Abstract
Background
Recently, a new subfamily of long-chain toxins with a Kunitz-type fold was found in scorpion venom glands. Functionally, these toxins inhibit protease activity and block potassium channels. However, the genomic organization and three-dimensional (3-D) structure of this kind of scorpion toxin has not been reported.
Principal Findings
Here, we characterized the genomic organization and 3-D nuclear magnetic resonance structure of the scorpion Kunitz-type toxin, LmKTT-1a, which has a unique cysteine pattern. The LmKTT-1a gene contained three exons, which were interrupted by two introns located in the mature peptide region. Despite little similarity to other Kunitz-type toxins and a unique pattern of disulfide bridges, LmKTT-1a possessed a conserved Kunitz-type structural fold with one α-helix and two β-sheets. Comparison of the genomic organization, 3-D structure, and functional data of known toxins from the α-KTx, β-KTx, γ-KTx, and κ-KTx subfamily suggested that scorpion Kunitz-type potassium channel toxins might have evolved from a new ancestor that is completely different from the common ancestor of scorpion toxins with a CSα/β fold. Thus, these analyses provide evidence of a new scorpion potassium channel toxin subfamily, which we have named δ-KTx.
Conclusions/Significance
Our results highlight the genomic, structural, and evolutionary diversity of scorpion potassium channel toxins. These findings may accelerate the design and development of diagnostic and therapeutic peptide agents for human potassium channelopathies.
Introduction
Over the last 400 million years, scorpions have evolved many peptide toxins that target different potassium channels [1]. Numerous potassium channel toxins have been isolated from scorpions including those identified by proteomic and transcriptome analysis of scorpion venom glands [2]–[4]. These toxins are divided into α-KTx, β-KTx, γ-KTx, and κ-KTx subfamilies based on their similarity [5], [6]. Some of the toxins are specific inhibitors that serve as useful pharmacological tools and candidate drugs that target various potassium channels [7]. Examples include charybdotoxin (ChTX), which is targeted toward Kv1.3 and BKCa channels [8], scyllatoxin (ScyTx), which inhibits SKCa channels [9], maurotoxin (MTX), which is targeted toward IKCa channels [10], and BeKm-1, which inhibits Herg channels [11].
Despite the molecular diversity of scorpion potassium channel toxins, only two structural scaffolds have been found [12]. One is the classical CSα/β fold, which comprises one or two short α-helices connected to a triple-stranded anti-parallel β-sheet stabilized by three or four disulfide bonds. The other is the unique cystine-stabilized α-helix-loop helix (CSα/α) fold, which comprises two α-helices [5]. Recently, a new kind of long-chain scorpion potassium channel toxin was functionally characterized, which has both protease and potassium channel inhibiting properties [4], [13], [14]. Amino acid sequence analyses showed that this kind of scorpion toxin might adopt a unique Kunitz-type fold [15]. However, the three-dimensional (3-D) structures and features of this kind of toxin remain unclear.
In this work, we report the nuclear magnetic resonance (NMR) structure and genomic organization of the scorpion Kunitz-type toxin, LmKTT-1a. The NMR experiments show that LmKTT-1a adopts a conserved Kunitz-type structural fold [16], which is different from other scorpion potassium channel toxins including α-, β-, and γ-potassium toxins (KTxs), which have a CSα/β fold, and κ-KTxs, which have a CSα/α fold [1]. Based on the genomic and functional data, we propose that scorpion Kunitz-type toxins are a new subfamily of potassium channels, which we have named δ-KTx. Our results demonstrate that scorpion potassium channel toxins have greater diversity than previously realized and highlight a new role for convergent evolution of animal toxins.
