Abstract
Background
Unsafe water supplies continue to raise public health concerns, especially in urban areas in low resource countries. To understand the extent of public health risk attributed to supply water in Dhaka city, Bangladesh, Escherichia coli isolated from tap water samples collected from different locations of the city were characterized for their antibiotic resistance, pathogenic properties and genetic diversity.
Methodology/Principal Findings
A total of 233 E. coli isolates obtained from 175 tap water samples were analysed for susceptibility to 16 different antibiotics and for the presence of genes associated with virulence and antibiotic resistance. Nearly 36% (n = 84) of the isolates were multi-drug(≥3 classes of antibiotics) resistant (MDR) and 26% (n = 22) of these were positive for extended spectrum β-lactamase (ESBL). Of the 22 ESBL-producers, 20 were positive for bla CTX-M-15, 7 for bla OXA-1-group (all had bla OXA-47) and 2 for bla CMY-2. Quinolone resistance genes, qnrS and qnrB were detected in 6 and 2 isolates, respectively. Around 7% (n = 16) of the isolates carried virulence gene(s) characteristic of pathogenic E. coli; 11 of these contained lt and/or st and thus belonged to enterotoxigenic E. coli and 5 contained bfp and eae and thus belonged to enteropathogenic E. coli. All MDR isolates carried multiple plasmids (2 to 8) of varying sizes ranging from 1.2 to >120 MDa. Ampicillin and ceftriaxone resistance were co-transferred in conjugative plasmids of 70 to 100 MDa in size, while ampicillin, trimethoprim-sulfamethoxazole and tetracycline resistance were co-transferred in conjugative plasmids of 50 to 90 MDa. Pulsed-field gel electrophoresis analysis revealed diverse genetic fingerprints of pathogenic isolates.
Significance
Multi-drug resistant E. coli are wide spread in public water supply in Dhaka city, Bangladesh. Transmission of resistant bacteria and plasmids through supply water pose serious threats to public health in urban areas.
Introduction
Diarrheal diseases account for an estimated 4.1% of the total daily global burden of disease and are responsible for the deaths of 1.8 million people every year, 90% of them are children under the age of 5 [1]. It was estimated that 88% of this burden is attributable to unsafe water supply, sanitation and hygiene, and is mostly concentrated in children in developing countries. Escherichia coli is widely used as an indicator organism for the microbiological quality of water is also an important causative agent of diarrhea and other enteric diseases. While most E. coli are generally harmless, certain strains of E. coli have virulence properties that may account for life threatening infections. Currently, six E. coli pathotypes are recognized that can cause diarrhea in humans [2]: enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), Shiga toxin-producing E. coli (STEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC) and diffusely adhering E. coli (DAEC). A very few studies have provided adequate information on the occurrence of pathogenic E. coli in household waters [3], [4].
Antimicrobial resistance among enteropathogens, including E. coli has been reported to be increasing in recent years [5], sometimes leading to point-break situations where no antibiotic treatment options remain [6]. These situations are of serious concern in developing countries where enteropathogens are frequently encountered and cause life-threatening infections, especially among children. The recent emergence and spread of a novel carbapenemase, New Delhi Metallo β-lactamase (NDM)-producing organisms is an example of that situation where available antibiotics are ineffective [7]. This novel enzyme along with other antibiotic resistance factors is carried by mobile genetic elements such as plasmids or transposons [8]. Horizontal gene transfer (HGT) is one of the most common mechanisms by which antibiotic resistance traits are transferred from one organism to another. In Enterobacteriaceae, plasmids are the major vectors for HGT. In vivo transfer of resistance traits between Enterobacteriaceae in natural ecosystem has been reported [9]. Generic E. coli are frequently used as indicator bacteria to monitor the trends in antimicrobial resistance because they are the prevalent commensal enteric bacteria in humans and animals, can be cultured easily and inexpensively [10], and they can acquire and preserve resistance genes from other organisms in the environment and in animal populations [11]. E. coli is also considered as a good indicator of the selective pressure imposed by antimicrobial use in food animals [12], [13].
Diarrheal diseases are endemic in Bangladesh. In 2008, an estimated 20,000 children less than 5 years old died of diarrheal diseases in Bangladesh [14]. E. coli is one of the leading causes of enteric infection in Bangladesh and ETEC is the predominant pathotype followed by EPEC, EAEC and STEC [15]. Moreover, the dissemination of ESBL and carbapenemases (CARBase) conferring resistance to life-saving β-lactams is of particular concern. Majority of the E. coli infections are waterborne as surface water is heavily contaminated with this organism. Poor sanitation and hygiene, overcrowded situation and lack of access to safe drinking water are the precipitating factors. In this study E. coli strains isolated from household water supply in Dhaka city were characterized for their antibiotic resistance, pathogenic types, ESBL phenotype, presence of major ESBL genes, and acquisition of transferrable plasmids.
Materials and Methods
Sampling site and sample collection
Water samples were collected from the south part of Dhaka city comprising an area of approximately 13 Km2 with an approximate population of 300,000. Dhaka is one of the fasted growing mega cities in the world which has an estimated population of 12.5 million living in an area of approximately 350 Km2. The areas that we covered under this study were mostly densely populated with low income people and have little water and sanitation facilities. A large urban slum was also included in the sampling area. Water supplied by the municipal authority was the only source of water for the population living in these areas. According to Dhaka Water Supply and Sewerage Authority (Dhaka WASA), around 87% of water supplied by the authority is from ground water abstraction using approximately 605 deep tube wells located in different places of the city and the remaining water comes from surface water treatments.
A total of 175 tap water samples were collected between November 2008 and July 2009 at points of uses in the community. From each point 500 ml of water sample were collected in pre-sterilized Nalgene sampling bottle and samples were transported to the laboratory within 3–4 h maintaining a cool chain.
Estimation of fecal coliform bacteria and isolation of E. coli
Number of fecal coliforms (FC) was estimated in water samples by membrane filtration method according to the procedures described earlier [16]. Briefly, a 100 ml aliquot of water sample was filtered through a 0.2 µm-pore-size membrane filter (Sartorius Stedim, Goettingen, Germany), and the filter was placed on a membrane fecal coliform (MFC) agar (BD, MD, USA) plates. MFC plates were incubated at 44°C for 18–24 h. After incubation, blue colonies which are typical of coliform bacteria were counted and expressed as colony forming units (CFU) per ml of water.
