Abstract
Specific small interfering RNAs (siRNAs) designed to silence different oncogenic pathways can be used for cancer therapy. However, non-modified naked siRNAs have short half-lives in blood serum and encounter difficulties in crossing biological membranes due to their negative charge. These obstacles can be overcome by using siRNAs complexed with bolaamphiphiles, consisting of two positively charged head groups that flank an internal hydrophobic chain. Bolaamphiphiles have relatively low toxicities, long persistence in the blood stream, and most importantly, in aqueous conditions can form poly-cationic micelles thus, becoming amenable to association with siRNAs. Herein, two different bolaamphiphiles with acetylcholine head groups attached to an alkyl chain in two distinct configurations are compared for their abilities to complex with siRNAs and deliver them into cells inducing gene silencing. Our explicit solvent molecular dynamics (MD) simulations showed that bolaamphiphiles associate with siRNAs due to electrostatic, hydrogen bonding, and hydrophobic interactions. These in silico studies are supported by various in vitro and in cell culture experimental techniques as well as by some in vivo studies. Results demonstrate that depending on the application, the extent of siRNA chemical protection, delivery efficiency, and further intracellular release can be varied by simply changing the type of bolaamphiphile used.
Keywords: bolaamphiphiles, cryo-EM, molecular dynamics simulations, FRET, poly-cationic micelles, RNA-based therapeutics, siRNA delivery, specific gene silencing
Introduction
In the past decade, efforts to develop RNA-based therapeutic technologies have significantly intensified.1,2,3,4,5 Triggering RNA interference (RNAi), in particular, has become one of the most widely used techniques for biomedical applications.1,2,3,4,5,6,7,8,9,10,11,12,13 RNAi uses a mechanism of posttranscriptional sequence specific gene silencing by processing double-stranded RNAs into small-interfering RNAs (siRNAs) used as part of the RNA-induced silencing complex to selectively cleave target mRNA.14 After the discovery that synthetic siRNAs can be exogenously introduced into cells to activate RNAi,15,16 this approach has become a powerful method for selective suppression of specific genes of interest in different species, showing potential for use in cancer therapeutics.3,5,6 However, the biomedical utility of the synthetic siRNAs is limited by several RNA structure-related factors such as the negative charge (uptake by cells that also have a negatively charged surface) and instability in the blood circulation (non-modified siRNAs have a very short half-life in the blood stream, mostly because of degradation by nucleases).4 These impediments can be overcome by using polymeric or lipid-based carriers to shield the negative charge and provide protection against nuclease activity,17,18,19,20,21,22 or by using a high-density lipoprotein.23
Novel synthetic bolaamphiphiles (bolas) have recently been shown to form complexes with a variety of small and large molecules including peptides,24,25 proteins,26 and plasmid DNAs25,27 delivering them across biological membranes28 including the blood brain barrier.18 Bolas also have relatively lower toxicity than cationic liposomes.29 Bolas have a hydrophobic alkyl chain flanked by positively charged head groups that can potentially interact with negatively charged nucleic acids and promote their delivery into cells. However, the nature of these interactions as well as the possibility to use bolas for delivery of therapeutic siRNAs remains largely unknown. To better understand the essence of bolas/siRNA interactions and to adjust the experimental conditions important for efficiencies of siRNA delivery and release inside cells, we used two structurally related bolas: GLH-19 and GLH-20 (Figure 1). These bolas have identical hydrophobic domains with acetylcholine head groups attached to an alkyl chain in two different configurations; via either the acetyl group of the acetylcholine (GLH-19), or the nitrogen atom of the choline moiety (GLH-20). These bolas have been shown to deliver their cargo in vivo with different efficiencies26,30 which can be due to their distinct interactions with the cargo molecules and the cell membranes. In this work, the interactions between these two structurally different bolas and siRNAs were theoretically studied by explicit solvent molecular dynamics (MD) simulations and then experimentally tested in vitro, in cell culture, and in vivo.
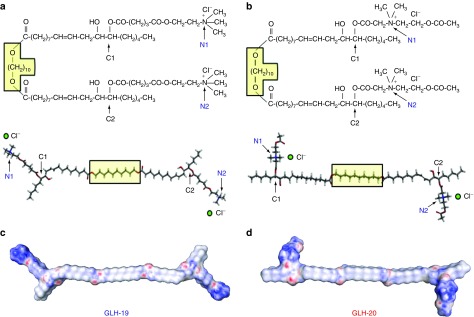
Figure 1.
Molecular structures of bolaamphiphiles (a) GLH-19 and (b) GLH-20. To indicate the correspondence to the 3D models presented in the lower half, a fragment common to both bolaamphiphiles (O-(CH2)10-O) was placed in the yellow box. Electrostatic surface potential maps of (c) GLH-19 and (d) GLH-20. Red, blue, and white colors indicate a negatively charged, positively charged, and neutral regions, respectively.
Results
MD simulation of bolaamphiphile micelles
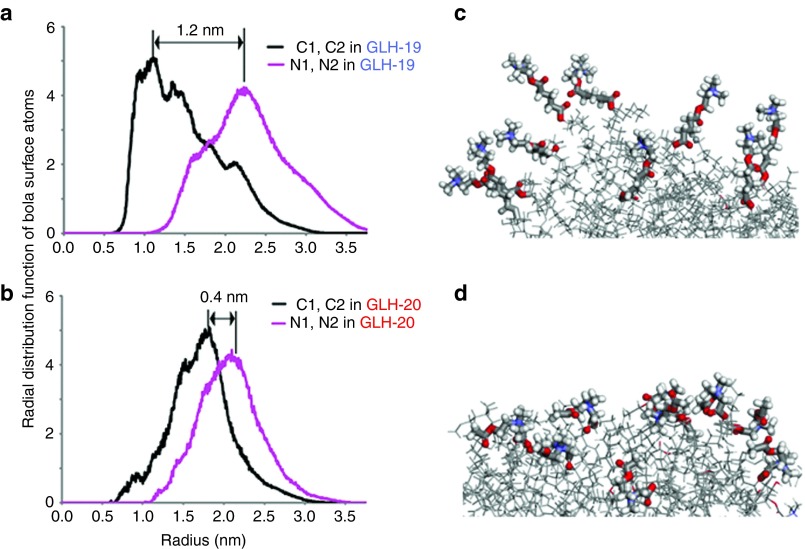
As detailed in Methods section, 20 individual bola molecules were aggregated into a single micelle in 0.15 mol/l NaCl solution during 50 nanoseconds simulations. Figure 2a,b show the radial distribution functions calculated for two nitrogen atoms (labeled as N1 and N2) representing the hydrophilic head groups of the micelle and two carbon atoms (labeled as C1 and C2) corresponding to the hydrophobic surface of the micelle. The radial distribution functions for these atoms were measured with respect to the center of the micelle. Thus, the different surface properties of the GLH-19 and GLH-20 micelles can be attributed to the different distances of their head groups from the center of each micelle (Figure 1). For example, the radial distribution function of GLH-19 indicated that the nitrogen atoms were distributed at ~12 Å above the hydrophobic surface of the micelle (Figure 2a), whereas for the case of GLH-20, the distribution was ~4 Å above the surface (Figure 2b). Hence, the hydrophilic head groups of GLH-19 protruded farther from the surface of the micelle and were therefore expected to be more flexible than those of the GLH-20 (Figure 2c,d). Consistent with this hypothesis, the mobilities of the head groups, presented as a mean square displacement (cm2) per unit of time (second), were calculated to be 1.8 × 10−7 cm2/s for GLH-19 and 1.0 × 10−7 cm2/s for GLH-20.
