- 17.Meehan P, Toomey KE, Drinnon J, Cunningham S, Anderson N, Baker E. Public health response for the 1996 Olympic Games. Jama. 1998 May 13;279(18):1469–73. doi: 10.1001/jama.279.18.1469. [DOI] [PubMed] [Google Scholar]
- 18.Mundorff MB, Gesteland P, Haddad M, Rolfs RT. Syndromic surveillance using chief complaints from urgent-care facilities during the Salt Lake 2002 Olympic Winter Games. MMWR. 2004 Sep 24;53(Supplement):254. [Google Scholar]
- 19.Dafni U, Tsiodras S, Panagiotakos D, Gkolfinopoulou K, Kouvatseas G, Tsourti Z, et al. Algorithm for statistical detection of peaks--Syndromic surveillance system for the Athens 2004 Olympics Games. MMWR. 2004 Sep 24;53(Supplement):86–94. [PubMed] [Google Scholar]
- 20.Evans RS. The HELP system: a review of clinical applications in infectious diseases and antibiotic use. MD Computing. 1991;8(5)(282–8):315. [PubMed] [Google Scholar]
- 21.Evans RS, Burke JP, Classen DC, Gardner RM, Menlove RL, Goodrich KM, et al. Computerized identification of patients at high risk for hospital-acquired infection. American Journal of Infection Control. 1992;20(1):4–10. doi: 10.1016/s0196-6553(05)80117-8. [DOI] [PubMed] [Google Scholar]
- 22.Evans RS, Gardner RM, Bush AR, Burke JP, Jacobson YA, Larsen RA, et al. Development of a computerized infectious disease monitor (CIDM) Computers & Biomedical Research. 1985;18(2):103–13. doi: 10.1016/0010-4809(85)90036-9. [DOI] [PubMed] [Google Scholar]
- 23.Evans RS, Larsen RA, Burke JP, Gardner RM, Meier FA, Jacobson JA, et al. Computer surveillance of hospital-acquired infections and antibiotic use. Jama. 1986;256(8):1007–11. [PubMed] [Google Scholar]
- 24.Evans RS, Pestotnik SL, Classen DC, Clemmer TP, Weaver LK, Orme JF, Jr, et al. A computer-assisted management program for antibiotics and other antiinfective agents. N Engl J Med. 1998 Jan 22;338(4):232–8. doi: 10.1056/NEJM199801223380406. [DOI] [PubMed] [Google Scholar]
- 25.Kahn MG, Steib SA, Fraser VJ, Dunagan WC. An expert system for culture-based infection control surveillance. Proceedings - the Annual Symposium on Computer Applications in Medical Care; 1993. pp. 171–5. [PMC free article] [PubMed] [Google Scholar]
- 26.Kahn MG, Steib SA, Spitznagel EL, Claiborne DW, Fraser VJ. Improvement in user performance following development and routine use of an expert system. Medinfo. 1995;8(Pt 2):1064–7. [PubMed] [Google Scholar]
- 27.Effler P, Ching-Lee M, Bogard A, Man-Cheng L, Nekomoto T, Jernigan D. Statewide system of electronic notifiable disease reporting from clinical laboratories: comparing automated reporting with conventional methods. JAMA. 1999;282(19):1845–50. doi: 10.1001/jama.282.19.1845. [DOI] [PubMed] [Google Scholar]
- 28.Panackal AA, M’ikanatha NM, Tsui F-C, McMahon J, Wagner MM, Dixon BW, et al. Automatic Electronic Laboratory-Based Reporting of Notifiable Infectious Diseases. Emerg Infect Dis. 2001;8(7):685–91. doi: 10.3201/eid0807.010493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overhage JM, Suico J, McDonald CJ. Electronic laboratory reporting: barriers, solutions and findings. J Public Health Manag Pract. 2001 Nov;7(6):60–6. doi: 10.1097/00124784-200107060-00007. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman M, Wilkinson T, Bush A, Myers W, Griffin R, Hoff G, et al. Multijurisdictional approach to biosurveillance, Kansas City. Emerg Infect Dis. 2003;9(10):1281–86. doi: 10.3201/eid0910.030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis M, Pavlin J, Mansfield J, O’Brien S, Boomsma L, Elbert Y, et al. Disease outbreak detection system using syndromic data in the greater Washington DC area. Am J Prev Med. 2002 Oct;23(3):180. doi: 10.1016/s0749-3797(02)00490-7. [DOI] [PubMed] [Google Scholar]
