Abstract
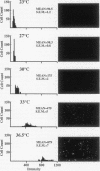
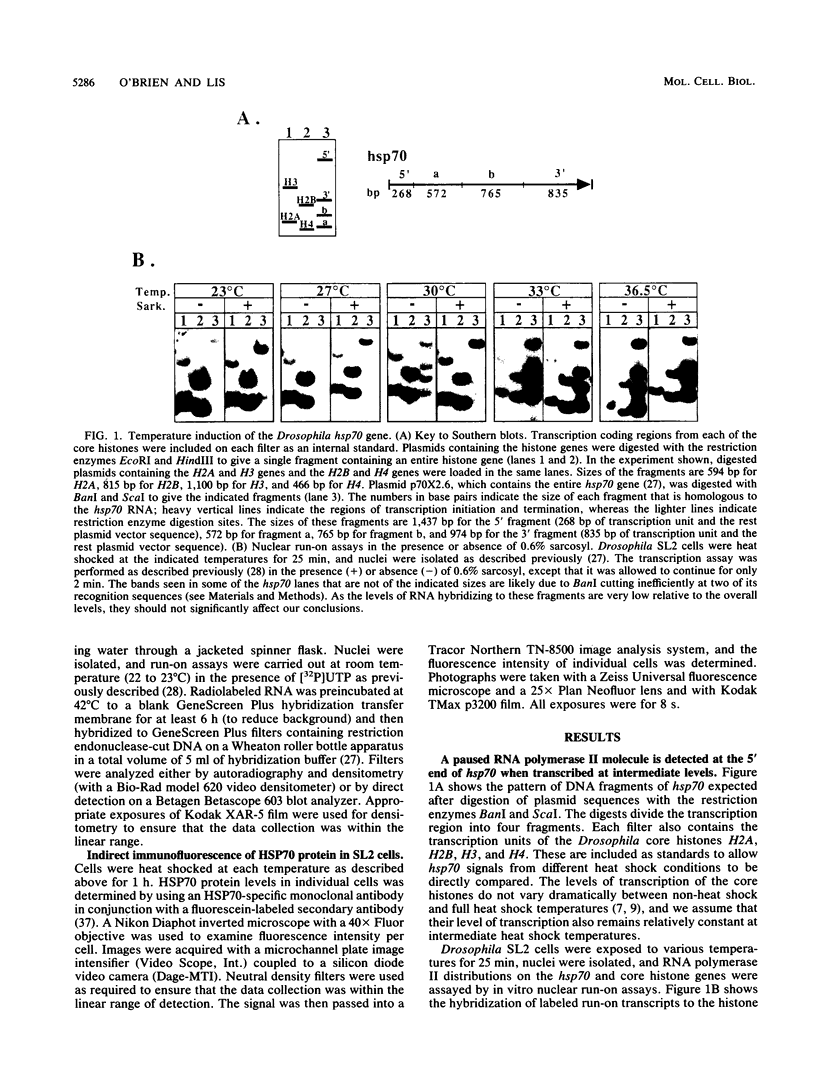
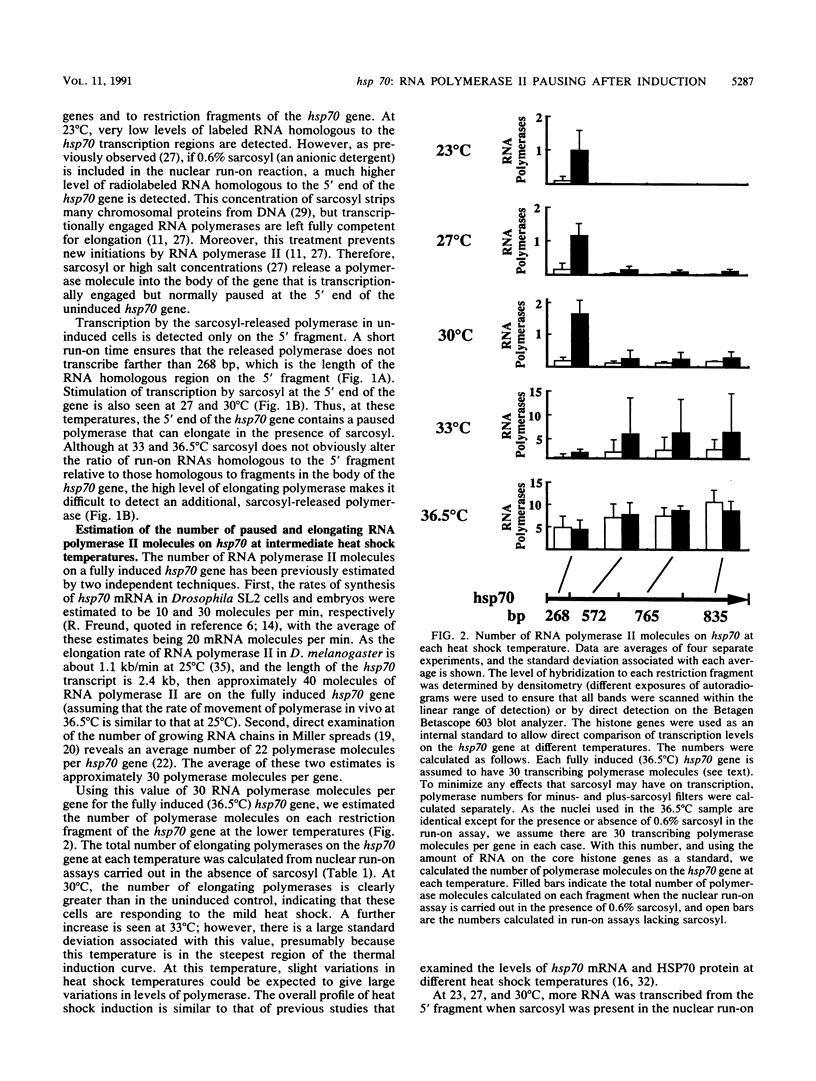
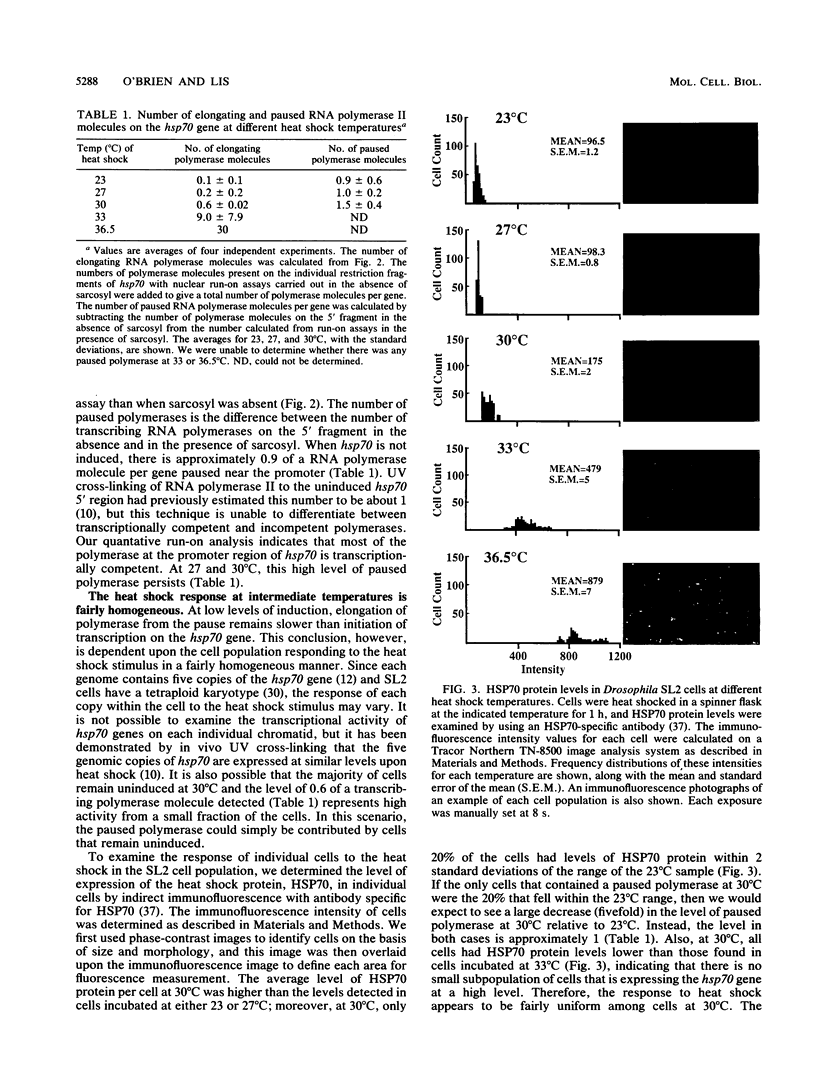
An RNA polymerase II molecule is associated with the 5' end of the Drosophila melanogaster hsp70 gene under non-heat shock conditions. This polymerase is engaged in transcription but has paused, or arrested, after synthesizing about 25 nucleotides (A. E. Rougvie and J. T. Lis, Cell 54:795-804, 1988). Resumption of elongation by this paused polymerase appears to be the rate-limiting step in hsp70 transcription in uninduced cells. Here we report results of nuclear run-on assays that measure the distribution of elongating and paused RNA polymerase molecules on the hsp70 gene in induced cells. Pausing of polymerase was detected at the 5' end of hsp70 in cells exposed to the intermediate heat shock temperatures of 27 and 30 degrees C. At 30 degrees C, each copy of hsp70 was transcribed approximately five times during the 25-min heat shock that we used. Therefore, once the hsp70 gene is induced to an intermediate level, initiation of transcription by RNA polymerase II remains more rapid than the resumption of elongation by a paused polymerase molecule.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Hay N. Attenuation may regulate gene expression in animal viruses and cells. CRC Crit Rev Biochem. 1985;18(4):327–383. doi: 10.3109/10409238509086785. [DOI] [PubMed] [Google Scholar]
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Chen Z., Harless M. L., Wright D. A., Kellems R. E. Identification and characterization of transcriptional arrest sites in exon 1 of the human adenosine deaminase gene. Mol Cell Biol. 1990 Sep;10(9):4555–4564. doi: 10.1128/mcb.10.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos J., Westwood J. T., Becker P. B., Wilson S., Lambert K., Wu C. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell. 1990 Nov 30;63(5):1085–1097. doi: 10.1016/0092-8674(90)90511-c. [DOI] [PubMed] [Google Scholar]