Figure 2.
Radial distribution functions from the center of each micelle to C1 and C2 hydrophobic surface atoms and N1 and N2 atoms of the hydrophilic head groups in (a) GLH-19 and (b) GLH-20 bolaamphiphiles (see also Figure 1). (c,d) Snapshots of GLH-19 and GLH-20 micelle surfaces respectively. Thicker bonds represent hydrophilic head groups.
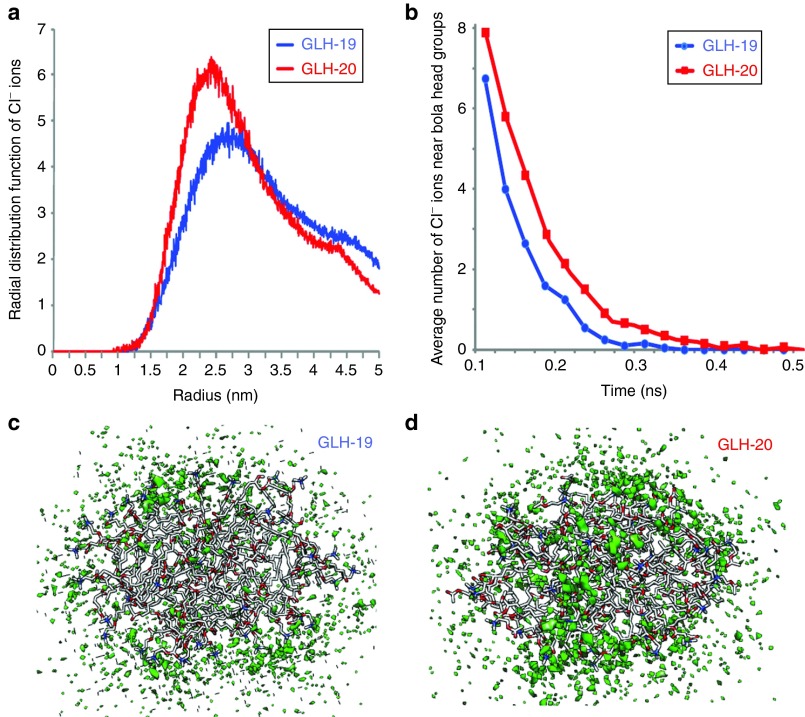
The differences between the properties of the GLH-19 and GLH-20 head groups affected the availability of the micelle surface for the chloride ions (Cl−) that are present in solution. The radial distribution function (Figure 3a) indicated a higher density of Cl− ions on the GLH-20 surface, compared with GLH-19 for longer periods of time (see Figure 3b). As a result, a more stable and solid cloud of Cl− ions was formed around the GLH-20 micelles (Figure 3d). Therefore, the electrostatic attractions between the GLH-20 micelle and the siRNA were expected to be more interfered with by the Cl− ion cloud than that of the GLH-19 micelle and the siRNA. Accordingly, a better association with siRNA was predicted for GLH-19.
Figure 3.
Chloride ion distribution around GLH-19 and GLH-20 micelles. (a) Radial distribution functions of Cl− ions from the center of the GLH-19 and GLH-20 micelles. (b) Average number of Cl− ions within a 7 Å range from each bolaamphiphile head group during 0.5 ns time range. 3D density plots of Cl− ions (green) around (c) GLH-19 and (d) GLH-20 micelles. ns, nanoseconds.
MD simulation of bolaamphiphile/siRNA complexes
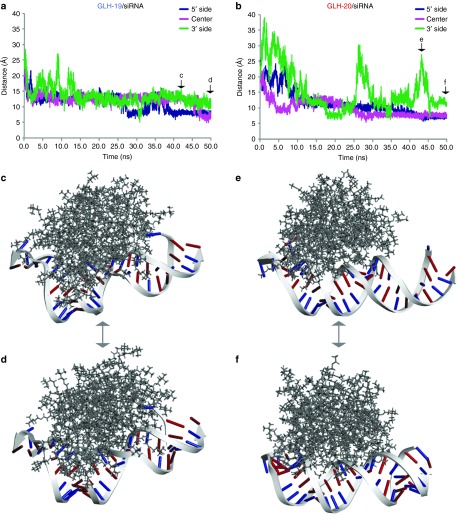
Each MD simulation was initiated by placing the center of mass of the individual siRNA duplex 40 Å away from the center of mass of the GLH-19 or GLH-20 micelle. This distance was sufficient to promote the electrostatic attraction between the positively charged hydrophilic head groups of the bolas and the negatively charged phosphates of the siRNA resulting in the formation of the bolas/siRNA complex. To track the behavior of bola/siRNA complexes during the simulation time, positions representing the 5′-side, middle, and 3′-side of the siRNA were selected. The centroids associated with the phosphate groups of C3, U14 and G25 (counted from the 5′-side of the sense strand) and their complements (G3, A14, and G24 of the antisense strand) were used. The relative distances between each centroid and the nearest bola head groups were continuously measured over the course of 50 nanoseconds (Figure 4). Within the first ~13 nanoseconds of the simulation, the siRNA duplexes formed stable complexes with both the GLH-19 and GLH-20 micelles (Supplementary Videos S1 and S2). However, during that time, the head groups of GLH-19 were positioned significantly closer to the selected phosphates than those of GLH-20. This suggested a faster and stronger binding interaction with GLH-19 (Figure 4a,b). Also, the extent of the interactions within the GLH-19/siRNA and GLH-20/siRNA complexes measured over the total simulation time was quite different. As can be seen from Figure 4a,b, all three phosphates of the siRNA were associated with GLH-19 during the entire simulation (Supplementary Video S1); whereas in the case of GLH-20, the 3′ side of the duplex (the right side of Figure 4c–f) was completely released from the interaction during the time periods of 25–30 nanoseconds and 40–45 nanoseconds (Supplementary Video S2). Snapshots of GLH-19/siRNA and GLH-20/siRNA complexes taken at 43 nanoseconds and at 50 nanoseconds (Figure 4d–f) clearly illustrate the difference in their binding affinities, with GLH-19 providing a higher association surface area and protection to the siRNA. These observations can be partly explained by the fact that the GLH-20 micelle was covered by a thicker cloud of Cl− ions (Figure 3d), which interferes with the interactions between the micelle and the siRNA consequently preventing complex formation. Due to the strong electrostatic interactions between the phosphates of the siRNAs, and the head groups of the bolas, the siRNA duplexes on the surface of each bola micelle were slightly bent (Figure 4c–f) with a higher relative deformation for the siRNA occurring for the case of GLH-19/siRNA (Figure 4c,d, Supplementary Video S1). The root mean square deviation of the siRNA duplex with respect to its initial structure reached 7.3 Å when it formed the GLH-19/siRNA complex, whereas the root mean square deviation of the siRNA duplex in the GLH-20/siRNA complex reached only 6.2 Å. Therefore, the siRNA that was associated with the GLH-19 micelle experienced a greater deformation due to the stronger interactions with GLH-19.
