Abstract
Background
MicroRNAs (miRNAs) are important diagnostic and prognostic markers in cancer. In the study reported here, we analyzed the potential relationship between miRNA expression and extrathyroidal invasion in papillary thyroid carcinoma (PTC).
Methods
Samples from 91 patients with PTC were collected from January 2008 to April 2012 at our hospital. To detect the levels of miRNA expression in fresh frozen tissues from patients with extrathyroidal invasion (n = 3) and non-extrathyroidal invasion (n = 3), miRNA array was carried out. The upregulated miRNAs between the two groups were confirmed by using real-time reverse transcriptase polymerase chain reaction.
Results
The levels of miR-146b, miR-221, miR-222, and miR-135b were significantly higher in the extrathyroidal invasion group than in the non-extrathyroidal invasion group (P = 0.001, 0.019, 0.004, and 0.006, respectively). In addition, the miR-146b expression level was significantly higher in the massive extrathyroidal invasion group than in the minimal extrathyroidal invasion group (P = 0.016). The expression levels of miR-146b, miR-222, and miR-135b were significantly associated with tumor size (P = 0.018, 0.008, and 0.024, respectively). The upregulation of miR-146b and miR-222 was significantly associated with higher tumor-node-metastasis stage (P = 0.004 and 0.0001, respectively). The expression level of miR-222 was also correlated with age and sex (P = 0.048 and 0.002, respectively).
Conclusion
Our results reveal that the expression of four miRNAs correlated with extrathyroidal invasion and other clinicopathologic features in PTCs, which may support their potential clinical value as prognostic biomarkers for PTCs.
Keywords: PTC, miR-146b, miR-221, miR-222, miR-135b
Introduction
Papillary thyroid carcinoma (PTC) is the most common type of thyroid malignancy, accounting for more than 80% of all thyroid cancers. Moreover, the incidence of PTC has increased dramatically in the past several decades.1 In general, the prognosis of patients with PTC is believed to be relatively good. However, certain clinicopathologic features have been associated with a poor prognosis, such as older age at diagnosis, sex, larger primary tumor (≥3 cm), extrathyroidal invasion, lymph node metastasis, advanced tumor-node-metastasis (TNM) stage, and multicentricity.2,3 In particular, studies have demonstrated extrathyroidal invasion as a significant independent prognostic factor for survival, with PTC with extrathyroidal invasion usually having incomplete resection for the invasion into other structures and a high incidence of recurrence and metastasis.4,5 The focus of the present research was the identification of biomarkers indicating the aggressiveness of PTC.
MicroRNAs (miRNAs) are conserved 18–25 nucleotide non-coding RNAs. They regulate protein expression at the post-transcriptional level mainly by annealing with the 3′ UTR of the target messenger RNA and interfering with its translation and/or stability.6 Some miRNAs regulate cellular proliferation, differentiation, and apoptotic processes that are important in cancer aggravation and serve as biomarkers for diagnosis and prognosis in many cancers.7,8 A few studies have reported the increased expression of some miRNAs in PTCs with aggressive properties and the association of these miRNA expressions with the clinicopathologic features.3,9,10 However, Ito et al11,12 further categorized the extrathyroid invasion into two types: minimal invasion (invasion to the sternothyroid muscle and/or perithyroid soft tissues) and massive invasion (invasion to the recurrent laryngeal nerve, trachea, esophagus, jugular vein, and/or subcutaneous soft tissues). They demonstrated that the disease-free survival of patients with minimal invasion was similar to that of those with no invasion, suggesting that minimal extrathyroidal invasion is not a risk factor for survival. Further investigation is needed to explore the miRNA biomarkers of different types of extrathyroidal invasion to guide the optimization of therapy.
The purpose of the study reported here was to investigate the upregulated miRNAs in PTCs with different types of extrathyroidal invasion, and to find the association between miRNA expression and the other clinicopathologic parameters of PTC using miRNA array and real-time polymerase chain reaction (PCR).
Materials and methods
Ethical approval and informed consent
The Ethics Committee of The First Affiliated Hospital of China Medical University, Shenyang, People’s Republic of China approved our study protocol and written informed consent was obtained from all study participants.