- Corces V., Pellicer A., Axel R., Meselson M. Integration, transcription, and control of a Drosophila heat shock gene in mouse cells. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7038–7042. doi: 10.1073/pnas.78.11.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell-Towt J., Sanders M. M. Noncoordinate histone synthesis in heat-shocked Drosophila cells is regulated at multiple levels. Mol Cell Biol. 1984 Dec;4(12):2676–2685. doi: 10.1128/mcb.4.12.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol. 1985 Aug;5(8):2009–2018. doi: 10.1128/mcb.5.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986 Nov;6(11):3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., Roeder R. G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985 Jul 5;260(13):8163–8172. [PubMed] [Google Scholar]
- Holmgren R., Livak K., Morimoto R., Freund R., Meselson M. Studies of cloned sequences from four Drosophila heat shock loci. Cell. 1979 Dec;18(4):1359–1370. doi: 10.1016/0092-8674(79)90246-0. [DOI] [PubMed] [Google Scholar]
- Lengyel J. A., Graham M. L. Transcription, export and turnover of Hsp70 and alpha beta, two Drosophila heat shock genes sharing a 400 nucleotide 5' upstream region. Nucleic Acids Res. 1984 Jul 25;12(14):5719–5735. doi: 10.1093/nar/12.14.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton R. P., Goldberg M. L., Karp R. W., Hogness D. S. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Varying patterns of protein synthesis in Drosophila during heat shock: implications for regulation. Dev Biol. 1980 Jun 15;77(2):463–479. doi: 10.1016/0012-1606(80)90488-1. [DOI] [PubMed] [Google Scholar]
- Maa M. C., Chinsky J. M., Ramamurthy V., Martin B. D., Kellems R. E. Identification of transcription stop sites at the 5' and 3' ends of the murine adenosine deaminase gene. J Biol Chem. 1990 Jul 25;265(21):12513–12519. [PubMed] [Google Scholar]