Materials and Methods
Gene Cloning of Representative Scorpion Potassium Channel Toxins
To identify the upstream region of the gene, we amplified genomic DNA using a genome walking kit (Takata, Japan). Net-polymerase chain reaction (PCR) was used to amplify the downstream and 3′ flanking regions of the gene. This method consists of four nested gene-specific primers and two PCR steps. The second PCR product was used for purification and was ligated into the pGEM-T Easy Vector (Promega, USA) for sequencing. Escherichia coli JM109 was used for plasmid propagation. Positive clones were sequenced.
Construction of Expression Vectors
We used the cDNA sequence of LmKTT-1a from a cDNA library of Lychas mucronatus venom glands as a template for PCR to generate the fragment. The PCR product was digested with NdeI and BamHI and inserted into expression vector pET-28a. After being confirmed by sequencing, plasmid pET-28a-LmKTT-1a was transformed into E. coli Rosetta (DE3) cells for expression. The QuikChange Site-Directed Mutagenesis Kit (Stratagene, USA) was used to generate mutants from the wild-type plasmid, pET-28a-LmKTT-1a, which were verified by DNA sequencing.
Expression and Purification of LmKTT-1a
To produce 13C/15N-labeled LmKTT-1a, recombinant cells containing the LmKTT-1a expression plasmid were cultured and induced in M9 medium containing 15N-NH4Cl as the only nitrogen source and 13C-glucose as the only carbon source. The refolding, separation, and identification of LmKTT-1a was performed as previously described [13]. Briefly, the recombinant LmKTT-1a protein was expressed in inclusion bodies and then refolded in vitro at 16°C. The refolded protein was finally purified by high-performance liquid chromatography (HPLC) on a C18 column (10 mm × 250 mm, 5 µm; Elite-HPLC, China). The fraction containing recombinant LmKTT-1a was eluted after 20 to 21 min and was further analyzed by MALDI-TOF-MS (Voyager-DESTR, Applied Biosystems). Unlabeled LmKTT-1a and its mutants were expressed in LB culture according to the same protocol described above.
Circular Dichroism (CD) Analysis of rLmKTT-1a and its Analogues
The secondary structures of Kunitz-type toxin LmKTT-1a and its mutants were analyzed by CD spectroscopy. All samples were dissolved in water at a concentration of 0.2 mg/ml. Spectra were recorded at 25°C from 250 to 190 nm with a scan rate of 50 nm/min on a Jasco-810 spectropolarimeter. CD spectra data were collected by subtracting the blank spectrum of water and averaging three scans.
Electrophysiological Recordings
The cDNAs encoding mKv1.1, hKv1.2, and mKv1.3 were generously provided by Professor Stephan Grissmer (University of Ulm, Ulm, Germany) and were subcloned into the pIRES2-EGFP vector (Clontech). Kv1.1, Kv1.2, and Kv1.3 plasmids were then transiently transfected into HEK293 cells. The whole-cell patch clamp was used to measure and record the channel currents according to a previously described procedure [17].
Serine Protease Inhibitor Activity Assay
The inhibitory activities of LmKTT-1a and its mutants were tested using methods that were previously described [18], [19]. The initial rate of every reaction was monitored continuously at 405 nm for 5 min at 25°C. The inhibitory constant (Ki) of the protease/inhibitor complex was determined by Lineweaver-Burk plots and slope re-plotting.