Patients
We performed a review of 91 PTC patient records from January 2008 to April 2012 at our hospital. Patients were aged from 13 to 77 years (mean 43.39 ± 12.88 years) and the sample included 30 men and 61 women. Fifty-nine patients had a total thyroidectomy or lobectomy with cervical dissection, 32 patients had had a total thyroidectomy or lobectomy with lateral lymph node dissection, and there had been histologic lymph node metastasis in 52 cases. The clinicopathological characteristics of the patients are shown in Table 1. All the pathological results for these patients were verified by the Department of Pathology in the First Affiliated Hospital of China Medical University. We classified the cancer stage according to the TNM staging system of the seventh edition of the AJCC Cancer Staging Manual (followed the NCCN guidelines on thyroid carcinoma).
Table 1.
Clinicopathological features of papillary thyroid carcinomas (PTCs) in this study
| Clinicopathological feature | Number |
|---|---|
| Sex (male:female) | 30:61 |
| TNM stage (AJCC) | |
| I | 41 |
| II | 15 |
| III | 12 |
| IV | 23 |
| Tumor size (cm) | 2.95 ± 1.26 |
| Microcarcinomaa | 14 |
| Multicentricityb | 14 |
| Extrathyroidal invasion (structures involved) | 25 |
| Perithyroid muscle and soft tissues only | 14 |
| Recurrent laryngeal nerve | 5 |
| Trachea | 4 |
| Esophagus | 2 |
| Carotid artery sheath | 2 |
| Jugular vein | 6 |
| Lymph node metastasis | 52 |
| Length of follow-up, mean years (range) | 1.81 (0.5–3.5) |
| Recurrence of lymph node metastasis | 5 |
| Distant metastasis | 3 |
Notes:
Tumors had a maximum diameter of ≤10 mm;
the number of PTCs was >2.
Abbreviations: AJCC, American Joint Committee on Cancer Staging; PTC, papillary thyroid carcinoma; TNM, tumor-node metastasis.
Tumors in our study were divided into two groups: extrathyroid invasion (EX) and non-extrathyroid invasion (NE). In addition, the EX group was further categorized into two subgroups: EX1 (minimal invasion, such as invasion to the perithyroid muscle and soft tissues) and EX2 (massive invasion, such as invasion to the recurrent laryngeal nerve, trachea, esophagus, jugular vein, and/or subcutaneous soft tissues). Shave resection was performed for three patients with trachea involvement and one with esophagus involvement. All remaining patients had had complete tumor resection.
Tissue samples
Eight fresh frozen tissues from six patients were used to undertake the miRNA array. Three samples were from the EX2 subgroup, three samples from the NE group, and two paired normal thyroid tissues (which were located more than 1 cm away from cancer margin and confirmed normal by pathology) were used. Samples of fresh frozen tissue from the remaining 85 PTC patients (14 in the EX1 subgroup, eight in the EX2 subgroup, and 63 in the NE group) were used for real-time PCR as the external validation cohort. All samples were frozen in liquid nitrogen until the extraction of RNA. None of the patients received radiation therapy or chemotherapy before the operation.
miRNA expression array
miRNA array was executed using an Agilent Human miRNA Microarray Kit (8*60 K, v 16.0; Agilent Technologies, Santa Clara, CA, USA), which contained capture probes targeting 1205 human miRNA sequences registered in the miRBase database (http://www.mirbase.org). Total RNA was extracted and purified using a mirVana™ miRNA Isolation Kit (Ambion, Austin, TX, USA) following the manufacturer’s instructions. The integrity of the obtained total RNA was checked by determining the RNA integrity number using an Agilent Bioanalyzer 2100 (Agilent Technologies). miRNA molecules in total RNA was labeled using an miRNA Complete Labeling and Hyb Kit (Agilent Technologies) following the manufacturer’s instructions for labeling. Each slide was hybridized with 100 ng Cy3-labeled RNA using the miRNA Complete Labeling and Hyb Kit in a hybridization oven (Agilent Technologies) at 55°C and 20 rpm for 20 hours, according to the manufacturer’s instructions for hybridization. Then the slides were washed in staining dishes (Cat#121, Thermo Scientific, Waltham, MA, USA) using a Gene Expression Wash Buffer Kit (Agilent Technologies). Slides were scanned using an Agilent Microarray Scanner (Agilent Technologies) with Feature Extraction software (v 10.7) using the default settings. Raw data were normalized using a quantile algorithm with GeneSpring Software (v 11.0; Agilent Technologies).