For E. coli, a 100 ml aliquot of water sample was filtered according to the procedure as described. The filter was immersed into EE broth (Oxoid ltd, Basingstoke, UK) and incubated at 37°C for 18–24 h. Enrichment broth was cultured on TBX agar medium (Oxoid) and incubated at 37°C for 18–24 h. Typical E. coli colonies from TBX plates were picked up and cultured on Eusine Methylene Blue agar (Oxoid) and MacConkey agar medium (Oxoid). From each sample, a maximum of 3 E. coli colonies were selected and stored at −70°C for further analysis.
Antimicrobial susceptibility tests
Susceptibility to antimicrobials was determined by an agar diffusion test using antimicrobial agents impregnated paper discs (Oxoid) as described by the Clinical Laboratory Standards Institute (CLSI) guidelines [17]. The antibiotics used in this study were ampicillin (10 µg), ceftriaxone (30 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), trimethoprim-sulfamethoxazole (25 µg), gentamicin (10 µg), mecillinam (25 µg), meropenem (10 µg), nalidixic acid (30 µg), tetracycline (30 µg), norfloxacin (10 µg), imipenem (10 µg), kanamycin (30 µg), erythromycin (15 µg), cefotaxime (30 µg), cefixime (5 µg), aztreonam (30 µg), ceftazidime (30 µg), cefoxitin (30 µg) and piperacillin-tazobactam (110 µg). E. coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were used as negative and positive controls, respectively. CLSI breakpoints were used to interpret the results [17]. Isolates that showed resistance or intermediate susceptibility to cephalosporins were tested for the presence of ESBL by doing double disc synergy test (DDST). The DDST was carried out on Mueller-Hinton agar (Difco Laboratories, Detroit, MI, USA) with discs containing 30 µg of ceftazidime, cefotaxime, or aztreonam, placed at a distance of 15 mm (center to center) from a disc containing amoxicillin-clavulanic acid (20 µg/10 µg) located in the center of the plate [17].
Detection of antibiotic resistance genes in ESBL-producing organisms
All ESBL-producing isolates were tested for the presence of bla ESBL genes (bla TEM, bla SHV, bla CTX-M-1-group, bla CTX-M-15, bla CTX-M-2-group, bla CTX-M-8-group, bla CTX-M-9-group), carbapenemase genes (bla OXA-1-group, bla OXA-47, and bla NDM-1) and ampC β-lactamase gene bla CMY-2 by PCR according to procedures described earlier [18]. PCR products of bla CTX-M-15 primers were sequenced using an ABI PRISM 310 sequencer (Applied Biosystems) in order to confirm the specificity of the gene. In addition, isolates were tested for 16sRNA methyltransferase genes (rmtA, rmtB and armA) and qnr genes (qnrA, qnrB and qnrS) according to procedures described earlier [18]. The primer sequences and corresponding annealing temperature used in the PCR reactions are listed in Table 1.
Table 1. PCR primers used in the study.
| Target gene | Primer | Nucleotide sequence (5′–3′) | Annealing temp (°C) | Product size (bp) |
| estA | ST-F | GCTAAACCAGTAG AGGTCTTCAAAA | 57 | 147 |
| ST-R | CCCGGTACAG AGCAGGATTACAACA | |||
| eltB | LT-F | CACACGGAGCTCCTCAGT C | 57 | 508 |
| LT-R | CCC CCA GCC TAG CTT AGT TT | |||
| bfpA | bfpA-F | GGAAGTCAAATTCATGGGGG | 57 | 300 |
| bfpA-R | GGAATCAGACGCAGACTGGT | |||
| eae | eae-F | CCCGAATTCGGCACAAGCATAAGC | 57 | 881 |
| eae-R | CCCGGATCCGTCTCGCCAGTATTCG | |||
| aaiC | aaiC-F | ATTGTCCTCAGGCATTTCAC | 57 | 215 |
| aaiC-R | ACGACACCCCTGATAAACAA | |||
| aat | Pcvd432-F | CTGGCGAAAGACTGTATCAT | 57 | 650 |
| Pcvd432-R | CAATGTATAGAAATCCGCTGTT | |||
| stx1 | stx1F | CACAATCAGGCGTCGCCAGCGCACTTGCT | 58 | 606 |
| stx1R | TGTTGCAGGGATCAGTGGTACGGGGATGC | |||
| stx2 | stx2F | CCACATCGGTGTCTGTTATTAACCACACC | 58 | 372 |
| stx2R | GCAGAACTGCTCTGGATGCATCTCTGGTC | |||
| iaa | ial upper | CTGGATGGTATGGTGAGG | 57 | 320 |
| ial lower | GGAGGCCAACAATTATTTCC | |||
| ipaH | Shig-1 | TGGAAAAACTCAGTGCCTCT | 57 | 424 |
| Shig-2 | CCAGTCCGTAAATTCATTCT | |||
| bla TEM | TEM-F | TCGGGGAAATGTGCGCG | 57 | 850 |
| TEM-R | TGCTTAATCAGTGAGGACCC | |||
| bla SHV | SHV-F | CACTCAAGGATGTATTGTG | 57 | 861 |
| SHV-R | TTAGCGTTGCCAGTGCTCG | |||
| bla CTX-M-1 group | M13-upper | GGTTAAAAAATCACTGCGTC | 54 | 866 |
| M13-lower | TTGGTGACGATTTTAGCCGC | |||
| bla CTX -M-2 group | M25-upper | ATGATGACTCAGAGCATTCG | 56 | 866 |
| M25-lower | TGGGTT ACGATTTTCGCCGC | |||
| bla CTX -M-8 group | CTXM8 -F | TCGCGTTAAGCGGATGATGC | 58 | 688 |
| CTXM8-R | AACCCACGATGTGGGTAGC | |||
| bla CTX-M-9 group | M9-upper | ATGGTGACAAAGAGAGTGCA | 56 | 870 |
| M9-lower | CCCTTCGGCGATGATTCTC | |||
| bla CTX-M-15 | CTX-M-15-SF | CACACGTGGAATTTAGGGACT | 56 | 996 |
| CTX-M-15-SR | GCCGTCTAAGGCGATAAACA | |||
| bla OXA-1 group | OXA-1F | ACACAATACATATCAACTTCGC | 56 | 814 |
| OXA-1R | AGTGTGTTTAGAATGGTGATC | |||
| bla OXA-47 | OXA-1A | TCAACTTTCAAGATCGCA | 48 | 609 |
| OXA-1B | GTGTGTTTAGAATGGTGA | |||
| bla CMY-2 | Forward | GACAGCCTCTTTCTCCACA | 50 | 1143 |
| Reverse | TGGAACGAAGGCTACGTA | |||
| bla NDM-1 | NDM-F | GGTTTGGCGATCTGGTTTTC | 57 | 465 |
| NDM-R | CGGAATGGCTCATCACGATC | |||
| rmtB | rmtBF | GCTTTCTGCGGGCGATGTAA | 55 | 173 |
