Abstract
Objective
CA 15-3 is a traditional biomarker for advanced breast cancer with limited sensitivity for early stage patients. In order to increase the sensitivity for early detection, in this study, we introduced novel tumor-associated autoantibodies that were measured concurrently with serum CA 15-3 to evaluate their diagnostic advantage in breast cancer.
Methods
We investigated a T7 breast cancer complementary deoxyribonucleic acid (cDNA) phage library for tumor-associated antigens using sera from normal and breast cancer patients. Identified novel tumor-associated antigens phage proteins were then used to develop enzyme-linked immunosorbent assays to measure corresponding autoantibodies in 150 breast cancer, 150 normal, and 40 other cancer (non breast) patient serum samples. Meanwhile, the same samples were measured for CA 15-3 concentrations. Receiver operating characteristic curve analysis was used to evaluate the predictive accuracies of single markers as well as combined markers.
Results
Sequencing analysis revealed that two phage-expressed proteins were within the open reading frame and had significant homology to proteins heterogeneous nuclear ribonucleoproteins F (hnRNPF) and ferritin heavy chain (FTH1). Autoantibodies against hnRNPF and FTH1 alone were significantly higher in patients than in control serum samples (P < 0.01), and the area under the curve for hnRNPF and FTH1 alone was 0.73 and 0.69, respectively. However, when the two autoantibody biomarkers were analyzed in combination with serum CA 15-3 values, the area under the curve increased to 0.93, and the optimal sensitivity and specificity became 89.3% and 93.8%, respectively. Further messenger ribonucleic acid (mRNA) analysis showed that hnRNPF and FTH1 were significantly upregulated in tumor tissues.
Conclusion
Our results indicated that combined serologic biomarkers of tumor-associated antigens with autoantibodies may improve the diagnostic accuracy of breast cancer.
Keywords: tumor-associate antigen, autoantibody, phage display, tumor marker, breast cancer, early detection
Introduction
Breast cancer is the most frequently diagnosed life-threatening cancer and the second leading cause of cancer deaths among women in the US.1 Further reduction in mortality will require successful strategies for early detection of this disease. Serological biomarkers that may accurately determine tumor onset are considered a promising approach for early cancer detection. Most of the efforts in the past have been centered on the discovery and characterization of circulating tumor-associated antigens (TAAs). Two clinically used breast cancer antigens, CA 15-3 and CA 27–29, are elevated in less than 10% of early stage and 75% of advanced stage breast cancer patients. Neither is recommended for screening nor diagnosis of breast cancer onset.2
In addition to the detection of serum antigens, humoral immune responses to tumor progression have recently been demonstrated to have reliable accuracy for early cancer detection.3–6 Autoantibodies to p53 have been reported in patients with early stage ovarian, colorectal, and breast cancers,7,8 and a panel of serum antibodies can detect non-small cell lung cancer 5 years prior to autoradiograph detection.9 Thirty percent of patients with ductal carcinoma in situ in which the proto-oncogene HER-2/neu is overexpressed have serum antibodies specific to this protein.10,11
Although autoantibodies have showed promising results as novel diagnostic biomarkers for the early detection of many cancers, the low sensitivities and specificities have limited rapid clinical application. The presence of p53 autoantibodies has been observed in only 15% of patients with breast cancer.12–14 In addition, p53 autoantibodies have also been found in patients with other malignancies and inflammatory conditions,15,16 thus the humoral response to p53 is nonspecific for breast cancer. A humoral response to the 90 kDa heat shock protein has been associated with early breast cancer existence,17 however, it has also been reported to be associated with other diseases as well. Therefore, it is logical and practical to combine both serological antigen and antibody biomarkers to improve sensitivity and specificity.
In this study, we measured serum CA 15-3 levels concurrently with novel autoantibodies from patients and control serum samples to evaluate their diagnostic advantage in breast cancer detection. The combined analysis will explore whether both novel autoantibodies and CA 15-3 would be complementary for better diagnostic accuracy over any single biomarker alone.