RNA extraction
Small RNA was extracted from snap-frozen PTC tissues using RNAiso for Small RNA (Takara, Dalian, People’s Republic of China) with some modifications to the manufacturer’s instructions. Briefly, the samples were centrifuged at 12,000 g for 10 minutes and the upper aqueous phase was transferred. Following a wash with 75% ethanol, the RNA was re-suspended in 30 μL diethylpyrocarbonate-treated water and stored at −80°C. The concentration and purity of the RNA was determined using a NanoDrop 1000 spectrophotometer (Thermo Scientific) at 260 nm and 280 nm. Only the RNA samples with a ratio of A260/A280 > 1.8 were considered suitable for further experimentation.
miRNA quantitative reverse-transcription PCR
According to the results of miRNA array, selected miRNAs were validated in a combined cohort comprising the original discovery cohort (n = 6) and an external cohort (n = 85) using SYBR Green I miRNA assays using a LightCycler® 480 Real-Time PCR System (Roche Applied Science, Penzberg, Germany). The miRNAs were selected based on the greatest fold change and the significant difference between probe and background signal. Briefly, 10 ng of small RNA was reverse transcribed using a miRNA cDNA Synthesis Kit (Takara Life Technologies). The cycle parameters for the reverse transcription (RT) reaction were 37°C for 60 minutes and 85°C for 5 seconds followed by maintenance at 4°C. Following the RT reactions, 2 μL of complementary DNA was used for PCR. The PCR was conducted at 95°C for 30 seconds, followed by 35 cycles of 95°C for 5 seconds and 60°C for 20 seconds in the LightCycler 480 Real-Time PCR System. The real-time PCR results were analyzed and expressed as the relative miRNA level using U6 small nuclear RNA for normalization purposes; the threshold cycle (Ct) range for the control U6 small RNA ranged from 11 to 12 cycles. The delta Ct (ΔCt) values in each sample represented the relative expression amount of miRNA: ΔCt = Ct (miRNA) − Ct (U6). The fold expression changes between groups were determined using the comparative Ct method (2−ΔΔCT).13 Tissues from group NE were used as calibrator samples. All experiments were performed in triplicate.
Statistical analysis
All calculations were performed with SPSS for Windows (v 16.0; IBM, Armonk, NY, USA). miRNA array data were analyzed using a two-sample Student’s t-test (carried out by Agilent Technologies). The same method was chosen to determine whether the difference in mean ΔCt values between the independent groups was statistically significant. Different miRNA expression levels according to clinicopathologic features were analyzed using the Mann-Whitney U test. Differences of P < 0.05 were considered statistically significant.
Results
miRNA array analysis
The relative expression levels of miRNA were initially evaluated with miRNA array in eight PTC tissues: three from patients in the EX group, three from patients in the NE group, and two paired normal thyroid tissues. A subset of upregulated miRNAs (miR-551b, miR-146b, miR-222, miR-221, miR-31, miR-96, miR-21, miR-135b, and miR-181b) and downregulated miRNAs (miR-7, miR-138, miR-150, miR-1275, and miR-199-5p) was found in all six tumors when compared with normal thyroid tissues. The fold-change data are shown in Table 2. In addition, we found miR-32, miR-146b, miR-221, miR-222, and miR-135b were significantly upregulated in EX PTCs compared with NE tumors (P = 0.03, 0.03, 0.07, 0.04, and 0.01, respectively), and miR-663, miR-214*, miR-299-5p, and miR-939 showed stronger downregulation in EX tumors than in NE PTCs (P = 0.0003, 0.0001, 0.02, and 0.01, respectively) (Figure 1 and Table 3).
Table 2.