Solution Structure Determination of LmKTT-1a
All of the NMR experiments were carried out at 298 K on a Bruker Avance-III 800 MHz spectrometer. The NMR sample contained 1 mM 13C/15N-labeled LmKTT-1a in 20 mM phosphate buffer with 10% deuterium oxide (D2O) (vol/vol) at pH 6.0. The backbone and side chain assignments of LmKTT-1a were determined from the following two- and three- dimensional NMR experiments: 1H-15N-HSQC, 1H-13C-HSQC, HNCO, HNCA, HN(CO)CA, HNCACB, CBCACONH, and 13C-HCCH-TOCSY. 3-D 15N-NOESY-HSQC and 13C-NOESY-HSQC spectra were recorded to generate inter-proton distance restraints. All of the spectra were processed with NMRPipe [20], and the assignments were accomplished using Computer-Aided Resonance Assignment [21]. Backbone dihedral angle restraints were derived from proton chemical shifts by TALOS (Torsion Angle Likelihood Obtained from Shift) [22]. The Cyana2.1 [23] and Assisted Model Building with Energy Restraint force field were used to calculate and refine the structures. Three pairs of disulfide bond restraints were added to the distance restraints based on the NOE evidences and position of cysteines from initial structure calculation (Fig. S4). Hydrogen bond restraints were obtained from the hydrogen-deuterium (H-D) exchange experiment after dissolving the lyophilized sample in D2O for 8 hours and adding it to the final run of the structure calculation (Fig. S3). The final 20 structures of LmKTT-1a with the lowest energies were assessed by PROCHECK-NMR [24] and graphically analyzed by MOLMOL [25]. It should be noted that recombinant LmKTT-1a contains 79 residues including a 20-residue expression tag at the N-terminus. The intact LmKTT-1a peptide contains 59 residues from Lys1 to Cys59. The protein structure was deposited into the Protein Data Bank (code 2M01) and the BioMagResBank (accession number 18789).
Results
Amino Acid Sequence Analysis of LmKTT-1a
Multiple sequence alignments showed that LmKTT-1a shared little homology with Kunitz-type toxins from other venomous animals, such as HWTX-XI from spider [26], APEKTx1 from sea anemone [27], α-DTX from snake [28], and conkunitzin-S1 from cone snail [29]. In addition, LmKTT-1a possessed a unique cysteine framework. It lacked the normal CysII–CysIV disulfide bridge but contained two cysteine residues at the C-terminus that might generate a new disulfide bridge (Fig. 1).