Materials and methods
Clinical samples
A total of 155 breast cancer patient, 155 normal control, and 40 other (non breast) cancer patient serum samples were collected from Shaoxing People’s Hospital, Zhejiang, People’s Republic of China. In addition, 40 specimens of breast tumor, 40 adjacent non tumor, and 32 benign breast tumor tissues were collected from the same hospital. Detailed information of the specimens is listed in Table 1. All the clinical samples were collected under approval by the Institutional Review Board of Shaoxing People’s Hospital.
Table 1.
Characteristics of serum and tissue specimens
| Age | Breast cancer sera (n = 155) |
Control sera (n = 155) |
Breast cancer tissues (n = 40) |
Benign tumor tissues (n = 32)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | Stage
|
Number | Number | Stage
|
Number | |||||
| I/II | III | IV | I/II | III | IV | |||||
| ≤45 | 30 | 5 | 16 | 9 | 35 | 7 | 4 | 3 | 0 | 5 |
| 45–54 | 43 | 7 | 24 | 12 | 41 | 10 | 5 | 4 | 1 | 9 |
| 55–64 | 46 | 6 | 25 | 15 | 44 | 15 | 6 | 7 | 2 | 12 |
| 65–75 | 24 | 4 | 11 | 9 | 25 | 8 | 4 | 3 | 1 | 6 |
| ≥75 | 12 | 1 | 6 | 5 | 10 | 0 | 0 | 0 | 0 | 0 |
Selection of TAA phage proteins
A T7 phage breast cancer not complementary deoxyribonucleic acid (cDNA) library (EMD Millipore Biosciences, Bellerica, MA, USA) was biopanned with sera from five pooled patients and five pooled normal donors to screen potential autoantigens recognized by circulating antibodies in patient sera as previously described.18 To confirm the selection of four rounds of biopanning, limiting dilutions of phages from biopan 4 were used to infect bacteria and grown on a lysogeny broth (LB) agar plate covered with 6% agarose (LB agar/agarose). Phage plaques were lifted twice by placing two nitrocellulose disk membranes (EMD Millipore) onto plaques formed on LB agar/agarose plates (4°C for 2 hours). One membrane was probed with the five pooled breast cancer patient sera, and the other membrane was probed with the five pooled normal sera (1:1000 dilution). The membranes were followed by anti-human horseradish peroxidase (HRP)-conjugated secondary antibody (1:10,000 dilution; Jackson ImmunoResearch, West Grove, PA, USA). The images were detected with electrogenerated chemiluminescence (TianGen Biotech Co, Ltd, Beijing, People’s Republic of China). Colonies corresponding to the highly immunoreactive spots on the membrane were harvested from the original LB agar/ agarose plate and amplified in E.coli as previously described.18
Sequencing and identification of TAA phage proteins
The cDNA inserts from the phage clones isolated above were polymerase chain reaction (PCR) amplified using commercially available T7 phage vector primer (EMD Millipore). The sequences are: T7 forward 5′-GGAGCTGTCGTATTCCAGTC-3′ and T7 reverse 5′-AACCCCTCAAGACCCGTTTA-3′. Sequences of unique clones were checked for the open reading frame (ORF) status in the T7 expression vector. Only the correct ORF encoded proteins were identified in the GenBank database using the BLAST search program.19
Measurement of autoantibodies against TAA phage proteins
Enzyme-linked immunosorbent assays (ELISAs) were developed using the identified phage proteins to evaluate their immunogenic reactivity with different serum samples. Ninety six well ELISA plates (Guangzhou Jet Bio-Filtration Products Co, Ltd, Guangzhou, People’s Republic of China) were separately coated with the identified ORF tumor associated proteins or T7 empty phages as a negative control (2.5 × 1010 phage/well in 1 × phosphate buffered saline [PBS]/0.1% bovine serum albumin [BSA] at 4°C o/n), blocked (PBS/1% BSA 37°C for 1 hour) and washed (PBS/Tween 20). Serum samples (1:200 diluted with 1 × PBS) from individual patients or controls were added to each well (37°C for 1 hour), the plates were washed, and then incubated with anti-human HRP secondary antibody (37°C for 1 h). Assays were developed with tetramethyl benzidine/H2O2 substrate (AMRESCO LLC, Solon, OH, USA) and stopped with 2 M H2SO4, and then read on a spectrophotometer at 450λ. Each individual serum was tested in triplicate. A total of 150 breast cancer patient, 150 normal control, and 40 cancer (non breast) patient serum samples were assayed.