Expression levels in papillary thyroid carcinoma (PTC) and normal group detected by microRNA (miRNA) array
| miRNA probe | Fold change (PTC/normal) |
|---|---|
| hsa-miR-551b | 30.30 |
| hsa-miR-146b | 16.47 |
| hsa-miR-222 | 11.47 |
| hsa-miR-221 | 11.14 |
| hsa-miR-31 | 5.86 |
| hsa-miR-96 | 4.78 |
| hsa-miR-21 | 3.08 |
| hsa-miR-135b | 2.09 |
| hsa-miR-181b | 2.06 |
| hsa-miR-7 | 0.17 |
| hsa-miR-138 | 0.21 |
| hsa-miR-150 | 0.25 |
| hsa-miR-1275 | 0.37 |
| hsa-miR-199-5p | 0.37 |
Figure 1.
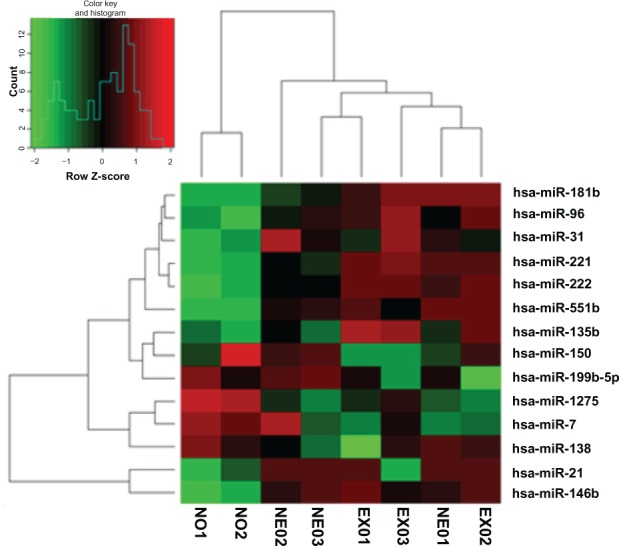
Cluster heat map of the relative expression of microRNAs (mRNAs) in papillary thyroid carcinoma (with extrathyroidal invasion [EX] and non-extrathyroidal invasion [NE]) and normal (N) samples.
Notes: Each row indicates a miRNA and each column indicates a sample. The miRNA-subset tree is on the left and the sample subset is at the top.
Table 3.
Expression levels in the EX (n = 3) and NE (n = 3) groups detected by microRNA (miRNA) array
| miRNA probe | Fold change (EX/NE) | P |
|---|---|---|
| hsa-miR-32 | 48.03 | 0.03 |
| hsa-miR-146b | 13.94 | 0.03 |
| hsa-miR-221 | 2.71 | 0.07 |
| hsa-miR-222 | 2.03 | 0.04 |
| hsa-miR-135b | 1.79 | 0.01 |
| hsa-miR-663 | 0.01 | 0.0003 |
| hsa-miR-214* | 0.01 | 0.0001 |
| hsa-miR-299-5p | 0.03 | 0.02 |
| hsa-miR-939 | 0.35 | 0.01 |
Abbreviations: EX, extrathyroidal invasion; NE, non-extrathyroidal invasion.
Real-time reverse-transcription PCR validation of selected miRNAs
Five upregulated miRNAs (miR-32, miR-146b, miR-221, miR-222, and miR-135b) were selected for real-time PCR validation in both the miRNA array (n = 6) and external cohorts (n = 85). Twenty-five PTC samples were collected from patients with EX and 66 from patients with NE. In concordance with the array data, we found that the expression levels of miR-146b, miR-221, miR-222, and miR-135b in group EX were significantly higher than in group NE. The fold-change values were 2.5, 2.1, 1.9, and 1.6, respectively (Figure 2). These results suggest that higher expression of these four miRNAs was associated with a higher risk of EX as compared with lower expression. However, we failed to find a significant difference in miR-32 expression between the EX and NE groups.
Figure 2.