- Maderious A., Chen-Kiang S. Pausing and premature termination of human RNA polymerase II during transcription of adenovirus in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5931–5935. doi: 10.1073/pnas.81.19.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O. L., Jr, Beatty B. R. Extrachromosomal nucleolar genes in amphibian oocytes. Genetics. 1969;61(1 Suppl):133–143. [PubMed] [Google Scholar]
- Miller O. L., Jr, Beatty B. R. Visualization of nucleolar genes. Science. 1969 May 23;164(3882):955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
- Petersen R., Lindquist S. The Drosophila hsp70 message is rapidly degraded at normal temperatures and stabilized by heat shock. Gene. 1988 Dec 10;72(1-2):161–168. doi: 10.1016/0378-1119(88)90138-2. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Phage lambda and the regulation of transcription termination. Cell. 1988 Jan 15;52(1):5–6. doi: 10.1016/0092-8674(88)90523-5. [DOI] [PubMed] [Google Scholar]
- Rosen C. A. Regulation of HIV gene expression by RNA-protein interactions. Trends Genet. 1991 Jan;7(1):9–14. doi: 10.1016/0168-9525(91)90015-i. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990 Nov;10(11):6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., Lis J. T. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988 Sep 9;54(6):795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Scheer U. Changes of nucleosome frequency in nucleolar and non-nucleolar chromatin as a function of transcription: an electron microscopic study. Cell. 1978 Mar;13(3):535–549. doi: 10.1016/0092-8674(78)90327-6. [DOI] [PubMed] [Google Scholar]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972 Apr;27(2):353–365. [PubMed] [Google Scholar]