Serum CA 15-3 measurement
In separate experiments, the same serum samples used for the autoantibody analysis were also tested for CA 15-3 levels using the CA 15-3 ELISA Kit from Invitrogen (Camarillo, CA, USA). The procedures were guided by the manufacturer’s manual and each serum sample was diluted 1:20 in 1 × PBS. Each sample was tested in triplicate, and the mean ± standard deviation (SD) for each sample was calculated for statistical analysis.
Analysis of TAA messenger ribonucleic acid (mRNA) expression by reverse transcription (RT)-PCR
Two novel TAAs, heterogeneous nuclear ribonucleoproteins F (hnRNPF) and ferritin heavy chain (FTH1) were identified in the sequencing analysis. To evaluate the correlation between protein expressions and antibody production, total RNA from 40 breast cancer, 40 cancer surrounding tissues, and 32 benign breast tumor tissues were extracted using the guanidinium thiocyanate-phenol-chloroform extraction (TRIZOL) reagent method. RNA of each sample was reverse transcribed to single-stranded cDNAs using Olig (dT) and M-MLV reverse transcriptase (Promega, Madison, WI, USA). The following primer sets for hnRNPF (forward: 5′-CCCTGGTCCTGCTCTGTT-3′; reverse: 5′-GGCAATGTGATCCCGTTT-3′), FTH1 (forward: 5′-TACGCCTCCTACGTTTAC-3′; reverse: 5′-GGCTTTCACCTGCTCATT-3′) and β-actin (forward: 5′-TTCCTTCTTGGGTATGGAAT-3′; reverse 5′-GAGCAATGATCTTGATCTTC-3′) were used to detect their molecular expression using semi-quantitative RT-PCR. The PCR products were subjected to 1% agarose gels and stained with ethidium bromide, and then visualized with ultraviolet light. Band intensity was analyzed using Quantity One Software (Bio-Rad Laboratories Inc, Hercules, CA, USA). All experiments were repeated three times.
Statistical analysis
To analyze the differences between patient and control samples, the absorbance of each serum sample on the ELISA plate was averaged from triplicate tests. An unequal variance t-test was run between cancer and control samples. Nonparametric receiver operating characteristic curves (ROC) were used to calculate sensitivity and specificity, and the area under the curve (AUC) with 95% confidence intervals was used for individual as well as combined markers. Logistic regression was used to calculate the optimal sensitivity and specificity of the combined analysis between CA 15-3 and autoantibody analysis. Relative mRNA expression values were normalized with β-actin expression. For all tests, P < 0.05 was considered statistically significant. All statistical analysis was performed using the SPSS software package version 16.0 (SPSS, Chicago, IL, USA).
Results
Identifying TAAs from the breast cancer phage library
In order to select tumor-associated phage-expressed proteins, a plaque-lift assay was carried out using the outputs of the biopanning. Comparisons between the duplicate plaque-lift membranes showed that the immunoreactivity of multiple phage-expressed clones exhibited higher affinity binding with antibodies in patient sera than in normal sera. Fifty putative TAA clones (darker spots) were selected for PCR and sequencing analysis. Among the 50 clones, only two were unique and expressed ORF proteins. DNA sequences of these two