| rmtBR | ATG CAA TGC CGC GCT CGT AT | |||
| rmtC | rmtC-F | CGA AGA AGT AAC AGC CAA AG | 55 | 711 |
| rmtC-R | ATC CCA ACA TCT CTC CCA CT | |||
| armA | armAF | ATT CTG CCT ATC CTA ATT GG | 55 | 315 |
| armAR | ACC TAT ACT TTA TCG TCG TC | |||
| qnrA | QnrAm-F | AGAGGATTTCTCACGCCAGG | 56 | 580 |
| QnrAm-R | TGCCAGGCACAGATCTTGAC | |||
| qnrB | QnrBm-F | GGMATHGAAATTCGCCACTG | 56 | 264 |
| QnrBm-R | TTTGCYGYYCGCCAGTCGAA | |||
| qnrS | QnrSm-F | GCAAGTTCATTGAACAGGGT | 56 | 428 |
| QnrSm-R | TCTAAACCGTCGAGTTCGGCG |
PCR for virulence genes
All isolates were examined for the presence of the heat labile (lt), heat stable (st), attaching and effacing gene (eae), bundle forming pilus (bfp), antiaggregation protein transporter gene (aat) and gene for AggR-activated island (aaiC) by multiplex PCR assay. DNA was prepared from overnight grown culture by boiling method. The respective 3 µl template DNA was suspended in 22 µl of reaction mix containing 2.5 µl of 10X PCR buffer with 0.75 µl MgCl2, 0.5 µl of 10 mM dNTPs, 0.4 µl each of lt, st, bfp, aat, aaiC primers, 0.44 µl of eae primers, together with 1 unit of Taq DNA polymerase (5 U/µl). PCR cycling conditions consisted of initial denaturation at 96°C for 4 min, followed by 34 cycles each of denaturation at 95°C for 20 s, annealing at 57°C for 20 s and extension at 72°C for 1 minute. A separate multiplex PCR for Shiga toxin genes (stx 1 and stx 2) was carried out according to the procedure described earlier [19]. PCR to demonstrate the presence of invasion associated locus (ial) and the invasion plasmid antigen H (ipaH) was performed according to published procedures [20], [21]. Primer sequences are listed in Table 1.
Plasmid profile analysis and conjugation experiment
Plasmid DNA was prepared using the rapid alkaline lysis method [22] and analysed by horizontal electrophoresis in 0.7% agarose gels. The molecular weight size of unknown plasmids was estimated by comparing with plasmids that have been used as size standards in the gel electrophoresis. The plasmids Sa (23 MDa), RP4 (34 MDa), R1 (62 MDa), pDK9 (140 MDa) and E. coli V517 plasmids (1.4, 1.8, 2.0, 2.6, 3.4, 3.7, 4.8 and 35.8 MDa) were used as standards [23]. Conjugation was carried out by both broth mating and filter mating assays at 30°C using MDR water isolates as donor and E. coli MC1061 (SmR, F−, non-lactose fermenting) and E. coli J53 (AziR, F−) as recipients. E. coli MC1061 transconjugants were selected on MacConkey agar containing ampicillin (50 mg/L), while the E. coli J53 transconjugants were selected on MacConkey agar containing sodium azide (100 mg/L) and cefotaxime (20 mg/L)/cefoxitin (16 mg/L). The duration of conjugation was 18 h. Transconjugant colonies were confirmed by antibiotic susceptibility tests. Plasmid DNA from transconjugants was extracted using alkaline lysis method as described previously [22]. Conjugation frequency per recipient was expressed by dividing the number of transconjugants by the initial number of recipients.
Genetic fingerprinting
All pathogenic isolates (n = 16) were selected for analysis by Pulsed-field gel electrophoresis (PFGE). Genomic DNA was prepared in agarose blocks and digested with the restriction enzyme XbaI (New England Biolabs). DNA fragments were separated by pulsed-field gel electrophoresis on a CHEF-MAPPER apparatus (Bio-Rad) according to the PulseNet program developed for E. coli [24]. Analysis of the TIFF images was carried out by the BioNumerics software (Applied Maths) using the dice coefficient and unweighted-pair group method using average linkages to generate dendrograms with 1.0% tolerance values.
Results
Enumeration of fecal coliform bacteria and isolation of E. coli
Around 80% (n = 139) of the water samples were positive for fecal coliform (FC) bacteria and 38% (n = 67) had a fecal coliform count of >100 CFU/ml of water. E. coli was isolated from 63% (n = 110) of samples. A total of 233 E. coli were isolated from 110 samples that were characterized in this study.
Antimicrobial susceptibility tests
Of the 233 isolates tested, 57% (n = 133) were resistant to ampicillin, followed by 45% (n = 105) to tetracycline, 37% (n = 87) to nalidixic acid, 36% (n = 83) to trimethoprim-sulfamethoxazole, 17% (n = 39) to ciprofloxacin, 9% (n = 22) to ceftriaxone, 9% (n = 20) to mecilinam, 8% (n = 18) to chloramphenicol and 1% (n = 2) to gentamicin. More than 73% (n = 171) of the isolates were resistant to at least one antibiotic and 36% of the isolates (n = 84) were resistant to three or more classes of antibiotics thus defined as multi-drug resistant (MDR). Further testing of the 22 ceftriaxone resistant isolates revealed that all were ESBL-producing as confirmed by the double disc synergy test. All the 22 isolates were resistant to cefotaxime and cefixime, 82% to erythromycin, 64% to aztreonam, 55% to ciprofloxacin/norfloxacin, 32% to kanamycin and ceftazidime, 14% to piperacillin-tazobactam and 9% to cefoxitin. None of the isolates were resistant to carbapenem antibiotics, including imipenem and meropenem.