Figure 4.
The distance measurement between the phosphates of C3 (5′-side of the sense strand), U14 (middle), and G25 (3′ side of the sense strand) of the siRNA and the nearest bolaamphiphile head groups of (a) GLH-19 and (b) GLH-20. The initial time points (0 ns) were chosen after the equilibration heating of the system. Snapshots of the GLH-19/siRNA complex (c) at 43 ns and (d) at 50 ns and snapshots of the GLH-20/siRNA complex (e) at 43 ns and (f) at 50 ns taken from corresponding MD trajectories (also see Supplementary Videos S1 and S2). Types of bases in the siRNA are color-coded (purines are in red and pyrimidines are in blue). The siRNA on the GLH-19 micelle surface experienced more deformation than that of GLH-20. ns, nanoseconds.
When the micelles formed stable complexes with the siRNAs (Figure 4d,f), a large number of bola head groups were found near the siRNA phosphates due to the electrostatic interactions. We monitored the occupancies of the bola head groups within a 7 Å range of each RNA phosphate during the 30–50 nanoseconds time range of the simulation. Then, the sum of the occupancies for each base pair was analyzed (see Supplementary Figure S1). Both the GLH-19 and GLH-20 head groups populated the 5′-end (nucleotides 1–8 in the sense and the corresponding bases in the antisense strands including the two base overhang at the 3′ end), the center of the siRNA (nucleotides 12–19 in the sense and antisense strand) and its 3′-side (nucleotides 22–25 in the sense and its complements in the antisense strand). Due to the helical nature of RNA, only these particular regions were accessible for interactions with the bolas. However, the occupancies of the GLH-19 head groups near the siRNA phosphates were higher than GLH-20 in most regions. (Supplementary Figure S1, upper panel). Thus, the greater number of GLH-19 head groups that were associated with the phosphates (Supplementary Figure S1, lower panel) provided better protection and higher binding affinity to the siRNA.
The hydrogen bond formations between the siRNAs and the bolas were also monitored (see Supplementary Table S1 and Figure S3). Hydroxyls positioned next to the C1 and C2 atoms of the bolas can form stable hydrogen bonds with the backbone phosphates of the siRNAs. Hydroxyls in GLH-20 located close to the positively charged head groups were brought in close proximity to the negatively charged siRNA phosphates thus, forming a higher number of hydrogen bonds than the hydroxyls in GLH-19 (Supplementary Figure S3a,b). Head groups located close to the minor groove formed stable hydrogen bonds with the H22 and N2 atoms of guanosines (Supplementary Figure S3c,d). In addition, the terminal bases at the 5′ and 3′ ends also formed hydrogen bond interactions with the bola head groups. Interestingly, the sugar edge hydroxyl groups of the siRNA formed very few and relatively weak hydrogen bonds with the bolas. For a 20 nanoseconds time range, the total number of hydrogen bond interactions for GLH-19/siRNA and GLH-20/siRNA were 8 and 11, respectively (Supplementary Table S1). Besides the electrostatic and hydrogen bond interactions between the bola head groups and the siRNA, we found that the overhang and end bases of the siRNA duplex also interacted with hydrophobic chains of the bolas.
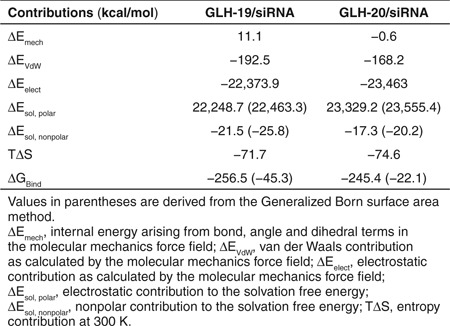
The free energies of binding for GLH-19/siRNA and GLH-20/siRNA were calculated using the molecular mechanics Poission Boltzmann/Generalized Born surface area (MM-PB/GBSA) methods (Table 1). The contribution of δEmech (which is (δEbond + δEangle + δEdihedral)) in the free energy of GLH-19/siRNA binding was higher due to the deformation of the siRNA. However, the contribution of the van der Waals interactions (δEvdW) in the free energy of binding was lower for the GLH-19/siRNA complexes. The GLH-20/siRNA complex had a lower contribution for the electrostatic interactions (δEele) in the binding energy than the GLH-19/siRNA. However, the solvation energy contributions in the binding free energies estimated by both PBSA and GBSA methods were more favorable in the case of GLH-19/siRNA. The difference in the entropy contribution for the GLH-19/siRNA complex and the GLH-20/siRNA complex was minimal. Therefore, the total binding free energies obtained by the PBSA (~11 kcal/mol) and GBSA (~23 kcal/mol) methods were stronger for the GLH-19/siRNA. Once more, these data indicated a greater binding affinity of the siRNA for GLH-19 than for GLH-20.