- 32.Gesteland PH, Gardner RM, Tsui FC, Espino JU, Rolfs RT, James BC, et al. Automated Syndromic Surveillance for the 2002 Winter Olympics. J Am Med Inform Assoc. 2003 Aug 4; doi: 10.1197/jamia.M1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yih WK, Caldwell B, Harmon R, Kleinman K, Lazarus R, Nelson A, et al. National Bioterrorism Syndromic Surveillance Demonstration Program. MMWR Morb Mortal Wkly Rep. 2004 Sep 24;53(Suppl):43–9. [PubMed] [Google Scholar]
- 34.Platt R, Bocchino C, Caldwell B, Harmon R, Kleinman K, Lazarus R, et al. Syndromic surveillance using minimum transfer of identifiable data: the example of the National Bioterrorism Syndromic Surveillance Demonstration Program. J Urban Health. 2003 Jun;80(2 Suppl 1):i25–31. doi: 10.1007/PL00022312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordin JD, Harpaz R, Harper P, Rush W. Syndromic surveillance for measleslike illnesses in a managed care setting. J Infect Dis. 2004 May 1;189(Suppl 1):S222–6. doi: 10.1086/378775. [DOI] [PubMed] [Google Scholar]
- 36.Heffernan R, Mostashari F, Das D, Karpati A, Kuldorff M, Weiss D. Syndromic surveillance in public health practice, New York City. Emerg Infect Dis. 2004 May;10(5):858–64. doi: 10.3201/eid1005.030646. [DOI] [PubMed] [Google Scholar]
- 37.DoD-GEIS. Electronic surveillance system for early notification of community-based epidemics (ESSENCE). 2000 [cited 2001 June 2]; Available from: http://www.geis.ha.osd.mil/getpage.asp?page=SyndromicSurveillance.htm&action=7&click=KeyPrograms.
- 38.Lazarus R, Kleinman KP, Dashevsky I, DeMaria A, Platt R. Using automated medical records for rapid identification of illness syndromes (syndromic surveillance): the example of lower respiratory infection. BMC Public Health. 2001;1(1):9. doi: 10.1186/1471-2458-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lombardo J, Burkom H, Elbert E, Magruder S, Lewis SH, Loschen W, et al. A systems overview of the Electronic Surveillance System for the Early Notification of Community-Based Epidemics (ESSENCE II) J Urban Health. 2003 Jun;80(2 Suppl 1):i32–42. doi: 10.1007/PL00022313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner M, Espino J, Tsui F-C, Gesteland P, Chapman BE, Ivanov O, et al. Syndrome and outbreak detection from chief complaints: The experience of the Real-Time Outbreak and Disease Surveillance Project. MMWR. 2004 Sep 24;53(Supplement):28–31. [PubMed] [Google Scholar]
- 41.Tsui F-C, Espino JU, Dato VM, Gesteland PH, Hutman J, Wagner MM. Technical description of RODS: A real-time public health surveillance system. J Am Med Inform Assoc. 2003;10(5):399–408. doi: 10.1197/jamia.M1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espino JU, Wagner M, Szczepaniak M, Tsui F-C, Su H, Olszewski R, et al. Removing a barrier to computer-based outbreak and disease surveillance: The RODS Open Source Project. MMWR. 2004 Sep 24;53(Supplement):32–9. [PubMed] [Google Scholar]
- 43.Chapman WW, Dowling JN, Wagner MM. Classification of emergency department chief complaints into seven syndromes: a retrospective analysis of 527,228 patients. Ann Emerg Med. 2005 doi: 10.1016/j.annemergmed.2005.04.012. (in press) [DOI] [PubMed] [Google Scholar]
- 44.Shortliffe EH, Cimino JJ. Biomedical informatics : computer applications in health care and biomedicine. 3rd ed. New York, NY: Springer; 2006. [Google Scholar]
- 45.Medow MA, Lucey CR. A qualitative approach to Bayes’ theorem. Evidence-based medicine. 2011. Aug 23, [DOI] [PubMed]
- 46.Ledley RS, Lusted LB. Reasoning foundation of medical diagnosis: symbolic logic, probability, and value theory aid our understanding of how physicians reason. Science. 1959;130(9):9–12. doi: 10.1126/science.130.3366.9. [DOI] [PubMed] [Google Scholar]