Detection of antibiotic resistance genes in ESBL-producing organisms
Of the 22 ESBL producing isolates, 20 (90%) were positive for bla CTX-M-1-group specific gene and bla CTX-M-15. Presence of bla CTX-M-15 gene was confirmed by sequencing the PCR product. One isolate was positive for bla CTX-M-9-group specific gene and none of the isolates was positive for bla CTX-M-2-group, bla CTX-M-8-group specific genes. Around 41% (n = 9) isolates were positive for bla TEM and none were positive for bla SHV. Among carbapenemase genes, bla OXA-1-group and bla OXA-47 were detected in 32% (n = 7) of the isolates. None of the isolates were positive for metallo-β-lactamase gene bla NDM-1. Plasmidic ampC-type β-lactamases bla CMY-2 was detected in 9% (n = 2) of the isolates. Among quinolone resistance genes, qnrS and qnrB were detected in 27% (n = 6) and 9% (n = 2) of the isolates, respectively (Table 2).
Table 2. Antibiotic resistance pattern, presence of antibiotic resistance genes and plasmid patterns of ESBL-producing E. coli isolated from water samples.
| Serial no. | Strain ID | Antibiotic resistance patterna | Presence of ESBL genes | Plasmid pattern (in MDa) |
| 1 | 4C3 | Amp, Cro, Cfm, Ctx | bla CTX-M-15, qnrS | 36 |
| 2 | 24C2 | Amp, Cip, Cro, Sxt, NA, Te, Cfm, Ctx, Nor, K, E | bla CTX-M-15, bla OXA-1, bla OXA-47 | 90,3.2,3 |
| 3 | 24C3 | Amp, Cip, Cro, Sxt, NA, Te, Mel, Atm, Cfm, Ctx, Nor, E, | bla CTX-M-15, bla TEM, | 90 |
| 4 | 28C2 | Amp, Cip, Cro, Sxt, NA, Te, C, Atm, Caz, Cfm, Ctx, Nor, K, E | bla CTX-M-15, bla TEM, bla OXA-1, bla OXA-47 | 105,90,17,2 |
| 5 | 88mf2 | Amp, Cro, Mel, Atm, Cfm, Ctx, Tzp | bla CTX-M-15, bla TEM | 105,90 |
| 6 | 90C1 | Amp, Cip, Cro, NA, C, Atm, Cfm, Ctx, Nor, E | bla CTX-M-15, bla TEM | 62 |
| 7 | 102C1 | Amp, Cip, Cro, Sxt, NA, Te, Cfm, Ctx, Nor, E | bla CTX-M-9 | 90,8.6,7.4,3.4 |
| 8 | 112C2 | Amp, Cip, Cro, Sxt, NA, Te, Fox, Atm, Caz, Cfm, Ctx, Nor, E | bla CTX-M-15, bla TEM, bla CMY-2, | 90,35.8,3.1 |
| 9 | 123C4 | Amp, Cip, Cro, Sxt, NA, Te, C, Mel, Atm, Caz, Cfm, Ctx, Nor, K, E | bla CTX-M-15, bla TEM, bla OXA-1, bla OXA-47 | 140,70 |
| 10 | 134C1 | Amp, Cro, Cfm, Ctx, E | bla CTX-M-15, qnrS | 140 |
| 11 | 145C2 | Amp, Cro, Cfm, Ctx, | bla CTX-M-15 | No Plasmid |
| 12 | 146C2 | Amp, Cro, Cfm, Ctx, E | bla CTX-M-15, qnrS | 100 |
| 13 | 156C1 | Amp, Cro, Cfm, Ctx, E | bla CTX-M-15, qnrS | 140, 62, 27 |
| 14 | 169C1 | Amp, Cro, Cfm, Ctx, E | bla CTX-M-15, | 70,2.7 |
| 15 | 169C3 | Amp, Cro, Sxt, Te, Atm, Cfm, Ctx, E | bla CTX-M-15, bla TEM, qnrS | 62 |
| 16 | 174TC1 | Amp, Cip, Cro, Sxt, NA, Te, C, Cn, Atm, Cfm, Ctx, Nor, K, E | bla CTX-M-15, bla OXA-1, bla OXA-47 | 105, 2.7,2.1,1.4,1.2 |
| 17 | 174FC1 | Amp, Cip, Cro, NA, Te, Atm, Caz, Cfm, Ctx, Nor, K, E, Tzp | bla CTX-M-15, bla OXA-1, bla OXA-47 | 140,62 |
| 18 | 177TC1 | Amp, Cro, Sxt, Te, Atm, Cfm, Ctx, E | bla CTX-M-15 | 62 |
| 19 | 185C2 | Amp, Cro, Te, Atm, Cfm, Ctx, E | bla CTX-M-15, qnrS | 200,100,35.8 |
| 20 | 186C2 | Amp, Cip, Cro, Sxt, NA, Te, C, Cn, Atm, Caz, Cfm, Ctx, Nor, K, E, Tzp | bla CTX-M-15, bla TEM, bla OXA-1, bla OXA-47, qnrB | 70 |
| 21 | 199C5 | Amp, Cip, Cro, NA, Mel, Fox, Atm, Caz, Cfm, Ctx, Nor, E | bla TEM, bla CMY-2, | 62,23,9 |
| 22 | 200C2 | Amp, Cip, Cro, Sxt, NA, Te, Atm, Caz, Cfm, Ctx, Nor, K, E | bla CTXM-15, bla OXA-1, bla OXA-47, qnrB | No Plasmid |
Amp, Ampicillin; Atm, Aztreonam; C, Cholramphenicol; Caz, Ceftazidime; Cfm, Cefixime; Cip, Ciprofloxacin; Cn, Gentamycin; Cro, Ceftriaxone; Ctx, Cefotaxime; E, Erythromycin; Fox, Cefoxitin; Imp, Imipenem; K, Kanamycin; Mel, Mecillinam; Mem, Meropenem; NA, Nalidixic acid; Nor, Norfloxacin; Sxt, Sulphamethoxazole-trimethoprim; Te, Tetracycline; and Tzp, Piperacillin-Tazobactam; All S, Sensitive to all antibiotics tested in the study.