- Spencer C. A., Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990 Jun;5(6):777–785. [PubMed] [Google Scholar]
- Spradling A., Pardue M. L., Penman S. Messenger RNA in heat-shocked Drosophila cells. J Mol Biol. 1977 Feb 5;109(4):559–587. doi: 10.1016/s0022-2836(77)80091-0. [DOI] [PubMed] [Google Scholar]
- Sugisaki H., Maekawa Y., Kanazawa S., Takanami M. New restriction endonucleases from Acetobacter aceti and Bacillus aneurinolyticus. Nucleic Acids Res. 1982 Oct 11;10(19):5747–5752. doi: 10.1093/nar/10.19.5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. H., Elgin S. C. Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988 Jul;7(7):2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C. S., Burtis K. C., Hogness D. S. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell. 1990 Apr 6;61(1):101–111. doi: 10.1016/0092-8674(90)90218-4. [DOI] [PubMed] [Google Scholar]
- Török I., Karch F. Nucleotide sequences of heat shock activated genes in Drosophila melanogaster. I. Sequences in the regions of the 5' and 3' ends of the hsp 70 gene in the hybrid plasmid 56H8. Nucleic Acids Res. 1980 Jul 25;8(14):3105–3123. doi: 10.1093/nar/8.14.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez J. M., Lindquist S. hsp70: nuclear concentration during environmental stress and cytoplasmic storage during recovery. Cell. 1984 Mar;36(3):655–662. doi: 10.1016/0092-8674(84)90345-3. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Wu C., Wilson S., Walker B., Dawid I., Paisley T., Zimarino V., Ueda H. Purification and properties of Drosophila heat shock activator protein. Science. 1987 Nov 27;238(4831):1247–1253. doi: 10.1126/science.3685975. [DOI] [PubMed] [Google Scholar]
- Zajac-Kaye M., Gelmann E. P., Levens D. A point mutation in the c-myc locus of a Burkitt lymphoma abolishes binding of a nuclear protein. Science. 1988 Jun 24;240(4860):1776–1780. doi: 10.1126/science.2454510. [DOI] [PubMed] [Google Scholar]