Figure 1. Amino acid sequence alignment of LmKTT-1a with Kunitz-type toxins from other venomous animals.
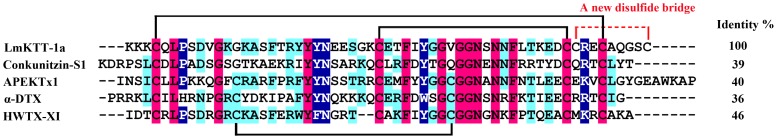
Representative Kunitz-type toxins are LmKTT-1a from scorpion, Conkunitzin-S1 (PDB Code: 1Y62) from conus, APEKTx1 (PDB Code: 1WQK) from sea anemone, α-DTX (PDB Code: 1DTX) from snake, and HWTXI-XI (PDB Code: 2JOT) from spider. The known disulfide bridges are labeled in black lines. The red dotted line suggests a possible new disulfide bridge.
Genomic Organization of LmKTT-1a
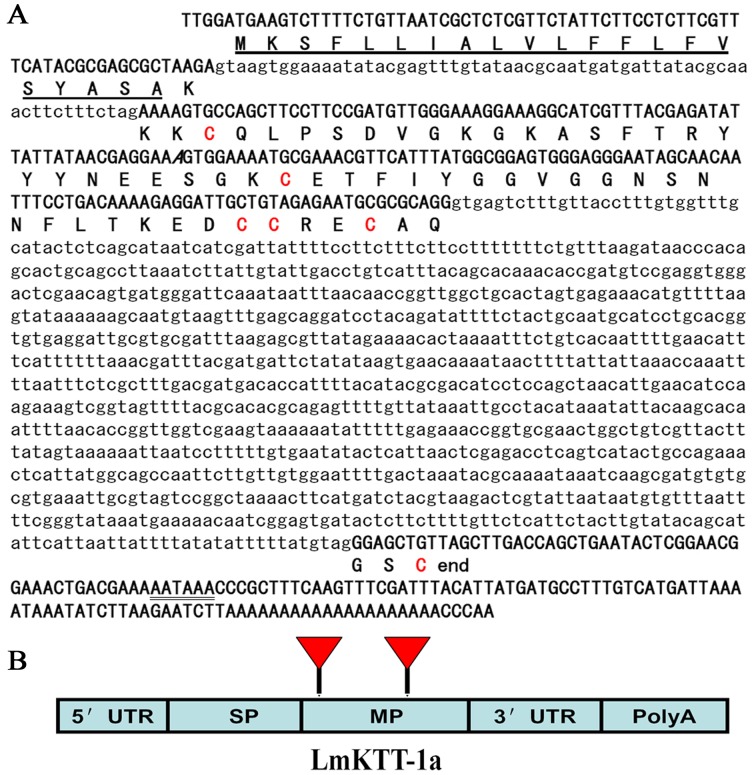
To determine the gene structure of LmKTT-1a, we first obtained the sequence of genomic DNA (Fig. 2A). A comparison of the LmKTT-1a genomic sequence with the corresponding cDNA sequence revealed that the gene contained three exons (a 5′ exon, an internal exon, and a 3′ exon) interrupted by two introns, which had a consensus GT-AG splice junction. Further analysis showed that the three exons corresponded to the signal peptide, mature peptide, and 3′-untranslated region (UTR), which are basic structural and functional domains of the LmKTT-1a precursor (Fig. 2B).
Figure 2. Genomic organization of scorpion Kunitz-type toxin, LmKTT-1a.
(A) The LmKTT-1a gene. The signal peptide sequence predicted from the nucleotide sequence is underlined. The putative polyadenylation signal (AATAAA) is underlined twice. (B) The gene structure of LmKTT-1a. The signal peptide (SP), mature peptide (MP), 5′-UTR, and 3′-UTR non-coding regions are shown. Introns are designated by triangles.
Pharmacological Activities of rLmKTT-1a
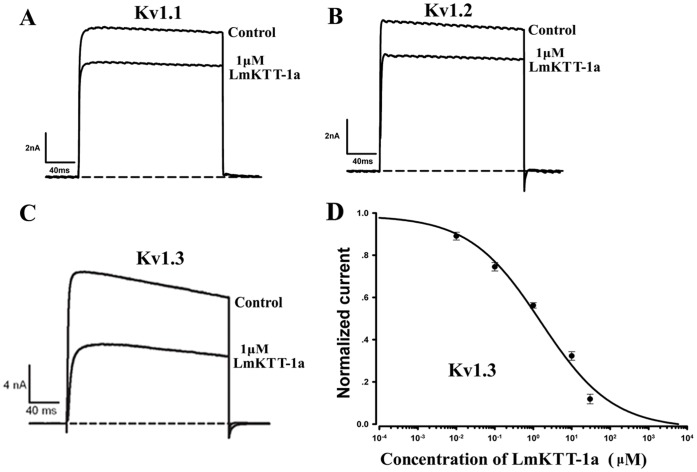
The biological effects of rLmKTT-1a on the Kv1.3 channel and trypsin protease were examined. The inhibitory effects of 1 µM rLmKTT-1a on the Kv1.1 channel, Kv1.2 channel, and Kv1.3 channel are shown (Fig. 3A–C). The results showed that 1 µM rLmKTT-1a inhibited the Kv1.3 current by approximately 50% with less effect on the Kv1.1 and Kv1.2 channels. The IC50 value of rLmKTT-1a on Kv1.3 channels was 1.58±0.73 µM (Fig. 3D). In addition, LmKTT-1a inhibited trypsin with a Ki value of about 0.14 µM. These results showed that LmKTT-1a is a unique bifunctional scorpion toxin with weak selectivity towards Kv1.3 channels.
Figure 3. Inhibition of Kv1 potassium channel activity by LmKTT-1a.