in frame proteins were matched to the proteins hnRNPF and FTH1. The function of hnRNPF is to provide the substrate for the processing events that pre-mRNAs undergo before becoming functional, translatable mRNAs in the cytoplasm, and hnRNPF also plays a role in the regulation of alternative splicing events.20 The FTH1 gene encodes the heavy subunit of ferritin, the major intracellular iron storage protein in prokaryotes and eukaryotes.21 Both proteins have been reported as overexpressed in adenocarcinoma of the pancreas, hepatocellular carcinoma, gastric carcinoma, and breast cancer tissues.20,21
Antibody affinities to the ORF phage proteins
To test the levels of autoantibodies against hnRNPF and FTH1 proteins, serum samples from 150 breast cancer patients, 150 normal controls, and 40 other cancer patients (non breast) were assayed using ELISAs, which were developed using the phage expressed hnRNPF and FTH1 proteins. The absorbance of OD450 (±SD) of autoantibodies against hnRNPF was 1.18 ± 0.41 in breast cancer patients and 0.86 ± 0.47 in normal controls, while FTH1 autoantibodies were 1.08 ± 0.48 in patients and 0.89 ± 0.39 in normal. Both autoantibodies were significantly higher in breast cancer patients than in normal controls (P < 0.01) determined by t-test, and were also found significantly higher in other cancer patients as well (Figure 1A). These results suggest that hnRNPF and FTH1 autoantibodies may not be breast cancer specific markers. When tested with different stage samples, these two autoantibodies also showed significant elevations both in late stage (stage II and IV, n = 50) and early stage (stage I and II, n = 23) breast cancer patients compared to the controls (n = 50), and the elevated levels were stage dependent (Figure 1B). These two autoantibodies were further tested for their abilities in distinguishing lymph node metastasis (LNM) patients and non-LNM patients. Although both antibodies showed significant higher levels in LNM patients (n = 30) than non-LNM patients (n = 30), the antibodies against hnRNPF were more significant (P < 0.01) than FTH1 (P < 0.05), suggesting that antibodies against hnRNPF might be a potential prognostic marker for breast cancer as well (Figure 1C).
Figure 1.
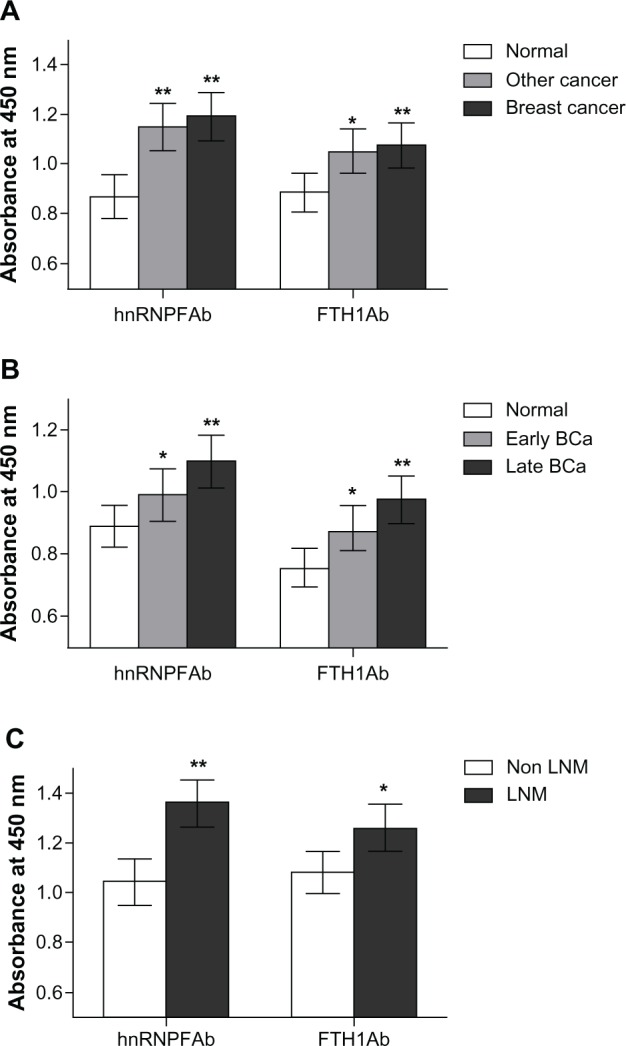
Autoantibodies against hnRNPF and FTH1 in cancer patient and normal serum samples. Antigen ELISAs were developed using hnRNPF and FTH1 phage proteins for testing corresponding autoantibodies. (A) A cohort of serum samples from 150 breast cancer patients, 150 normal controls, and 40 other cancer patients were tested. Both antibodies against hnRNPF and FTH1 were significantly higher in breast cancer and other cancer patients than in the normal controls. (B) When tested with different stage samples, these two autoantibodies also showed significant elevations both in late stage (stage II and IV, n = 50) and early stage (stage I and II, n = 23) breast cancer patients compared to controls (n = 50), and the elevated levels were stage dependent. (C) Although both antibodies showed significant higher levels in LNM patients (n = 30) than non-LNM patients (n = 30), the antibodies against hnRNPF were more significant than FTH1.