- 47.Warner HR, Toronto AF, Veasey G, Stephenson R. A mathematical approach to medical diagnosis: application to congenital heart disease. J Am Med Assoc. 1961;177(3):177–83. doi: 10.1001/jama.1961.03040290005002. [DOI] [PubMed] [Google Scholar]
- 48.Chapman WW, Christensen LM, Wagner MM, Haug PJ, Ivanov O, Dowling JN, et al. Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artif Intell Med. 2005 Jan;33(1):31–40. doi: 10.1016/j.artmed.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Que J, Tsui FC, Wagner MM. Timeliness study of radiology and microbiology reports in a healthcare system for biosurveillance. AMIA Annu Symp Proc. 2006:1068. [PMC free article] [PubMed] [Google Scholar]
- 50.Mnatsakanyan ZR, Burkom HS, Coberly JS, Lombardo JS. Bayesian information fusion networks for biosurveillance applications. J Am Med Inform Assoc. 2009 Nov-Dec;16(6):855–63. doi: 10.1197/jamia.M2647. [Multicenter StudyResearch Support, Non-U.S. Gov’tResearch Support, U.S. Gov’t, P.H.S.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cooper GF, Dash DH, J.D. L, Wong WKH, W.R., Wagner MM, editors. Bayesian biosurveillance of disease outbreaks. 2004. Uncertainty in Artificial Intelligence.
- 52.Cooper GF, Dowling JN, Levander JD, Sutovsky PA, editors. A Bayesian algorithm for detecting CDC category A outbreak diseases from emergency department chief complaints. International Society for Disease Surveillance (ISDS) conference; 2006. [Google Scholar]
- 53.Jiang X, Cooper GF. A Bayesian spatio-temporal method for disease outbreak detection. J Am Med Inform Assoc. 2010 Jul-Aug;17(4):462–71. doi: 10.1136/jamia.2009.000356. [Research Support, U.S. Gov’t, Non-P.H.S.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsui F-C, Espino JU, Weng Y, Choudary A, Su H-D, Wagner MM. Key design elements of a data utility for national biosurveillance: event-driven architecture, caching, and web service model. Proc AMIA Symp (in press) 2005 [PMC free article] [PubMed] [Google Scholar]
- 55.Huang C, Darwiche A. “Inference in belief networks: a procedural guide. Intl J Approximate Reasoning. 1996;15(3):225–63. [Google Scholar]
- 56.Jensen FV, Nielsen TD. Bayesian networks and decision graphs. 2nd ed. Springer; 2007. [Google Scholar]
- 57.Tsui F-C, Su H, Dowling J, Voorhees RE, Espino JU, Wagner M. An automated influenza-like-illness reporting system using freetext emergency department reports. ISDS; Park City, Utah: 2010. [Google Scholar]
- 58.Two more people die from H1N1 In Pittsburgh area. KDKA; 2009; Available from: http://kdka.com/health/H1N1.flu.deaths.2.1321133.html.
- 59.GeNie. Graphical Network Interface. [cited 2008 Jan. 22]; Available from: http://genie.sis.pitt.edu/.
- 60.Druzdzel M. GENIE Homepage. 2001. Available from: http://www2.sis.pitt.edu/~genie/about_genie.html.
- 61.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004 Sep-Oct;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.CDC. PHIN Vocabulary access and distribution system (VADS). 2011 [9/21/2011]; Available from: https://phinvads.cdc.gov/vads/ViewCodeSystemConcept.action?oid=2.16.840.1.114222.4.5.274&code=RCMT.
- 63.Ganesan S. Influenza - Lab Tests & Results - RCMT. 2011. [cited 2011 9/22/2011]; Available from: http://www.phconnect.org/group/rcmt/forum/topics/influenza-lab-tests-results.
- 64.Hutwagner LC, Maloney EK, Bean NH, Slutsker L, Martin SM. Using laboratory-based surveillance data for prevention: an algorithm for detecting Salmonella outbreaks. Emerging Infectious Diseases. 1997;3(3):395–400. doi: 10.3201/eid0303.970322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol. 2004 Feb;121(2):176–86. doi: 10.1309/E6K3-3GBP-E5C2-7FYU. [DOI] [PubMed] [Google Scholar]