Table 1. Results from molecular mechanics Poission-Boltzmann/Generalized Born surface area methods.
To demonstrate the generality of the approach, two additional 40 nanoseconds MD simulations were performed for each bola/siRNA complex. One simulation set used different initial velocities and the other used different micelle surfaces. In all arrangements, a higher number of siRNA phosphates populated the bola head groups corresponding to the GLH-19 micelles (Supplementary Figure S4). The binding affinities calculated by MM-PBSA for GLH-19/siRNA were lower by ~4 kcal/mol (for MD simulations with different initial velocities) and ~13 kcal/mol (for MD simulations with a different initial micelle surface) compared with GLH-20/siRNA. The value of δδG (δG (GLH-19/siRNA) − δG (GLH-20/siRNA)) derived from three independent trajectories was calculated to be −9.3 ± 2.8 (SEM) kcal/mol. This confirms a higher affinity of binding to siRNA for GLH-19 micelles.
As an alternative simulation to illustrate relative propensity of unconstrained bolas to interact with RNA, a shorter duplex (12-mer) was surrounded by individual bolas evenly distributed in the 3D simulation space (Supplementary Figure S5 and S6). To increase the statistical sampling, we placed 6, 15, and 24 single bola molecules around the RNA duplex and analyzed their interactions using 30 nanoseconds MD simulations. For all three cases, GLH-19 head groups showed higher population densities near the RNA phosphate groups. Interestingly, in the case of 24 individual bolas, the relatively short distances between them promoted their aggregation resulting in small micelle formations which in turn interacted with the RNA duplex (Supplementary Figure S6). All these MD simulations additionally prove a better binding affinity of GLH-19/RNA complexes.
Experimental concentration dependence and structural characterization of GLH-19/siRNA and GLH-20/siRNA complex formations
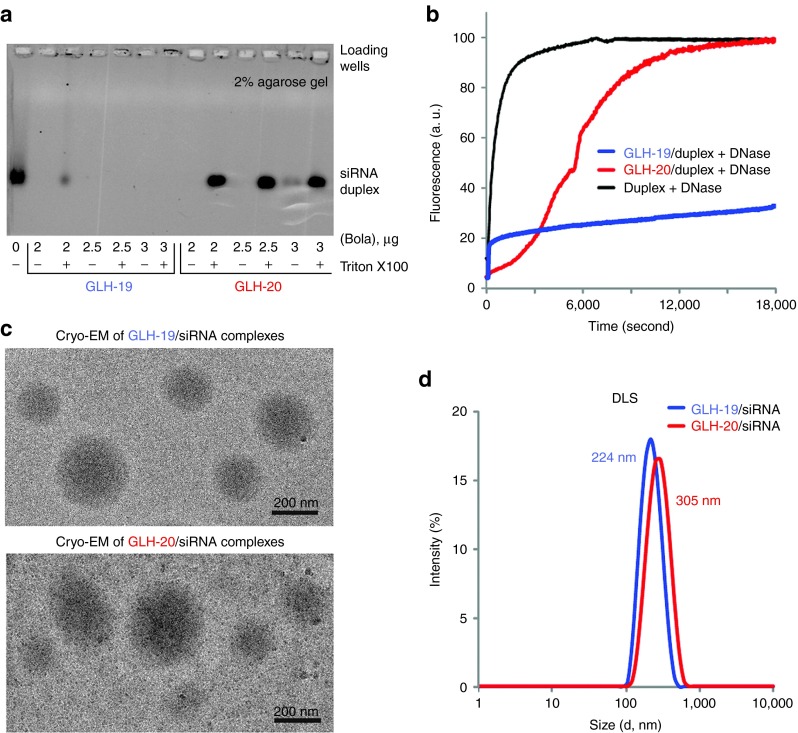
Binding affinity. To assess the abilities of GLH-19 and GLH-20 bolas to interact with nucleic acids, bolas at different concentrations (2, 2.5, 3, and 3.5 µg/ml) were introduced to fixed amounts of siRNAs (400 nmol/l final). Bolas at higher concentrations (10 µg/ml) were also tested and demonstrated consistent results (data not shown). After mixing and incubating, samples were analyzed by agarose gel electrophoresis as described in methods (Figure 5a). Upon staining with ethidium bromide, free siRNAs, not associated with bolas, appeared as single bands. However, the association of the positively charged bolas with the negatively charged siRNAs resulted in the formation of a complex too big to enter the gel thereby, preventing staining. To recover the siRNAs, a commonly used detergent Triton X100 was added to the bola/siRNA complexes. All the conditions tested show the formation of a complex between the bolas and the siRNAs because no free siRNA labeling was observed in the absence of detergent. Results indicated however that those complexes were very different in their sensitivity to detergent extraction of siRNA. For all the concentrations tested, the GLH-20/siRNA complexes were disrupted by the addition of Triton X100 leading to a complete recovery of the RNAs. Conversely, a very tight binding between GLH-19 and siRNAs was observed as the complex that could only be partially disrupted by the detergent at the lowest concentration (2 µg/ml), whereas at the higher concentrations (2.5, 3, and 3.5 µg/ml), they remained intact in the presence of Triton X100. These results were in a good agreement with the MD simulations that predicted that GLH-19/siRNA complexes have a stronger binding free energy than the GLH-20/siRNA complexes.
Figure 5.
In vitro characterizations of GLH-19/siRNA and GLH-20/siRNA complexes. (a) Formation of complexes was confirmed by agarose gel electrophoresis. siRNA (400 nmol/l final concentration) was mixed with bolaamphiphiles (bola) at final amounts indicated below the gel (in µg). For each amount of bola, equal amounts of detergent Triton X100 were added, aiming to prevent complex formation. Please note that in the case of GLH-19, binding is affected by detergent only at a very low concentration of bola. (b) Relative stabilities of DNA duplexes associated with either GLH-19 (blue line) or GLH-20 (red line) in the presence of DNase. Quenched DNA duplex (100 nmol/l final) labeled with Alexa488 and IowaBlack FQ was mixed with bolaamphiphiles (10 µg final) and DNase was added after 2 minutes of incubation at 37 °C. As the control, naked DNA duplex was completely digested by DNase (black line). Excitation was set at 460 nm and fluorescence signal (arbitrary units (a. u.)) was measured at 520 nm. Please note that there was no significant degradation observed for GLH-19/DNA after 3 hours of incubation. (c) Cryo-EM images of GLH-19/siRNA and GLH-20/siRNA complexes. (d) Size histograms from dynamic light scattering (DLS) experiments indicate average diameters for GLH-19/siRNA (blue) and GLH-20/siRNA (red) to be 224 and 305 nm respectively.