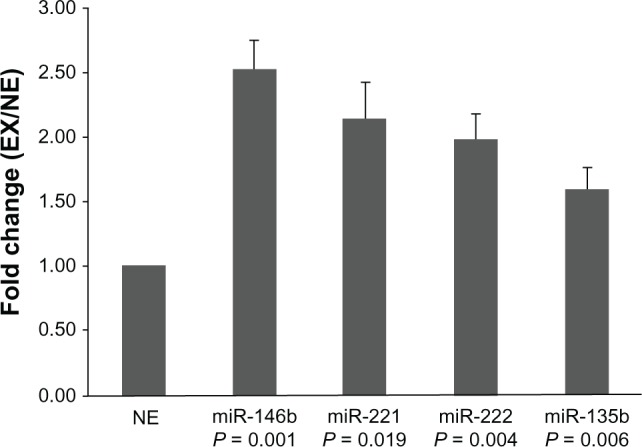
Expression of miR-146b, miR-221, miR-222, and miR-135b.
Notes: These four microRNAs (miRNAs) were significantly more highly expressed in the extrathyroid invasion (EX) group (n = 25) than in the non-extrathyroid invasion (NE) group (n = 66) (P < 0.05). The fold-change values of these miRNAs were calculated by comparing the mean ΔCt in the EX and NE groups, assuming that the mean ΔCt value in the NE group was equal to 1. ΔCt = Ct (miRNA) − Ct (U6).
The relative expression levels of these four miRNAs in groups EX1 and EX2 were compared. Statistical analyses showed that the expression of miR-146b was significantly higher in group EX2 than in group EX1 (P = 0.016) (Figure 3). The differences of the other miRNAs were not significant. Further analysis revealed significant associations between miR-146b, miR-221, miR-222, and miR-135b expression and other clinicopathologic features. Dividing the patients into two groups according to tumor maximal diameter < 3 cm and ≥3 cm,3 we found that the expression levels of miR-146b, miR-222, and miR-135b were significantly associated with tumor size (P = 0.018, 0.008, and 0.024, respectively). The expression levels of miR-146b and miR-222 were significantly higher in the TNM stage III to IV group than in the I to II group (P = 0.004 and 0.0001, respectively). We also found the expression level of miR-222 was correlated with age and sex (P = 0.048 and 0.002, respectively). The expression of miRNAs was not related to other clinicopathological features such as microcarcinoma, multicentricity, or lymph node metastasis (Table 4).
Figure 3.
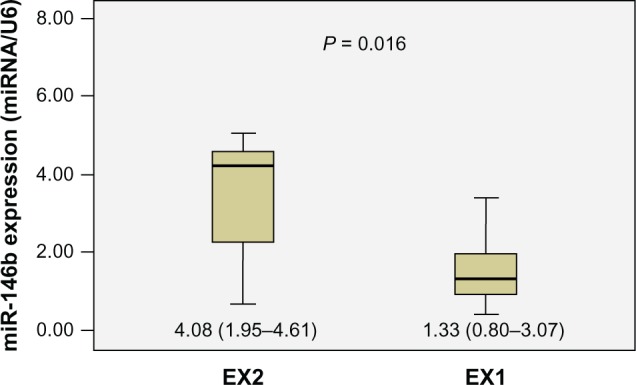
Diagrammatic representation of Mann–Whitney U test results, showing that the relative expression of miR-146b in the EX2 group (n = 11, median = 4.08, interquartile range 1.95–4.61) was significantly higher than that in the EX1 group (n = 14, median = 1.33, interquartile range 0.80–3.07) (P = 0.016).
Abbreviations: EX1, minimal extrathyroid invasion; EX2, massive extrathyroid invasion.
Table 4.