(A) Current traces of the Kv1.1 channel in the absence (control) or presence of 1 µM LmKTT-1a. (B) Current traces of the Kv1.2 channel in the absence (control) or presence of 1 µM LmKTT-1a. (C) Current traces of the Kv1.3 channel in the absence (control) or presence of 1 µM LmKTT-1a. (D) Concentration-dependent inhibition of Kv1.3 channel currents by LmKTT-1a. Data represent the mean ± S.D. of at least three experiments.
Solution Structure of LmKTT-1a
Due to its novel cysteine patterns and genomic organization, the NMR structure of LmKTT-1a was determined. Multi-dimensional NMR spectra were obtained using 13C- and 15N-labeled LmKTT-1a, and the signals were fully assigned (Figs. S1 and S2). NMR structural statistics are summarized in Table 1. The structure of LmKTT-1a was determined using 709 NOE distance restraints, 30 dihedral angle restraints, and 26 H-bond restraints. A family of 20 accepted structures with the lowest energies and the best Ramachandran plots was selected to represent the three-dimensional solution structure of LmKTT-1a. The Ramachandran plot revealed that 86.5% of the residues were in the most favored regions, 10.9% were in additionally allowed regions, and 2.6% were in generously allowed regions. A diagram showing overlay between the backbone atoms of the 20 lowest energy structures of LmKTT-1a is presented in Fig. 4A. The root mean square deviation from the mean structure was 0.97+/−0.24 Å for the backbone and 1.56+/−0.30 Å for heavy atoms (Fig. 4A).
Table 1. Structural statistics for the family of 20 structures of LmKTT-1a.
| Experimental constraints | |
| Intra-residue NOE (i = j) | 426 |
| Sequential NOE (|i-j| = 1) | 163 |
| Medium range (1<|i-j|<5) | 41 |
| Long range (|i-j|≥5) | 79 |
| Dihedral angle | 30 |
| Hydrogen bonds | 26 |
| Disulfide constraints | 9 |
| Target functiona (Å2) | 3.15+/−0.63 |
| RMSD Valuesa | |
| Average backbone RMSD to mean | 0.97+/−0.24 |
| Average heavy atom RMSD to mean | 1.56+/−0.30 |
| Ramachandran plotb | |
| Residues in most favored regions | 86.5% |
| Residues in additional regions | 10.9% |
| Residues in generously allowed regions | 2.6% |
| Residues in disallowed regions | 0.0% |
Data from Cyana.
Data from PROCHECK-NMR.
Figure 4. NMR solution structure of LmKTT-1a.
A) Superposition of the 20 structures with lowest total energy. (B) Ribbon presentation of the backbone of LmKTT-1a from scorpion. (C) Ribbon presentation of the backbone of ConK-S1 from snail. (D) Ribbon presentation of the backbone of α-dendrotoxin from snake. (E) Ribbon presentation of the backbone of HWTX-XI from spider. (F) Ribbon presentation of the backbone of APEKTX1 from sea anemone.
The solution structure of LmKTT-1a resembled a typical Kunitz-type fold (Fig. 4B), which contains an N-terminal helix from Lys2 to Cys4, double-stranded anti-parallel β-sheets from Phe17 to Asn23 and Lys28 to Tyr34, and a C-terminal helix from Asp49 to Ala55. The C-terminal helix contained a unique disulfide link between Cys51 and Cys59, which was confirmed by their direct Nuclear Overhauser Enhancement (NOE) contacts and long-range NOEs of the adjacent residues (Fig. S4C). The structural integrity of LmKTT-1a was also maintained by two additional disulfide bridges located at Cys4–Cys54 and Cys29–Cys50 (Fig. S4A and B). The Cys4–Cys54 linkage shortened the distance between the N-terminal and C-terminal helices (Fig. S1). The β-sheet was stabilized by the connection between Cys29 and the C-terminal Cys50, which was the most rigid region of the structure.