Detection of virulence genes
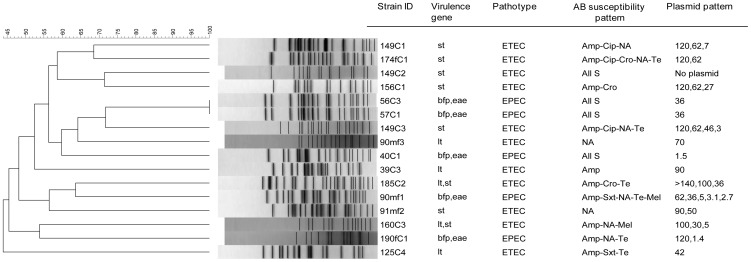
Around 7% (n = 16) of the isolates were found to be positive for at least one of the 10 pathogenic genes specific to E. coli pathotypes. Majority of the isolates (n = 11) possessed either lt (n = 3) or st (n = 6) or both (n = 2), and thus belonged to ETEC. The remaining 5 isolates belonged to EPEC as all of these contained both bfp and eae genes (Fig. 1).
Figure 1. Dendrogram of PFGE fingerprints from pathogenic E. coli isolates isolated from water samples.
The percentage of genetic homology between banding patterns is indicated. Presence of virulence genes, pathotypes, antibiotic susceptibility pattern and plasmid pattern are plotted next to dendrogram.
Plasmid profile analysis and conjugation experiment
Of the 233 isolates, 186 (80%) carried plasmids of varying sizes generating heterogeneous inter-isolate plasmid profiles. Molecular weights of plasmids ranged from 1.2 MDa to >120 MDa and the number of plasmid ranged from 2 to 8. Conjugation assays with representative isolates (n = 15) having different plasmid patterns revealed that plasmids of 50 to 105 MDa were self-transmissible to E. coli recipient strain carrying ampicillin resistance. Antibiotic susceptibility tests of transconjugant strains demonstrated that ampicillin resistance conferring plasmids co-transferred trimethoprim-sulfamethoxazole, tetracycline and ceftriaxone resistance. However, resistance to cefoxitin was not transferred via conjugative plasmid. Quinolone resistance determinants were not transferred via conjugative plasmids. Plasmid transfer frequency was nearly the same for all conjugative plasmids expect for a 50 MDa plasmid carrying ampicillin, trimethoprim-sulfamethoxazole and tetracycline resistance, which showed a relatively higher transfer frequency (1.95×10−2) than that of the other plasmids (4.4−9.0×10−4) (Table 3).
Table 3. Results of conjugation assays between antibiotic resistant E. coli isolates obtained from water samples and the recipient E. coli MC-1061 strain.
| Strain no. | Parent strain | Transconjugant | Transfer frequency | ||
| Resistance patterna | Plasmid pattern (MDa) | Resistance patterna | Plasmid pattern (MDa) | ||
| 25C3 | Amp-Sxt | 140,62,45,2.3,2.0 | Amp-Sxt | 62 | 9×10−4 |
| 51C1 | Amp-Cip-NA-Sxt-Mel | 90,62,4.8,3.7 | Amp | 62 | 5.2×10−4 |
| 88mf2 | Amp-Cro-Mel | 105, 70 | Amp-Cro | 70 | 4.8×10−4 |
| 133C4 | Amp-Sxt-Te | 50 | Amp-Sxt-Te | 50 | 1.95×10−2 |
| 174TC1 | Amp-Cip-Cro-Sxt-NA-Te-C-Cn | 105,2.7,2.1,1.4,1.2 | Amp-Cro | 105 | 4.4×10−4 |
See footnote a of Table 2 for definitions of abbreviations.
Genetic fingerprinting
PFGE analysis of pathogenic E. coli isolates revealed diverse banding patterns with similarity indices ranging from 72% to <45%. Two ETEC isolates had identical PFGE patterns and were members of a single clone (Fig. 1).
Discussion
Household water supply provided by the municipal authority is an important shared resource for millions of people living in Dhaka metropolitan area. E. coli is commonly isolated from water sources, including the municipal water supply of Dhaka city [25]. In this study, we found that around 38% of the water samples were contaminated with high counts of fecal coliform bacteria (>100 CFU/ml) and E. coli was isolated from 63% of the samples. The presence of E. coli in the water sample indicates the presence of microorganisms that might be potentially hazardous for human health and also indicates fecal contamination in water supply system. It becomes a serious threat when these E. coli exhibit resistance to multiple antibiotics and pathogenic properties that cause enteric diseases in people who consume this contaminated water. Studies conducted in other countries demonstrated the presence of MDR pathogenic bacteria in water sources including rivers, ponds and lakes [26], [27]. One study from India and another from Canada also reported the presence of antibiotic resistant E. coli in drinking water [28], [4].
In the present study, we found that more than 73% of the E. coli isolates were resistant to at least one of the 10 antibiotics tested and almost half (49%) of these isolates were multidrug resistant, defined as resistant to three or more classes of antibiotics. Akin to other studies, a higher frequency of resistance against β-lactam, quinolone and floroquinolone antibiotics was observed among the isolates in this study [28], [4]. A significant proportion (9%) of E. coli isolates tested in the study was ESBL-producing. This might be due to the residual effect of these antibiotics, which have been used extensively in human population as well as in the food chain creating a selective antibiotic pressure in the environment. A study carried out in 2004 reported that around 43% of E. coli isolates obtained from an urban hospital in Dhaka city were ESBL-producing [29].