Expression levels of miR-146b, miR-221, miR-222, and miR-135b in papillary thyroid carcinomas with different clinicopathologic features
| Clinicopathologic feature | N | miR-146b (mean ΔCt) | P | miR-221 (mean ΔCt) | P | miR-222 (mean ΔCt) | P | miR-135b (mean ΔCt) | P |
|---|---|---|---|---|---|---|---|---|---|
| Age (years) | |||||||||
| <45 | 51 | 0.88 (0.43–1.95) | 0.114 | 0.06 (0.03–0.10) | 0.612 | 0.06 (0.03–0.09) | 0.048* | 0.09 (0.07–0.17) | 0.761 |
| ≥45 | 40 | 1.22 (0.79–1.96) | 0.06 (0.04–0.09) | 0.07 (0.04–0.12) | 0.10 (0.06–0.14) | ||||
| Sex | |||||||||
| Male | 30 | 1.21 (0.78–2.11) | 0.173 | 0.07 (0.04–0.13) | 0.053 | 0.09 (0.06–0.13) | 0.002* | 0.10 (0.07–0.17) | 0.455 |
| Female | 61 | 1.04 (0.46–1.88) | 0.05 (0.03–0.09) | 0.05 (0.03–0.08) | 0.09 (0.06–0.14) | ||||
| TNM staging (AJCC) | |||||||||
| I, II | 56 | 0.83 (0.44–1.67) | 0.004* | 0.05 (0.03–0.08) | 0.075 | 0.05 (0.03–0.08) | 0.0001* | 0.09 (0.06–0.14) | 0.230 |
| III, IV | 35 | 1.55 (0.90–2.86) | 0.06 (0.04–0.15) | 0.10 (0.05–0.15) | 0.10 (0.07–0.20) | ||||
| Tumor size (cm) | |||||||||
| <3 | 56 | 0.91 (0.48–1.67) | 0.018* | 0.05 (0.04–0.09) | 0.108 | 0.05 (0.03–0.08) | 0.008* | 0.08 (0.06–0.13) | 0.024* |
| ≥3 | 35 | 1.31 (0.75–2.96) | 0.08 (0.04–0.12) | 0.07 (0.06–0.16) | 0.09 (0.07–0.21) | ||||
| Microcarcinoma | |||||||||
| Yes | 14 | 1.08 (0.54–1.67) | 0.652 | 0.06 (0.04–0.10) | 0.746 | 0.05 (0.03–0.08) | 0.187 | 0.10 (0.07–0.13) | 0.403 |
| No | 77 | 1.08 (0.62–1.96) | 0.06 (0.03–0.10) | 0.09 (0.04–0.11) | 0.13 (0.06–0.15) | ||||
| Multicentricity | |||||||||
| Yes | 14 | 0.85 (0.42–2.60) | 0.648 | 0.05 (0.02–0.11) | 0.478 | 0.07 (0.03–0.12) | 0.826 | 0.07 (0.06–0.10) | 0.096 |
| No | 77 | 1.16 (0.63–1.95) | 0.06 (0.04–0.10) | 0.07 (0.04–0.10) | 0.10 (0.07–0.15) | ||||
| Lymph node metastasis | |||||||||
| Yes | 52 | 1.03 (0.68–1.95) | 0.639 | 0.05 (0.03–0.09) | 0.267 | 0.07 (0.03–0.11) | 0.968 | 0.09 (0.06–0.13) | 0.109 |
| No | 39 | 1.28 (0.27–1.95) | 0.07 (0.04–0.11) | 0.07 (0.04–0.10) | 0.11 (0.07–0.23) | ||||
Notes: All data were calculated using the Mann-Whitney U test The data are shown as median (interquartile range). ΔCt = Ct (miRNA) − Ct (U6).
Statistically significant (P < 0.05).
Abbreviations: AJCC, American Joint Committee on Cancer Staging; PTC, papillary thyroid carcinoma; TNM, tumor-node metastasis.