Notes: *P < 0.05; **P < 0.01.
Abbreviations: ELISA, enzyme-linked immunosorbent assay; FTH1, ferritin heavy chain; hnRNPF, heterogeneous nuclear ribonucleoproteins F; LNM, lymph node metastasis.
Diagnostic value of CA 15-3 levels combined with novel autoantibodies
The same serum samples used in the autoantibody testing were also assayed for CA 15-3 protein levels using a commercial ELISA kit. The mean OD450 (±SD) values of serologic CA 15-3 proteins were 1.130 ± 0.152 in breast cancer patients and 0.871 ± 0.141 in control individuals (P < 0.01). ROC curves were plotted to identify an optimal cutoff value that would distinguish case from control samples. According to the ROC curve, the AUC for CA 15-3 was 0.792, whereas the optimal sensitivity was 69.1% and the specificity was 89.4% when a cutoff value was set at OD450 = 0.986. The results were then analyzed combined with autoantibody values against hnRNPF and FTH1 using logistic regression and ROC analysis to determine the optimal cutoffs for the maximal sensitivity and specificity (Table 2). Autoantibodies against FTH1 alone had an AUC of 0.686 with an optimal sensitivity of 81.2% and a specificity of 56.1% when the cutoff was set at OD450 = 0.98, and hnRNPF alone AUC was 0.725 with an optimal sensitivity of 84.2% and a specificity of 60.8% when the cutoff was set at 1.09. However, when the two autoantibody analysis combined with CA15-3, the AUC increased to 0.931, and the optimal sensitivity and specificity improved to 89.3% and 93.8%, respectively (Table 2). This diagnostic value was much greater than any single biomarker could achieve alone, indicating that CA 15-3 tumor antigen levels are complementary to autoantibody levels against hnRNPF and FTH1 in breast cancer patient sera.
Table 2.
Logistic regression analysis
| Biomarker | AUC | 95% confidence interval
|
|
|---|---|---|---|
| Sensitivity (%) | Specifcity (%) | ||
| FTH1 | 0.686 | 0.812 | 0.561 |
| hnRNPF | 0.725 | 0.842 | 0.608 |
| CA 15-3 | 0.792 | 0.691 | 0.894 |
| FTH1 + hnRNPF | 0.816 | 0.911 | 0.720 |
| FTH1 + CA 15-3 | 0.834 | 0.851 | 0.927 |
| hnRNPF + CA 15-3 | 0.862 | 0.874 | 0.910 |
| FTH1 + hnRNPF + CA 15-3 | 0.931 | 0.893 | 0.938 |
Notes: AUC indicates diagnostic accuracy of biomarkers. The highest AUC is 1.
Abbreviations: AUC, area under the curve; FTH1, ferritin heavy chain; hnRNPF, heterogeneous nuclear ribonucleoproteins F.