Digestion. To investigate the effect of complex formation on the nucleic acids' chemical stability, digestion assays with nucleases were carried out (Figure 5b). In these experiments, for reason of economy, we used a DNA duplex as a model system, with sequences that are identical to those used in the siRNA. The main difference between DNA and RNA is the presence of hydroxyl at the sugar edge. Our MD simulation showed that hydrogen bond interactions between the bolas and the hydroxyls in the siRNA were minor (see Supplementary Table S1). Therefore, the fundamental interactions between the DNA and bolas can be presumably very similar to that of the siRNAs and the corresponding bolas. The 3′-end of the DNA antisense and the 5′-end of the DNA sense strands were tagged with Alexa488 and IowaBlack fluorescence quencher respectively (see Supplementary Data). Therefore, the fluorescence of Alexa488 is quenched in a DNA duplex due to the proximity of the IowaBlack fluorescence quencher. However, upon digestion of the DNA duplex with DNase, IowaBlack gets separated from Alexa488 restoring its emission (Figure 5b, black line). Fluorescently quenched DNA duplexes (100 nmol/l final) were associated with GLH-19 and GLH-20 (10 µg/ml final) and DNase was added after 2 minutes of incubation at 37 °C. The results demonstrated some protection for the GLH-20/DNA (red line) complex with complete digestion of DNA only after 5 hours, whereas for GLH-19/DNA (blue line), no significant degradation of the DNA duplexes was observed during the same time period. It should be noticed that a significant part of the siRNA helix was exposed to water in the GLH-20/siRNA complex during the MD simulation, whereas the GLH-19 head groups maintained stable interactions with the entire siRNA under the same conditions. Thus, the experimental results are in a good agreement with the computationally predicted considerably higher accessibility of the duplex in the case of the GLH-20 complexes.
Cryogenic electron microscopy imaging and dynamic light scattering. We further studied the bola/siRNA complexes using cryogenic electron microscopy (cryo-EM) and to the best of our knowledge, this was one of the first investigations of siRNA/lipid micelle complexes by this technique. The cryo-EM images (Figure 5c) showed that a minority of GLH-19/siRNA complexes (upper image) was buried in thick ice and a majority was located on the carbon grid, whereas the GLH-20/siRNA complexes (lower image) were mostly found on the carbon grid. Interactions of bolas with the carbon surface thus emphasize their hydrophobic properties (Figure 1) with GLH-20/siRNA complexes being more hydrophobic. The size of the GLH-19/siRNA and GLH-20/siRNA complexes varied from 100 nm to 600 nm with the majority of the complexes being ~250 nm in diameter.
Using dynamic light scattering, the diameters of individual micelles were measured to be 135 nm for GLH-19 and 250 nm for GLH-20 (Supplementary Figure S7). The sizes of bola/siRNA complexes were measured to be slightly higher: 224 and 305 nm for GLH-19/siRNA and GLH-20/siRNA, respectively (Figure 5d) and are in a good agreement with the cryo-EM observations. The slight increases in size upon siRNA addition can be attributed to the partial growth of the poly-cationic micelles in the presence of multiple negatively charged siRNAs which is consistent with computational predictions (Supplementary Figure S5 and S6).
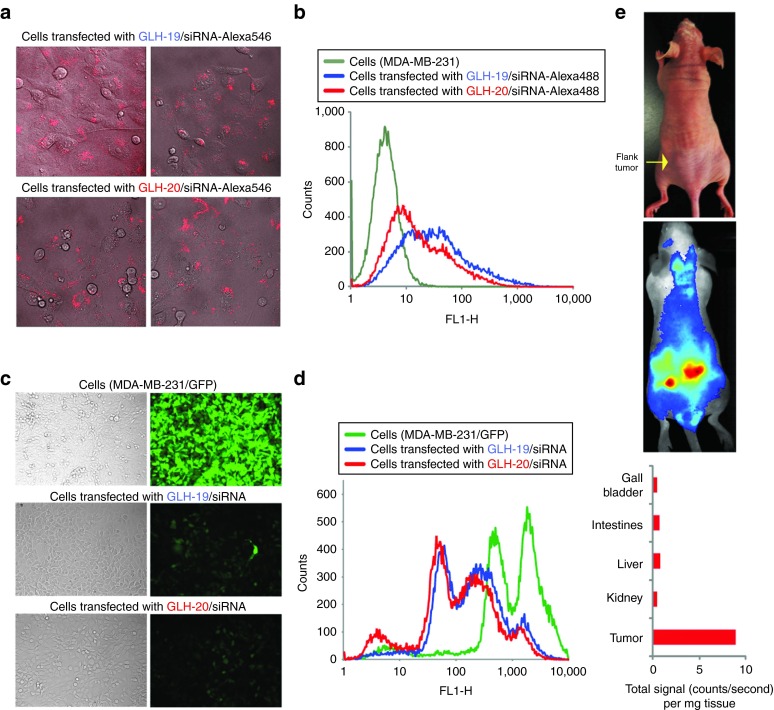
Transfection and gene silencing efficiencies for bola/siRNA complexes. To study the potential of bolas for siRNA delivery, GLH-19 and GLH-20 were associated with fluorescently labeled (RNA/DNA) duplexes. As fluorophore modified RNA strands are considerably more expensive than DNAs, RNA/DNA hybrids with labeled DNAs were used. The 3′-end of the DNA antisense strands were fluorescently labeled with Alexa488 and duplexed with the sense siRNA strand with a sequence capable of silencing enhanced green fluorescent protein (eGFP). These complexes were then transfected into human breast cancer cells MDA-MB-231 as described in methods. On the next day, transfection efficiencies were visualized by confocal fluorescence microscopy (Figure 6a) and statistically analyzed by fluorescence-activated cell sorting, revealing significant intracellular uptake with slightly higher efficiency for the GLH-19 complexes (Figure 6b). The lesser uptake for the GLH-20/siRNA complexes can be explained by the relatively lower binding affinity of GLH-20 which may promote partial dissociation of the complexes in the transfection media thus, preventing some fractions of the siRNAs from entering the cells.
Figure 6.