Among ESBL producers, the majority were positive for clinically significant class A β-lactamases, including bla CTX-M-1-group, particularly the bla CTX-M-15. With the beginning of the twenty-first century, E. coli strains producing bla CTX-M-15 have emerged and disseminated worldwide and are now important cause of both nosocomial and community-onset urinary tract and bloodstream infections in humans [30], [31]. The prevalence of CTX-M type β-lactamases in Enterobacteriaceae is increasing and in some geographic locations they are now-a-days more prevalent than TEM and SHV types [32]. Both TEM and SHV types have been reported mostly from clinical samples and from some environmental samples like farm animals and estuarine waters [33], [34]. Interestingly, majority of isolates in our study were positive for bla TEM, while none were positive for bla SHV. Plasmid mediated quinolone resistance gene, qnr has been identified worldwide in different enterobacterial species, including E. coli [35], [36], [37]. The prevalence of qnr genes, especially qnrB, has been reported from clinically important K. pneumoniae and other Enterobacteriaceae species in Asian countries [38], [39]. Among non-clinical sources, qnr has been detected in E. coli isolates from livestock, swine and poultry [40], [41]. A qnrS gene was identified in a water-borne bacterial species, Aeromonas, isolated from the River Seine in Paris [42] and from a Swiss lake [43]. In the present study, we found that two strains were positive for plasmid mediated qnr genes of qnrS and qnrB types. The isolate carrying the qnrB co-harbored different classes of β-lactamase genes, including bla CTX-M-15 and bla OXA-47 and were resistant to 13 antibiotics, including ciprofloxacin. In contrast, the isolate carrying qnrS gene co-harbored bla CTX-M-15 and bla TEM and were resistant to 8 antibiotics, excluding ciprofloxacin. Therefore, the presence of qnrS alone may not confer resistance to fluroquinolones as also discussed in previous studies [42].
Many reports have been published on E. coli isolates from clinical samples carrying multiple classes of β-lactamases, and metallo β-lactamases [30]. The versatility and fitness of clinically important E. coli are proven to acquire most of the variants of β-lactamase genes and the recent acquisition is the New Delhi metallo β-lactamase. A recent study has shown that E. coli from environmental sources, including public tap water from New Delhi area, India were positive for multiple classes of β-lactamase, including the NDM-1 [44]. Enterobacteriaceae containing NDM-1 gene were also found from the clinical samples in Bangladesh [18]. However, none of the isolates in the present study was positive for NDM-1 gene. Isolates in this study were collected during 2008–2009, the period preceding the emergence of NDM-1. Nevertheless, it is not unlikely that the recent isolates will carry this gene and hence a continued surveillance is warranted. Pathogenic E. coli contributes significantly to the burden of infectious diseases in parts of the world where enteric diseases are endemic. Although water is considered as an important route of transmission of pathogenic E. coli, only a few published reports are available that describe its transmission via household water supply. In this study, we found that a significant percentage (7%) of E. coli isolates from supply water sources belonged to the pathogenic types, including EPEC and ETEC. In Bangladesh, diarrheal diseases are a major health problem, and pathogenic E. coli are the second leading causes of diarrhea next to rota virus. ETEC accounts for about 20% of all diarrheal cases in children under 2 years of age [15]. It has been shown previously that ETEC is present in drinking water and environmental water in Dhaka and viable after long-term water incubation which suggests that water might be an important route of transmission [45], [46]. In a recent study it has been shown that ETEC form biofilms in household drinking water which can be found during all months of the year and an increase during summer and rainy season [47]. At present, there is no vaccine available for E. coli diarrhea and the treatment modalities, including antibiotic therapy are not very efficient due to the emergence of MDR organisms. It is likely that multiple exposure pathways are involved in transmitting the MDR pathogenic E. coli to humans but household water supply play a significant role as it is highly contaminated and people get exposed to contaminated water very easily.
Plasmid profile analysis revealed that the majority (80%) of isolates contained multiple plasmids and there was a little similarity of patterns among the isolates indicating their clonal diversity. Around 14% of the isolates (n = 32) contained a large plasmid of >120 MDa. It is established that plasmids of this size carry invasive properties for certain enteropathogens, including Shigella spp., and Enteroinvasive E. coli (EIEC) [48]. In general, all invasive Shigella spp. and EIEC strains are positive for ipaH and ial genes, which are considered as surrogate markers for the test of invasiveness. None of the large plasmid-containing isolates in the study was positive for ipaH and ial genes. This accentuates the need for further studies to understand the role of large plasmids in E. coli isolates. Analysis of plasmid profile revealed that a large number of isolates (n = 122) contained plasmids in the range of 50 to 100 MDa (middle-ranged). It has been demonstrated previously that plasmids of these sizes in Enterobacteriaceae, particularly in Shigella spp. and E. coli are generally self-transmissible and carry the antimicrobial resistance factors [49], [50]. In this study, we also found correlation between the presence of middle ranged plasmids and multi-drug resistance phenotypes among the isolates. We identified self-transmissible plasmids that carry ampicillin resistance in different strains ranging in weight size from 50–105 MDa. Although we selected the conjugative plasmids based on ampicillin resistance, other antibiotic resistance particularly trimethoprim-sulfamethoxazole, tetracycline and ceftriaxone were co-transferred by these plasmids with a different transfer frequency (Table 3). The resistance to cefoxitin in two isolates (112C2 and 199C5) was not transferred by conjugation to the recipient E. coli J53, although both isolates were positive for bla CMY-2, a plasmid-mediated AmpC beta-lactamase. No transfer of ciprofloxacin or nalidixic acid was observed indicating that quionolone groups are not transferrable by conjugative plasmid. Transfer of resistance plasmids by conjugation was not successful for a number of isolates. The plasmid carrying the resistance gene in these isolates may be in non-conjugative plasmids or chromosomally encoded. Further studies are needed to determine the location of resistance genes in these isolates.
A high degree of polymorphism was observed in PFGE patterns of the isolates. A total of 15 distinct profiles were obtained among 16 pathogenic isolates indicating their genetic diversity. Only two ETEC isolates had identical PFGE patterns. Interestingly, both isolates had identical plasmid and antibiotic susceptibility patterns (Fig. 1). Tracing back to the source, we found that these organisms were isolated from water samples obtained from two different points-of-use within the same area where water is supplied from the same point-of-source through a single pipeline. This result indicates the clonal transmission of pathogenic organisms in the community through supply water system.
One of the limitations of the study is that we could not collect water samples from the entire areas of the city. Therefore, the level of contamination of water might not representative of other areas of the city. However, most Dhaka residents rely on the municipal water, which is mainly abstracted from underground sources and circulated to households following the same system. As such, the risk of exposure to MDR pathogenic organisms via water supply system in Dhaka residents would not be significantly different from one area to the other.