mRNA expressions of hnRNPF and FTH1 in breast cancer and non tumor tissues
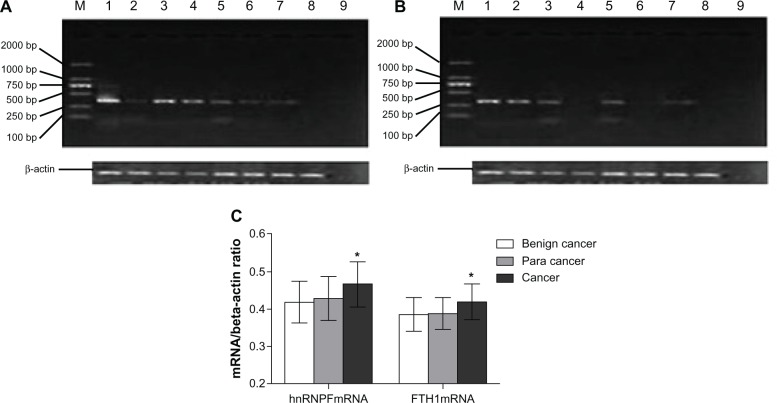
Expression of hnRNPF and FTH1 in 40 breast cancer, 40 cancer surrounding, and 32 benign breast tumor tissues were measured using semi-quantitative RT-PCR and analyzed with Quantity One software (Bio-Rad Laboratories Inc). The ratio of mRNA of hnRNPF/β-actin (±SD) was 0.419 ± 0.038 in benign tumor tissues, 0.422 ± 0.039 in cancer surrounding tissues, and 0.457 ± 0.047 in breast cancer tissues; while the ratio of mRNA of FTH1/ β-actin was 0.368 ± 0.031 in benign tumor tissues, 0.369 ± 0.029 in cancer surrounding tissues, and 421 ± 0.032 in breast cancer tissues. Both hnRNPF and FTH1 showed significant overexpression (P < 0.05) in breast cancer tissues compared to benign breast tumor tissues, while the cancer surrounding tissues showed slightly elevated mRNA levels of hnRNPF only (Figure 2). These results suggest that the increased autoantibody levels for hnRNPF and FTH1 might be triggered by the overexpression of their corresponding proteins in tumor tissues.
Figure 2.
Analysis of hnRNPF and FTH1 mRNA from tumor tissues. Expression of mRNA of (A) hnRNPF and (B) FTH1 were measured using semi-quantitative RT-PCR in tissues from 40 breast cancer, 40 cancer surrounding tissues, and 32 benign breast tumors. In panels (A) and (B), lanes 1–4 are breast cancer samples; lanes 5–6 are cancer surrounding tissue samples; lanes 7–8 are benign breast tumor samples; lane 9 is a negative control. (C) Both hnRNPF and FTH1 showed significant overexpression in the breast cancer tissues than in the benign breast tumor tissues, while the cancer surrounding tissues showed slightly elevated mRNA level of hnRNPF only.
Note: *P < 0.05.
Abbreviations: FTH1, ferritin heavy chain; hnRNPF, heterogeneous nuclear ribonucleoproteins F; mRNA, messenger ribonucleic acid; RT-PCR, reverse transcriptase polymerase chain reaction.
Discussion
Serum tumor markers have the potential of being incorporated into diagnostic, prognostic, and therapeutic practice in breast cancer.22–26 This potential has generated considerable interest with respect to identifying predictive tumor markers over the past three decades.27,28 However, efforts have only been focused on searching for either novel serological tumor-associated antigens or autoantibodies as biomarkers for breast cancer; they have not been tested in combination for breast cancer detection. So far, insignificant sensitivity and specificity have made clinical usage of these markers less dependable.
In this study, we identified two novel autoantibody biomarkers, hnRNPF and FTH1, and tested them in combination with the CA 15-3 from breast cancer patient and control sera. Although testing the two autoantibodies alone revealed significant differences between patient and control sera, the specificities remained relatively low (56.1% for FTH1 and 60.8% for hnRNPF). Even when we combined the two antibody biomarkers, the specificity only increased to 72.0%, while the sensitivity increased to 91.0%. On the other hand, analyzing CA 15-3 alone with the same serum samples, the sensitivity achieved was only 59.1% while the specificity was 89.4%. Interestingly, when the two autoantibodies were analyzed combined with CA 15-3, sensitivity and specificity increased to 89.3% and 93.8%, respectively; the predictive accuracy was much greater than using any single biomarker alone.
Although the results shown here appear promising, this high accuracy in the laboratory may not hold true when increasing the sample size. In addition, most of the patient serum samples collected in this study were from patients with advanced disease. Therefore, a large cohort of well characterized patient samples, especially with early stage patient samples, may be needed for further validation. Furthermore, a user friendly platform may be required when the assay is translated into a clinical routine. In this study, we measured CA 15-3 and the two autoantibodies separately using ELISA. It would be ideal if we could incorporate these two types of markers together on one plate or slide, so that the serum sample could be measured for these markers simultaneously rendering the results more consistent.