Discussion
Invasion of surrounding structures is rare (nearly 10%) in differentiated thyroid carcinoma, and patients with EX show considerable postoperative mortality as a result of the high incidence of residual cancer.4,5 Thus, to guide the optimization of therapy and long-term follow-up, identifying novel biomarkers indicating EX in PTC is becoming a focus of research. Cancer-specific miRNAs in PTC have been identified in many studies.14–16 Not only is the spectrum of miRNA expressed in tumor tissues/cells significantly different from that of paired normal tissues/cells, but it also has significantly different clinicopathologic features.3,10
With this in mind, we performed a miRNA array analysis in a cohort of EX and NE tumor samples. The validation of selected miRNAs using real-time PCR confirmed the results of the miRNA array. We revealed that the expression levels of miR-146b, miR-221, miR-222, and miR-135b were significantly higher in PTCs with EX than in those without EX. This is in agreement with the results of previous studies.3,10 Although the difference in expression of miR-221 was not significant in the miRNA array results, a significant difference was found using real-time PCR. The different results may be attributed to the different sample sizes; the mean miRNA expression level of a relatively large sample (ie, in this study, n = 91) may be more reliable than that of a small sample (ie, n = 6). The mean fold changes of miR-146b detected by real-time quantitative reverse-transcription PCR were far smaller than the corresponding mean fold changes obtained by miRNA microarray. The remaining miRNA expression trends, as detected by both methods, were similar. By further comparing the miRNA expression with various clinicopathologic features, we found that overexpression of miR-146b was associated with tumor size, and there was an association between miR-222 expression and sex, age, TNM stage, and tumor size in PTCs, which was not reported in Chou et al’s study.3 However, the association of miR-146b expression with sex and of miR-221 expression with TNM stage and lymph node metastasis that has been shown in previous reports was not found.3,10
Ito et al11,12 reported that massive extrathyroidal invasion is a risk factor for lymph node metastasis and an indicator for prophylactic lateral neck dissection; however, minimal extrathyroid invasion was not found to be a prognostic factor and the disease-free survival of patients with minimal extension was similar to that of those with no extension. There was no need to expand the resection region in minimal extension PTCs. Accordingly, we compared the expression levels of four miRNAs in PTCs with minimal invasion (EX1) and with massive invasion (EX2). The results showed that the expression of miR-146b was significantly higher in group EX2 than in group EX1. To our knowledge, this is the first study to have demonstrated the association between miRNA expression and different types of invasion. Our results have potential value for application in the identification of the degree of extrathyroid invasion, which would help guide practitioners to the optimal operation strategy and decrease the incidence of complications.
Several studies have reported the function and mechanism of deregulated miRNAs in PTC. Recently, Chou et al17 demonstrated that miR-146b is a novel prognostic factor of PTC. They found that patients with higher miR-146b expression had significantly lower overall survival rates compared with those with lower miR-146b expression. The associated hazard ratio was 3.92. In vitro gain-of-function experiments in a BCPAP human papillary thyroid cancer cell line showed that miR-146b significantly promoted cell migration and invasiveness. Kim et al18,19 have investigated the function and mechanism of miR-221. Microarray analysis and a Gaussia luciferase reporter system were used to find the target genes that are directly and indirectly regulated by miR-221. They found that messenger RNA levels of HOXB5 were significantly downregulated by miR-221. In addition, in cells and nude mice, Gaussia luciferase activities using Cytomegalovirus/Gaussia luciferase 3′UTR of HOXB5 systems were downregulated dose dependently by endogenous or exogenous miR-221.
miR-135b has been studied in other cancers. Using miRNA array and real-time PCR, Wang et al20 found that the expression level of miR-135b was significantly upregulated in colonic cancer tissues compared with paracancerous tissues. Singh et al21 reported that the overexpression of miR-135b was associated with the reduction of tumor suppressive E-cadherin expression in colon carcinoma cell lines. Munding et al22 identified miR-135b as one of the new miRNA biomarkers for pancreatic ductal adenocarcinoma in human pancreas acinar and ductal cells by using miRNA array and real-time PCR, which has improved early diagnosis modalities and treatment strategies. Our study has shown the significant overexpression of miR-135b in PTCs with EX and the association between the expression level of miR-135b and tumor size, demonstrating the potential importance of miR-135b in determining the aggressive features of PTCs.
Limitations
The present study has some limitations. First, the mechanisms by which the selected miRNAs contribute to the EX of PTCs were not investigated. Further, the relatively small sample size and short-term follow-up meant that the prognosis information was insufficient. Large-scale studies with long-term follow-up will be necessary to confirm our results.
Conclusion
Our study has revealed that the expression of a subset of miRNAs is correlated with extrathyroidal invasion and other clinicopathologic features in PTCs. In particular, miR-146b expression was associated with different types of extrathyroidal invasion, which may indicate that it has potential clinical value as a prognostic biomarker for PTC. Further studies are required to explore the mechanism underlying the upregulation of the expression of these miRNAs with aggressive features in PTCs.
Author contributions
Hao Zhang designed the study and Zhihong Wang performed research and wrote the first draft. Zhihong Wang, Liang He, and Wenwu Dong collected and analyzed the data. Jing Li, Zhongyan Shan, and Weiping Teng contributed to the design and interpretation of the study and to further drafts. Hao Zhang is the guarantor.