The household water supply is normally consumed by people without any pre-treatment, although boiling of water before consumption is often advised. Hence, the presence of multi-drug resistant ESBL-producing pathogenic E. coli in household water supply in Dhaka has important implications for health of the urban population. The MDR E. coli may represent an important reservoir of genetic determinants of antimicrobial resistance that can easily be transferred to other microorganisms in the environment through HGT, including the potential human pathogens. Effective prevention strategies are needed to limit the widespread circulation of these bacteria in the community and to contain the threat of emerging drug resistance among various enteric bacterial pathogens.
Funding Statement
This research study was funded by icddr,b and its donors which provide unrestricted support to icddr,b for its operations and research. Current donors providing unrestricted support include: Australian Agency for International Development (AusAID), Government of the People's Republic of Bangladesh, Canadian International Development Agency (CIDA), Swedish International Development Cooperation Agency (Sida), and the Department for International Development, UK (DFID). The authors gratefully acknowledge these donors for their support and commitment to icddr,b ′s research efforts. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO (2012) Facts and figures: Water, sanitation and hygiene links to health.
- 2. Turner SM, Scott-Tucker A, Cooper LM, Henderson IR (2006) Weapons of mass destruction: virulence factors of the global killer enterotoxigenic Escherichia coli . FEMS Microbiology Letters 263: 10–20. [DOI] [PubMed] [Google Scholar]
- 3. Higgins JA, Belt KT, Karns JS, Russell-Anelli J, Shelton DR (2005) tir- and stx-positive Escherichia coli in stream waters in a metropolitan area. Applied and Environmental Microbiology 71: 2511–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pathak SP, Gopal K (2008) Prevalence of bacterial contamination with antibiotic-resistant and enterotoxigenic fecal coliforms in treated drinking water. Journal of Toxicology and Environmental Health Part? A 71: 427–433. [DOI] [PubMed] [Google Scholar]
- 5. Pitout JD, Laupland KB (2008) Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infectious Diseases 8: 159–166. [DOI] [PubMed] [Google Scholar]
- 6. Lynch JP 3rd, Clark NM, Zhanel GG (2013) Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum β-lactamases and carbapenemases). Expert Opinion on Pharmacotherapy 14: 199–210. [DOI] [PubMed] [Google Scholar]
- 7. Moellering RC Jr (2010) NDM-1--a cause for worldwide concern. New England Journal of Medicine 363: 2377–9. [DOI] [PubMed] [Google Scholar]
- 8. Nordmann P, Poirel L, Walsh TR, Livermore DM (2011) The emerging NDM carbapenemases. Trends in Microbiology 19: 588–95. [DOI] [PubMed] [Google Scholar]
- 9. Martinez JL (2009) The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proceedings Biological Sciences/The Royal Society 276: 2521–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Den Bogaard AE, London N, Stobberingh EE (2000) Antimicrobial resistance in pig faecal samples from the Netherlands (five abattoirs) and Sweden. The Journal of Antimicrobial Chemotherapy 45: 663–671. [DOI] [PubMed] [Google Scholar]
- 11. Murray BE (1997) Antibiotic resistance. Advances in Internal Medicine 42: 339–367. [PubMed] [Google Scholar]
- 12. Aarestrup FM, Bager F, Jensen NE, Madsen M, Meyling A, et al. (1998) Resistance to antimicrobial agents used for animal therapy in pathogenic-, zoonotic- and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP). APMIS: Acta Pathologica, Microbiologica, et Immunologica Scandinavica 106: 745–770. [DOI] [PubMed] [Google Scholar]
- 13. van den Bogaard AE, Stobberingh EE (2000) Epidemiology of resistance to antibiotics. Links between animals and humans. International Journal of Antimicrobial Agents 14: 327–335. [DOI] [PubMed] [Google Scholar]
- 14. Huda TM, Unicomb L, Johnston RB, Halder AK, Yushuf Sharker MA, et al. (2012) Interim evaluation of a large scale sanitation, hygiene and water improvement programme on childhood diarrhea and respiratory disease in rural Bangladesh. Social Science & Medicine 75: 604–611. [DOI] [PubMed] [Google Scholar]
- 15. Qadri F, Svennerholm AM, Faruque AS, Sack RB (2005) Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clinical Microbiology Reviews 18: 465–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.APHA (1998) Standard Methods for the Examination of Water and Wastewater, 19th edn. Washington, DC: American Public Health Association.
- 17.CLSI (2009) Performance Standards for Antimicrobial Disk Susceptibility Test; Approved Standard. Wayne, PA: Clinical and Laboratory Standard Institute.
- 18. Islam MA, Talukdar PK, Hoque A, Huq M, Nabi A, et al. (2012) Emergence of multidrug-resistant NDM-1-producing Gram-negative bacteria in Bangladesh. European Journal of Clinical Microbiology & Infectious Diseases 31: 2593–600. [DOI] [PubMed] [Google Scholar]
- 19. Islam MA, Heuvelink AE, de Boer E, Sturm PD, Beumer RR, et al. (2007) Shiga toxin-producing Escherichia coli isolated from patients with diarrhoea in Bangladesh. Journal of Medical Microbiology 56: 380–385. [DOI] [PubMed] [Google Scholar]
- 20. Talukder KA, Islam MA, Khajanchi BK, Dutta DK, Islam Z, et al. (2003) Temporal shifts in the dominance of serotypes of Shigella dysenteriae from 1999 to 2002 in Dhaka, Bangladesh. Journal of Clinical Microbiology 41: 5053–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thong KL, Hoe SL, Puthucheary SD, Yasin RM (2005) Detection of virulence genes in Malaysian Shigella species by multiplex PCR assay. BMC Infectious Diseases 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kado CI, Liu ST (1981) Rapid procedure for detection and isolation of large and small plasmids. Journal of Bacteriology 145: 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM (1978) A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1: 417–420. [DOI] [PubMed] [Google Scholar]
- 24. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, et al. (2006) Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathogens and Disease 3: 59–67. [DOI] [PubMed] [Google Scholar]
- 25.Islam S, Begum HA, Mili NY (2010) Bacteriological Safety Assessment of Municipal Tap Water and Quality of Bottle Water in Dhaka City: Health Hazard Analysis. Bangladesh Journal of Medical Microbiology: 9–13.