Cell culture and in vivo characterizations of GLH-19/siRNA and GLH-20/siRNA complexes. Transfection and silencing efficiencies for GLH-19/siRNA and GLH-20/siRNA complexes. (a,b) Transfection efficiencies of human breast cancer cells (MDA-MB-231) with GLH-19/siRNA and GLH-20/siRNA were (a) visualized by confocal fluorescence microscopy and (b) statistically analyzed with flow cytometry experiments. (c,d) eGFP knockdown assays for human breast cancer cells (MDA-MB-231/eGFP) which stably express enhanced GFP (eGFP). Three days after the transfection of cells with siRNAs, eGFP expression was (c) observed by fluorescence microscopy and (d) statistically analyzed with flow cytometry experiments. Please note that the initial fluorescent cells had two populations represented by two maximums. (e) In vivo live fluorescence imaging showing significant accumulation of GLH-19/siRNA-IRDye700 complexes in the left flank tumor 1 hour after systemic tail-vein injection.
To assess the ability of bola/siRNA complexes to release siRNA inside the cells, experiments with human breast cancer cells stably expressing enhanced green fluorescent protein (MDA-MB-231/eGFP) were carried out (Figure 6c,d). Cells were transfected with siRNAs against eGFP27 complexed with GLH-19 and GLH-20. After 3 days, the levels of eGFP expression were analyzed with fluorescence microscopy (Figure 6c) and fluorescence-activated cell sorting (Figure 6d). The results demonstrated significant and comparable levels of silencing in eGFP production for both types of bolas/siRNA complexes, which taken together with the higher levels of intracellular uptake for the GLH-19 complexes, confirmed the MD predictions of lower binding affinity for GLH-20/siRNA, which thus allows for a better release of the siRNA inside the cell.
In vivo experiments. To assess the ability of bolas to be used for siRNA delivery in vivo, experiments with tumor bearing athymic nude mice were carried out (Figure 6e). siRNAs fluorescently labeled with IRDye700 and associated with GLH-19 were systemically delivered to the mouse by tail-vein injections. Next, in vivo siRNA tumor uptake was evaluated by whole body fluorescence imaging. Fluorescent signals of major organs (spleen, liver, kidney, intestines, and gall bladder) were measured and normalized to their weights. Fluorescent signals in the spleen, lungs, heart, and brain were not detected (data not shown). The results demonstrated a relatively high uptake of the siRNA within the tumor compared with the other major organs within the time course of 3 hours thus, suggesting the potential for using bolas for in vivo siRNA delivery.
Discussion
The purpose of this study was to investigate the potential use of bolaamphiphile micelles as a carrier for siRNA delivery. We characterized the interactions between siRNA and two types of bolaamphiphiles, GLH-19 and GLH-20 using MD simulations and various experimental techniques. With the MD simulations, we predicted better protection against nuclease degradation for siRNAs associated with the GLH-19 micelles. The MM-PBSA and MM-GBSA methods predicted a higher binding affinity for the GLH-19/siRNA complex. Consistent with the computational results, gel experiments indicated stronger binding and more stable interactions for the GLH-19/siRNA complexes, which in addition showed almost no degradation in the presence of nucleases. Our cryo-EM studies helped to characterize the bolaamphiphile/siRNA complexes indicating that GLH-19/siRNA and GLH-20/siRNA have different relative hydrophobicities with GLH-19/siRNA being less hydrophobic. On the basis of the cell culture transfections and the in vivo live fluorescence imaging results, we can conclude that both GLH-19 and GLH-20 bolaamphiphiles have a great potential to be used as carriers for siRNA delivery with GLH-19 being a better candidate. Bola/siRNA complexes significantly increase the chemical stability of siRNAs and provide excellent intracellular uptake followed by specific gene silencing. Moreover, depending on the application, the extent of chemical protection of the siRNA can be altered by simply changing the carrier.
Materials and Methods
MD simulations. All MD simulations and experimental methods are detailed in the Supplementary Data. Explicit solvent MD simulations were used to study the interactions between a bola micelle and an siRNA duplex in the presence of 0.15 mol/l NaCl. Sequences for the siRNA duplex in our MD and experimental studies were derived from previous studies.1,7,31,32,33 The initial structure of the siRNA molecule was equilibrated by 30 nanoseconds MD simulations before it was used to examine the interactions with the bola micelle. Bola micelle structures were prepared as follows. MD simulations of a single GLH-19 and a single GLH-20 bola were performed for 40 nanoseconds, and 20 different snapshots of the structure were taken from the MD trajectories of each bola. The 20 structures of the same bola were placed 5 Å from each other, and used for an additional 50 nanoseconds of MD simulations. During these MD simulations, the 20 bolas gradually formed a micelle structure in which the hydrophobic chains aggregated, whereas the positive head groups were distributed on the micelle surface. These equilibrated GLH-19 and GLH-20 micelles were used to examine the interactions with the siRNA duplex (Supplementary Figure S2). The initial separation distance between the center of mass of the equilibrated bola and the center of mass of the equilibrated siRNA duplex was set to 40 Å. Water and ions were introduced and allowed to be equilibrated with a gradual release of harmonic constraints on the siRNAs and bolas followed by a heating step bringing the system to 300 K (see Supplementary Data for more details). Two additional configurations, one with different initial velocities and another with different initial bola micelle surfaces (achieved by rotating the micelles around the siRNA), were tested for 40 nanoseconds of MD simulations. Also, we examined the interactions between individual bola molecules and RNAs by placing 6, 15, and 24 individual bolas around a 12-mer duplex, mimicking the process of RNA coating with bolaamphiphiles.
More details of the MD simulations are discussed in the Supplementary Data. For all MD simulations, the sizes of the bola micelles (~10 nm) were smaller compared with the experimentally found sizes (~200 nm). This discrepancy is a result of the limitations in computational power and computing time, but it is believed that the simulations are reasonable approximations of larger systems.
Hydrogen bond interactions between the bolas and the siRNA were monitored using 3.5 Å and 120° cutoffs. The population density of the bola head groups and the Cl− ions on the micelle surfaces were calculated by a radial distribution function. The binding free energies of the siRNA-bola complexes were calculated by molecular mechanics MM-PB/GBSA methods.
Synthesis of bolaamphiphiles. The bolas, GLH-19 and GLH-20, were synthesized in Ben Gurion University as previously described25 and detailed in the Supplementary Data.
Duplex assemblies, bolaamphiphiles/siRNA complex formations, and qualitative agarose gel electrophoresis. Sequences for the siRNA duplex were used from previous studies1,7,31,32,33 and were designed to silence eGFP. Prepared siRNA-bolas complexes were analyzed at room temperature on 2% agarose gels in the presence of 89 mmol/l Tris-borate, pH 8.3. A Hitachi FMBIO II Multi-View Imager was used to visualize ethidium bromide stained oligos.