To date, most autoantibody biomarkers identified by us and others were found to have known tumor associated functions in different cancers or roles in signaling pathways.26,29–31 However, none of these autoantibody biomarkers were derived from antibodies against traditional TAAs, such as carcinoembryonic antigen (CEA), prostate-specific antigen (PSA), CA 15-3, or others. The reason for this phenomenon might be due to the balance between antigens and antibodies in the plasma. Since both antigens and the corresponding antibodies coexist in plasma, they would be bound to each other to form immunoprecipitates. Therefore, if the antigen titer is greater than the antibody levels, the antigen would likely be detected and used as an antigen biomarker, eg, CEA, PSA, and CA 15-3, or vice versa for the autoantibody biomarkers. Hirasawa et al reported that the KL-6/MUC1 antigen has an almost reciprocal relationship to levels of anti-KL-6/MUC1 antibodies in non-small cell lung cancer patient sera.32 In our study, the two identified TAAs are overexpressed in breast cancer tissue compared to non tumor tissues, suggesting that the overexpression of tissue restricted gene products is a factor in the development of the humoral immune response in breast cancer patients.
In summary, our results showed that serologic TAAs and antibodies can be complementary to each other and combined measurements using these two types of biomarkers may improve the predictive accuracy for cancer detection.
Acknowledgments
This study was supported by research grants from Strategic Projects of Science and Technology of Shaoxing City (GJSX-010-003), Health Science Research Foundation of Zhejiang Province (2009A208), Public Charity Projects of Zhejiang Science and Technology Department (2010C33010), and Key Science and Technology Innovation Team of Zhejiang Province (2010R50048), and also supported by an NSFC grant (81071795).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Harris L, Fritsche H, Mennel R, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 3.Hanash S. Disease proteomics. Nature. 2003;422(6928):226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 4.Belousov PV, Kuprash DV, Nedospasov SA, Shebzukhov YV. Autoantibodies to tumor-associated antigens as cancer biomarkers. Curr Mol Med. 2010;10(2):115–122. doi: 10.2174/156652410790963259. [DOI] [PubMed] [Google Scholar]
- 5.Tan HT, Low J, Lim SG, Chung MC. Serum autoantibodies as biomarkers for early cancer detection. FEBS J. 2009;276(23):6880–6904. doi: 10.1111/j.1742-4658.2009.07396.x. [DOI] [PubMed] [Google Scholar]
- 6.Desmetz C, Cortijo C, Mange A, Solassol J. Humoral response to cancer as a tool for biomarker discovery. J Proteomics. 2009;72(6):982–988. doi: 10.1016/j.jprot.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Finn OJ. Immune response as a biomarker for cancer detection and a lot more. N Engl J Med. 2005;353(12):1288–1290. doi: 10.1056/NEJMe058157. [DOI] [PubMed] [Google Scholar]
- 8.Sahin U, Tureci O, Schmitt H, et al. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA. 1995;92(25):11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong L, Coe SP, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J Thorac Oncol. 2006;1(6):513–519. [PubMed] [Google Scholar]
- 10.Old LJ, Chen YT. New paths in human cancer serology. J Exp Med. 1998;187(8):1163–1167. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minenkova O, Pucci A, Pavoni E, et al. Identification of tumor-associated antigens by screening phage-displayed human cDNA libraries with sera from tumor patients. Int J Cancer. 2003;106(4):534–544. doi: 10.1002/ijc.11269. [DOI] [PubMed] [Google Scholar]
- 12.Lenner P, Wiklund F, Emdin SO, et al. Serum antibodies against p53 in relation to cancer risk and prognosis in breast cancer: a population-based epidemiological study. Br J Cancer. 1999;79(5–6):927–932. doi: 10.1038/sj.bjc.6690148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soussi T. p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer Res. 2000;60(7):1777–1788. [PubMed] [Google Scholar]