Acknowledgment
This research was supported by the Innovation Team Project of the Education Department of Liaoning Province (no LT2010102) and the Liaoning Millions of Talents Project (no 2010921070).
Footnotes
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Chen AY, Jemal A, Ward EM. Increasing incidence of differentiated thyroid cancer in the United States, 1988–2005. Cancer. 2009;115(16):3801–3807. doi: 10.1002/cncr.24416. [DOI] [PubMed] [Google Scholar]
- 2.Voutilainen PE, Multanen MM, Leppäniemi AK, Haglund CH, Haapiainen RK, Franssila KO. Prognosis after lymph node recurrence in papillary thyroid carcinoma depends on age. Thyroid. 2001;11(10):953–957. doi: 10.1089/105072501753211028. [DOI] [PubMed] [Google Scholar]
- 3.Chou CK, Chen RF, Chou FF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20(5):489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 4.Nishida T, Nakao K, Hashimoto T. Local control in differentiated thyroid carcinoma with extrathyroidal invasion. Am J Surg. 2000;179(2):86–91. doi: 10.1016/s0002-9610(00)00275-0. [DOI] [PubMed] [Google Scholar]
- 5.McCaffrey TV, Bergstralh EJ, Hay ID. Locally invasive papillary thyroid carcinoma: 1940–1990. Head Neck. 1994;16(2):165–172. doi: 10.1002/hed.2880160211. [DOI] [PubMed] [Google Scholar]
- 6.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yip L, Kelly L, Shuai Y, et al. Mi-croRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol. 2011;18(7):2035–2041. doi: 10.1245/s10434-011-1733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou YL, Liu C, Dai XX, Zhang XH, Wang OC. Overexpression of miR-221 is associated with aggressive clinicopathologic characteristics and the BRAF mutation in papillary thyroid carcinomas. Med Oncol. 2012;29(5):3360–3366. doi: 10.1007/s12032-012-0315-8. [DOI] [PubMed] [Google Scholar]
- 11.Ito Y, Tomoda C, Uruno T, et al. Prognostic significance of extrathyroid extension of papillary thyroid carcinoma: massive but not minimal extension affects the relapse-free survival. World J Surg. 2006;30(5):780–786. doi: 10.1007/s00268-005-0270-z. [DOI] [PubMed] [Google Scholar]
- 12.Ito Y, Tomoda C, Uruno T, et al. Minimal extrathyroid extension does not affect the relapse-free survival of patients with papillary thyroid carcinoma measuring 4 cm or less over the age of 45 years. Surg Today. 2006;36(1):12–18. doi: 10.1007/s00595-005-3090-8. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93(5):1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13(2):497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 17.Chou CK, Yang KD, Chou FF, et al. Prognostic Implications of miR-146b Expression and Its Functional Role in Papillary Thyroid Carcinoma. J Clin Endocrinol Metab. 2013;98(2):E196–E205. doi: 10.1210/jc.2012-2666. [DOI] [PubMed] [Google Scholar]
- 18.Kim HJ, Kim YH, Lee DS, Chung JK, Kim S. In vivo imaging of functional targeting of miR-221 in papillary thyroid carcinoma. J Nucl Med. 2008;49(10):1686–1693. doi: 10.2967/jnumed.108.052894. [DOI] [PubMed] [Google Scholar]
- 19.Kim HJ, Chung JK, Hwang do W, Lee DS, Kim S. In vivo imaging of miR-221 biogenesis in papillary thyroid carcinoma. Mol Imaging Biol. 2009;11(2):71–78. doi: 10.1007/s11307-008-0188-6. [DOI] [PubMed] [Google Scholar]
- 20.Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11(1):50–54. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 21.Singh N, Liu G, Chakrabarty S. Isolation and characterization of calcium sensing receptor null cells: A highly malignant and drug resistant phenotype of colon cancer. Int J Cancer. 2013;132(9):1996–2005. doi: 10.1002/ijc.27902. [DOI] [PubMed] [Google Scholar]
- 22.Munding JB, Adai AT, Maghnouj A, et al. Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma. Int J Cancer. 2012;131(2):E86–E95. doi: 10.1002/ijc.26466. [DOI] [PubMed] [Google Scholar]