- 26. Hu J, Shi J, Chang H, Li D, Yang M, et al. (2008) Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environmental Science & Technology 42: 3415–3420. [DOI] [PubMed] [Google Scholar]
- 27. Ram S, Vajpayee P, Tripathi U, Singh RL, Seth PK, et al. (2008) Determination of antimicrobial resistance and virulence gene signatures in surface water isolates of Escherichia coli . Journal of Applied Microbiology 105: 1899–1908. [DOI] [PubMed] [Google Scholar]
- 28. Coleman BL, Salvadori MI, McGeer AJ, Sibley KA, Neumann NF, et al. (2012) The role of drinking water in the transmission of antimicrobial-resistant E. coli . Epidemiology and Infection 140: 633–642. [DOI] [PubMed] [Google Scholar]
- 29. Rahman MM, Haq JA, Hossain MA, Sultana R, Islam F, et al. (2004) Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in an urban hospital in Dhaka, Bangladesh. International Journal Antimicrobial Agents 24: 508–510. [DOI] [PubMed] [Google Scholar]
- 30. Oteo J, Perez-Vazquez M, Campos J (2010) Extended-spectrum beta-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Current Opinion in Infectious Diseases 23: 320–326. [DOI] [PubMed] [Google Scholar]
- 31. Pitout JDD (2010) Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs 70: 313–333. [DOI] [PubMed] [Google Scholar]
- 32. Falagas ME, Karageorgopoulos DE (2009) Extended-spectrum beta-lactamase-producing organisms. The Journal of Hospital Infection 73: 345–354. [DOI] [PubMed] [Google Scholar]
- 33. Henriques IS, Fonseca F, Alves A, Saavedra MJ, Correia A (2006) Occurrence and diversity of integrons and beta-lactamase genes among ampicillin-resistant isolates from estuarine waters. Research in Microbiology 157: 938–947. [DOI] [PubMed] [Google Scholar]
- 34. Hiroi M, Harada T, Kawamori F, Takahashi N, Kanda T, et al. (2011) A survey of beta-lactamase-producing Escherichia coli in farm animals and raw retail meat in Shizuoka Prefecture, Japan. Japanese Journal of Infectious Diseases 64: 153–155. [PubMed] [Google Scholar]
- 35. Poirel L, Cattoir V, Nordmann P (2008) Is plasmid-mediated quinolone resistance a clinically significant problem? Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases 14: 295–297. [DOI] [PubMed] [Google Scholar]
- 36. Robicsek A, Jacoby GA, Hooper DC (2006) The worldwide emergence of plasmid-mediated quinolone resistance. The Lancet Infectious Diseases 6: 629–640. [DOI] [PubMed] [Google Scholar]
- 37. Takasu H, Suzuki S, Reungsang A, Pham HV (2011) Fluoroquinolone (FQ) contamination does not correlate with occurrence of FQ-resistant bacteria in aquatic environments of Vietnam and Thailand. Microbes and Environments/JSME 26: 135–143. [DOI] [PubMed] [Google Scholar]
- 38. Shin JH, Jung HJ, Lee JY, Kim HR, Lee JN, et al. (2008) High rates of plasmid-mediated quinolone resistance QnrB variants among ciprofloxacin-resistant Escherichia coli and Klebsiella pneumoniae from urinary tract infections in Korea. Microbial Drug Resistance 14: 221–226. [DOI] [PubMed] [Google Scholar]
- 39. Teo JW, Ng KY, Lin RT (2009) Detection and genetic characterisation of qnrB in hospital isolates of Klebsiella pneumoniae in Singapore. International Journal of Antimicrobial Agents 33: 177–180. [DOI] [PubMed] [Google Scholar]
- 40. Ma J, Zeng Z, Chen Z, Xu X, Wang X, et al. (2009) High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrobial Agents and Chemotherapy 53: 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yue L, Jiang HX, Liao XP, Liu JH, Li SJ, et al. (2008) Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli . Veterinary Microbiology 132: 414–420. [DOI] [PubMed] [Google Scholar]
- 42. Cattoir V, Poirel L, Aubert C, Soussy CJ, Nordmann P (2008) Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerging Infectious Diseases 14: 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Picao RC, Poirel L, Demarta A, Silva CS, Corvaglia AR, et al. (2008) Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. The Journal of Antimicrobial Chemotherapy 62: 948–950. [DOI] [PubMed] [Google Scholar]
- 44. Walsh TR, Weeks J, Livermore DM, Toleman MA (2011) Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. The Lancet Infectious Diseases 11: 355–362. [DOI] [PubMed] [Google Scholar]
- 45. Begum YA, Talukder KA, Nair GB, Qadri F, Sack RB, et al. (2005) Enterotoxigenic Escherichia coli isolated from surface water in urban and rural areas of Bangladesh. Journal of Clinical Microbiology 43: 3582–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lothigius A, Janzon A, Begum YA, Sjöling A, Qadri F, et al. (2008) Enterotoxigenic Escherichia coli is detectable in water samples from an endemic area by real-time PCR. Journal of Applied Microbiology 104: 1128–1136. [DOI] [PubMed] [Google Scholar]
- 47. Ahmed D, Islam MS, Begum YA, Janzon A, Qadri F, et al. (2012) Presence of enterotoxigenic Escherichia coli in biofilms formed in water containers in poor households coincides with epidemic seasons in Dhaka. Journal of Applied Microbiology doi: 10.1111/jam.12109. [DOI] [PubMed] [Google Scholar]
- 48. Honma Y, Sasakawa C, Tsuji T, Iwanaga M (2003) Comparison of antimicrobial susceptibility between invasive and non-invasive Shigella organisms. International Journal of Antimicrobial Agents 21: 262–266. [DOI] [PubMed] [Google Scholar]
- 49. Talukder KA, Islam MA, Dutta DK, Hassan F, Safa A, et al. (2002) Phenotypic and genotypic characterization of serologically atypical strains of Shigella flexneri type 4 isolated in Dhaka, Bangladesh. Journal of Clinical Microbiology 40: 2490–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Talukder KA, Islam Z, Dutta DK, Islam MA, Khajanchi BK, et al. (2006) Antibiotic resistance and genetic diversity of Shigella sonnei isolated from patients with diarrhoea between 1999 and 2003 in Bangladesh. Journal of Medical Microbiology 55: 1257–1263. [DOI] [PubMed] [Google Scholar]