- 14.Kulić A, Sirotković-Skerlev M, Jelisavac-Cosić S, Herceg D, Kovac Z, Vrbanec D. Anti-p53 antibodies in serum: relationship to tumor biology and prognosis of breast cancer patients. Med Oncol. 2010;27(3):887–893. doi: 10.1007/s12032-009-9301-1. [DOI] [PubMed] [Google Scholar]
- 15.Winter SF, Minna JD, Johnson BE, Takahashi T, Gazdar AF, Carbone DP. Development of antibodies against p53 in lung cancer patients appears to be dependent on the type of p53 mutation. Cancer Res. 1992;52(15):4168–4174. [PubMed] [Google Scholar]
- 16.Raedle J, Oremek G, Welker M, Roth WK, Caspary WF, Zeuzem S. p53 autoantibodies in patients with pancreatitis and pancreatic carcinoma. Pancreas. 1996;13(3):241–246. doi: 10.1097/00006676-199610000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Conroy SE, Sasieni PD, Fentiman I, Latchman DS. Autoantibodies to the 90 kDa heat shock protein and poor survival in breast cancer patients. Eur J Cancer. 1998;34(6):942–943. [PubMed] [Google Scholar]
- 18.Zhong L, Peng X, Hidalgo GE, Doherty DE, Stromberg AJ, Hirschowitz EA. Identification of circulating antibodies to tumor-associated proteins for combined use as markers of non-small cell lung cancer. Proteomics. 2004;4(4):1216–1225. doi: 10.1002/pmic.200200679. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honoré B, Baandrup U, Vorum H. Heterogeneous nuclear ribonucleo-proteins F and H/H’ show differential expression in normal and selected cancer tissues. Exp Cell Res. 2004;294(1):199–209. doi: 10.1016/j.yexcr.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Shpyleva SI, Tryndyak VP, Kovalchuk O, et al. Role of ferritin alterations in human breast cancer cells. Breast Cancer Res Treat. 2011;126(1):63–71. doi: 10.1007/s10549-010-0849-4. [DOI] [PubMed] [Google Scholar]
- 22.Chapman C, Murray A, Chakrabarti J, et al. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol. 2007;18(5):868–873. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]
- 23.Sugita Y, Wada H, Fujita S, et al. NY-ESO-1 expression and immunogenicity in malignant and benign breast tumors. Cancer Res. 2004;64(6):2199–2204. doi: 10.1158/0008-5472.can-03-3070. [DOI] [PubMed] [Google Scholar]
- 24.Scanlan MJ, Gout I, Gordon CM, et al. Humoral immunity to human breast cancer: antigen definition and quantitative analysis of mRNA expression. Cancer Immun. 2001;1:4. [PubMed] [Google Scholar]
- 25.Qian F, Odunsi K, Blatt LM, et al. Tumor associated antigen recognition by autologous serum in patients with breast cancer. Int J Mol Med. 2005;15(1):137–144. [PubMed] [Google Scholar]
- 26.Jäger D, Stockert E, Güre AO, et al. Identification of a tissue-specific putative transcription factor in breast tissue by serological screening of a breast cancer library. Cancer Res. 2001;61(5):2055–2061. [PubMed] [Google Scholar]
- 27.Volkmann M, Sinn HP, Gaugel D, et al. Anti-p53 in breast cancer: concordance of different assay procedures and association with p53 antigen expression. Oncology. 2002;63(3):297–305. doi: 10.1159/000065472. [DOI] [PubMed] [Google Scholar]
- 28.Laessig D, Nagel D, Heinemann V, et al. Importance of CEA and CA 15-3 during disease progression in metastatic breast cancer patients. Anticancer Res. 2007;27(4A):1963–1968. [PubMed] [Google Scholar]
- 29.Lin HS, Talwar HS, Tarca AL, et al. Autoantibody approach for serum-based detection of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2396–2405. doi: 10.1158/1055-9965.EPI-07-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Yu J, Sreekumar A, et al. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353(12):1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 31.Zhong L, Hidalgo GE, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. Using protein microarray as a diagnostic assay for non-small cell lung cancer. Am J Respir Crit Care Med. 2005;172(10):1308–1314. doi: 10.1164/rccm.200505-830OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirasawa Y, Kohno N, Yokoyama A, Kondo K, Hiwada K, Miyake M. Natural autoantibody to MUC1 is a prognostic indicator for non-small cell lung cancer. Am J Respir Crit Care Med. 2000;161(2 Pt 1):589–594. doi: 10.1164/ajrccm.161.2.9905028. [DOI] [PubMed] [Google Scholar]