Dynamic light scattering experiments. For dynamic light scattering experiments,34 sample solutions containing either bola micelles prepared RNA/bola complexes were used. The samples were measured at 25 °C with a Zetasizernano (Malvern Instruments, Worcestershire, UK) equipped with a 633 nm laser.
Stability assays with RQ1 DNase fluorescent studies. To study the stability of the preformed duplex/bola complexes in the presence of nucleases, DNA duplexes containing one 3′ antisense strand modified with Alexa488 and a 5′ sense strand modified with IowaBlack FQ (see Supplementary Data) were preincubated with bolas (GLH-19 or GLH-20) and the dequenching of Alexa488 upon digestion with RQ1 DNase (Promega, Madison, WI) was followed by fluorescence resonance energy transfer experiments.
Cryo-EM experiments. Quantifoil Copper 200 mesh R 3.5/1 grids were washed overnight with acetone. To prepare a frozen, hydrated grid, 2.5 µl of sample was applied to the grid, blotted, and plunged into liquid ethane using Vitrobot III (FEI, Hillsboro, OR). Images were collected at liquid nitrogen temperature (~100 K) on a JEM-2010F (JEOL, Tokyo, Japan) transmission electron cryo-microscope equipped with a field emission gun. JEM-2010F was operating at 200 kV and was equipped with a Gatancryo-holder (model 626) (Gatan, Pleasanton, CA). Images were recorded on DE-12, a 12.6 megapixel (3,072 ×4,096) Direct Detection Device sensor (Direct Electron LP, San Diego, CA). Samples were imaged at 13,900× effective magnification targeted at 3–4 µm under focus. We used a total specimen exposure for each image of 30 e−/Å2 second.
Transfection of human breast cancer cells with bolaamphiphile/siRNA complexes. All siRNA transfections in this project were performed with the human breast cancer cell line MDA-MB-231 (with or without eGFP) using bolas (GLH-19 or GLH-20).
Fluorescent light microscopy. To assess the silencing efficiency, cells were imaged after the transfection with a LSM 710 confocal microscope (Carl Zeiss) and a Nikon 200 TE inverted microscope (Nikon, Melville, NJ) respectively.
Flow cytometry experiments. For statistical analysis of the flow cytometry experiments, at least 30,000 MDA-MB-231 cells (with or without eGFP) were analyzed by fluorescence-activated cell sorting analysis.
In vivo experiments. Animal studies were performed according to the Frederick National Laboratory for Cancer Research (Frederick, MD) Animal Care and Use Committee guidelines. Imaging studies were performed on MDA-MB-231 tumor bearing athymic nude mice (Charles River Laboratories, Frederick, MD).
SUPPLEMENTARY MATERIAL Figure S1. Total occupancy of bola head groups near the siRNA phosphate groups in terms of base pairs for the 30–50 nanoseconds range of the simulation. Figure S2. Structures and nomenclature of siRNA duplex and bolas used. Figure S3. Examples of typical hydrogen bond interactions between hydroxyls of one bola (other bola molecules in each micelle are not shown) and the phosphates of the siRNA backbone for (a) GLH-19 and (b) GLH-20. Figure S4. The number of bola head groups that are associated with the siRNA phosphate groups in the additional MD simulations, which used different velocities (upper panel) and different micelle surfaces (lower panel). Figure S5. MD simulations of individual bolas interaction with RNA duplex. Figure S6. MD simulations of individual bolas interaction with RNA duplex. Figure S7. Size histograms from dynamic light scattering experiments indicate average diameters for GLH-19 (blue) and GLH-20 (red) micelles to be 135 and 246 nm respectively. Table S1. Hydrogen bond interactions between the siRNAs and bolaamphiphiles (bolas). Video S1. Video S2. Data.
Acknowledgments
This study used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD and the National Cancer Institute's Advanced Biomedical Computing Center (ABCC) of the Frederick National Laboratory for Cancer Research, Frederick, MD. The authors thank Nimit Patel, Lisa Riffle, and Joseph Kalen in the Small Animal Imaging Program at the Frederick National Laboratory for Cancer Research for their guidance and support in animal imaging. They also thank Matthew Dougherty for help in preparing the movies and EckartBindewald for technical assistance. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute. This work has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the National Institutes of Health P41GM103832 and RC2GM092599 and the Postdoctoral Training Fellowship from the Keck Center Computational Cancer Biology Training Program of the Gulf Coast Consortia to A.Y.K. (CPRIT grant no. RP101489).
Supplementary material
Total occupancy of bola head groups near the siRNA phosphate groups in terms of base pairs for the 30–50 nanoseconds range of the simulation.
Structures and nomenclature of siRNA duplex and bolas used.
Examples of typical hydrogen bond interactions between hydroxyls of one bola (other bola molecules in each micelle are not shown) and the phosphates of the siRNA backbone for (a) GLH-19 and (b) GLH-20.
The number of bola head groups that are associated with the siRNA phosphate groups in the additional MD simulations, which used different velocities (upper panel) and different micelle surfaces (lower panel).
MD simulations of individual bolas interaction with RNA duplex.
MD simulations of individual bolas interaction with RNA duplex.
Size histograms from dynamic light scattering experiments indicate average diameters for GLH-19 (blue) and GLH-20 (red) micelles to be 135 and 246 nm respectively.
Hydrogen bond interactions between the siRNAs and bolaamphiphiles (bolas).
References
- Afonin, KA, Grabow, WW, Walker, FM, Bindewald, E, Dobrovolskaia, MA, Shapiro, BA et al. (2011). Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 6: 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, ME (2009). The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm 6: 659–668. [DOI] [PubMed] [Google Scholar]
- Davis, ME, Zuckerman, JE, Choi, CH, Seligson, D, Tolcher, A, Alabi, CA et al. (2010). Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464: 1067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole, R, Krainer, AR and Altman, S (2012). RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 11: 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot, CV, Calin, GA, Coleman, RL, Lopez-Berestein, G and Sood, AK (2011). RNA interference in the clinic: challenges and future directions. Nat Rev Cancer 11: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout, B and Sanders, RW (2011). Molecular strategies to design an escape-proof antiviral therapy. Antiviral Res 92: 7–14. [DOI] [PubMed] [Google Scholar]
- Grabow, WW, Zakrevsky, P, Afonin, KA, Chworos, A, Shapiro, BA and Jaeger, L (2011). Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett 11: 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, P (2010). The emerging field of RNA nanotechnology. Nat Nanotechnol 5: 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, Y, Cinier, M, Shu, D and Guo, P (2011). Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 54: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, GC, Haque, F, Tor, Y, Wilhelmsson, LM, Toulmé, JJ, Isambert, H et al. (2011). A boost for the emerging field of RNA nanotechnology. ACS Nano 5: 3405–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, GJ and Rossi, JJ (2004). Unlocking the potential of the human genome with RNA interference. Nature 431: 371–378. [DOI] [PubMed] [Google Scholar]
- Bramsen, JB and Kjems, J (2012). Development of Therapeutic-Grade Small Interfering RNAs by Chemical Engineering. Front Genet 3: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin, KA, Kireeva, M, Grabow, WW, Kashlev, M, Jaeger, L and Shapiro, BA (2012). Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett 12: 5192–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A, Xu, S, Montgomery, MK, Kostas, SA, Driver, SE and Mello, CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Elbashir, SM, Lendeckel, W and Tuschl, T (2001). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, SM, Martinez, J, Patkaniowska, A, Lendeckel, W and Tuschl, T (2001). Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foged, C (2012). siRNA delivery with lipid-based systems: promises and pitfalls. Curr Top Med Chem 12: 97–107. [DOI] [PubMed] [Google Scholar]
- Vader, P, van der Aa, LJ, Storm, G, Schiffelers, RM and Engbersen, JF (2012). Polymeric carrier systems for siRNA delivery. Curr Top Med Chem 12: 108–119. [DOI] [PubMed] [Google Scholar]
- Schroeder, A, Levins, CG, Cortez, C, Langer, R and Anderson, DG (2010). Lipid-based nanotherapeutics for siRNA delivery. J Intern Med 267: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliabadia, HM, Landrya, B, Sunb, C, Tangb, T and Uludăg, H (2012). Supramolecular assemblies in functional siRNA delivery: Where do we stand? Biomaterials 33: 2546–2569. [DOI] [PubMed] [Google Scholar]
- Ouyang, D, Zhang, H, Parekh, HS and Smith, SC (2010). Structure and dynamics of multiple cationic vectors-siRNA complexation by all-atomic molecular dynamics simulations. J Phys Chem B 114: 9231–9237. [DOI] [PubMed] [Google Scholar]
- Pavan, GM, Posocco, P, Tagliabue, A, Maly, M, Malek, A, Danani, A et al. (2010). PAMAM dendrimers for siRNA delivery: computational and experimental insights. Chemistry 16: 7781–7795. [DOI] [PubMed] [Google Scholar]
- Nakayama, T, Butler, JS, Sehgal, A, Severgnini, M, Racie, T, Sharman, J et al. (2012). Harnessing a physiologic mechanism for siRNA delivery with mimetic lipoprotein particles. Mol Ther 20: 1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg, S, Kipnis, N, Linder, C, Kolot, V, and Heldman, E (2010). Asymmetric bolaamphiphiles from vernonia oil designed for drug delivery. Eur J Lipid Sci Technol 112: 137–151. [Google Scholar]
- Grinberg, S, Linder, C, Kolot, V, Waner, T, Wiesman, Z, Shaubi, E et al. (2005). Novel cationic amphiphilic derivatives from vernonia oil: synthesis and self-aggregation into bilayer vesicles, nanoparticles, and DNA complexants. Langmuir 21: 7638–7645. [DOI] [PubMed] [Google Scholar]
- Dakwar, GR, Abu Hammad, I, Popov, M, Linder, C, Grinberg, S, Heldman, E et al. (2012). Delivery of proteins to the brain by bolaamphiphilic nano-sized vesicles. J Control Release 160: 315–321. [DOI] [PubMed] [Google Scholar]
- Wiesman, Z, Dom, NB, Sharvit, E, Grinberg, S, Linder, C, Heldman, E et al. (2007). Novel cationic vesicle platform derived from vernonia oil for efficient delivery of DNA through plant cuticle membranes. J Biotechnol 130: 85–94. [DOI] [PubMed] [Google Scholar]
- Linder, C, Grinberg, S and Heldman, E (2010). Nano-sized particles comprising multi-headed amphiphiles for targeted drug delivery. Patent application US20120164072; EP2427175 A2 (text in WO2010/128504 A2).
- Popov, M, Linder, C, Deckelbaum, RJ, Grinberg, S, Hansen, IH, Shaubi, E et al. (2010). Cationic vesicles from novel bolaamphiphilic compounds. J Liposome Res 20: 147–159. [DOI] [PubMed] [Google Scholar]
- Popov, M, Grinberg, S, Linder, C, Waner, T, Levi-Hevroni, B, Deckelbaum, RJ et al. (2012). Site-directed decapsulation of bolaamphiphilic vesicles with enzymatic cleavable surface groups. J Control Release 160: 306–314. [DOI] [PubMed] [Google Scholar]
- Bindewald, E, Afonin, K, Jaeger, L and Shapiro, BA (2011). Multistrand RNA secondary structure prediction and nanostructure design including pseudoknots. ACS Nano 5: 9542–9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, DH, Behlke, MA, Rose, SD, Chang, MS, Choi, S and Rossi, JJ (2005). Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol 23: 222–226. [DOI] [PubMed] [Google Scholar]
- Rose, SD, Kim, DH, Amarzguioui, M, Heidel, JD, Collingwood, MA, Davis, ME et al. (2005). Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res 33: 4140–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonin, KA, Bindewald, E, Yaghoubian, AJ, Voss, N, Jacovetty, E, Shapiro, BA et al. (2010). In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 5: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total occupancy of bola head groups near the siRNA phosphate groups in terms of base pairs for the 30–50 nanoseconds range of the simulation.
Structures and nomenclature of siRNA duplex and bolas used.
Examples of typical hydrogen bond interactions between hydroxyls of one bola (other bola molecules in each micelle are not shown) and the phosphates of the siRNA backbone for (a) GLH-19 and (b) GLH-20.
The number of bola head groups that are associated with the siRNA phosphate groups in the additional MD simulations, which used different velocities (upper panel) and different micelle surfaces (lower panel).
MD simulations of individual bolas interaction with RNA duplex.
MD simulations of individual bolas interaction with RNA duplex.
Size histograms from dynamic light scattering experiments indicate average diameters for GLH-19 (blue) and GLH-20 (red) micelles to be 135 and 246 nm respectively.
Hydrogen bond interactions between the siRNAs and bolaamphiphiles (bolas).