The solution structure of LmKTT-1a was very similar to other Kunitz-type toxins, such as ConK-S1 from snail [29], α-dendrotoxin from snake [16], HWTX-XI from spider [26], and APEKTX1 from sea anemone [27] (Fig. 4). Although LmKTT-1a showed low amino acid sequence homologies with classical Kunitz-type peptides, it still adopted a conserved Kunitz-type fold. These findings suggest that peptides with a Kunitz-type fold maintain structural conservation yet show molecular diversity.
Effects of the Unique Cys51–Cys59 Disulfide Bridge on Functions of LmKTT-1a
LmKTT-1a was a Kunitz-type toxin that adopted unique disulfide bridges (Figs. 1 and 4). To evaluate the function of characteristic disulfide bridges in LmKTT-1a, a mutant LmKTT-1a-C51A/C59A was designed (Fig. 5A), which had the same kind of disulfide bridge as the sea anemone ConK-S1 Kunitz-type toxin. Circular dichroism spectroscopy indicated that rLmKTT-1a-C51A/C59A had a secondary structure similar to LmKTT-1a (Fig. 5D). Enzyme and inhibitor reaction kinetics experiments showed that recombinant LmKTT-1a-C51A/C59A inhibited trypsin with 5-fold lower activity than wild-type LmKTT-1a (Fig. 5A). Electrophysiological experiments further demonstrated that LmKTT-1a-C51A/C59A inhibited the Kv1.3 channel to a similar degree as LmKTT-1a. Our results indicated that the unique Cys51–Cys59 disulfide bridge of LmKTT-1a had weak effects on its abilities to inhibit potassium channels and proteases, highlighting the evolutionary diversity and functional conservation of Kunitz-type toxins that have different disulfide bridge patterns.
Figure 5. Functional evaluation of the unique disulfide bridge, Cys51–Cys59, in LmKTT-1a. (.
A) A mutant LmKTT-1a-C51A/C59A lacking the unique disulfide bridge Cys51–Cys59 was designed from LmKTT-1a. Ki values for trypsin are labeled in bold font. (B) Current traces in the absence (control) or presence of 1 µM LmKTT-1a-C51A/C59A and LmKTT-1a. (C) Structural stability of the LmKTT-1a mutant, LmKTT-1a-C51A/C59A.
Discussion
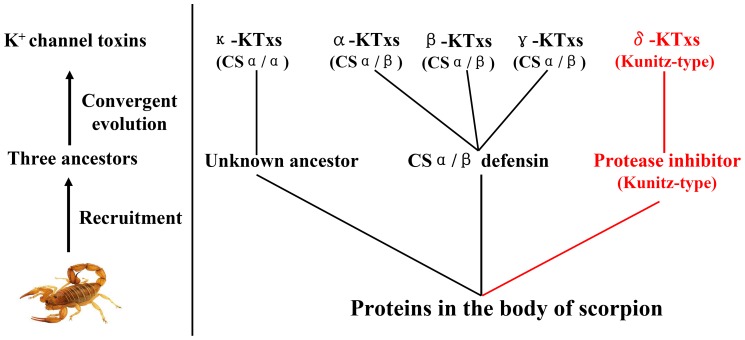
Kunitz-type peptides are ubiquitous in numerous organisms including plants, animals, and microbes [30], [31]. The first Kunitz-type toxin to be reported was α-dendrotoxin, which was isolated from snake venom in 1974 [32]. Subsequently, additional Kunitz-type toxins were isolated from the venom glands of snakes, lizards, cone snails, spiders, and sea anemones [26]. Recently, scorpion Kunitz-type toxins were found and functionally characterized as trypsin and Kv1.3 channel inhibitors [13]. Based on previous findings, we determined the 3-D structure and genomic organization of a representative scorpion Kunitz-type toxin, LmKTT-1a, and discovered unique structural and evolutionary features of scorpion potassium channel toxins (Fig. 6). Our work highlights the third evolutionary ancestor of scorpion potassium channel toxins [33], [34].
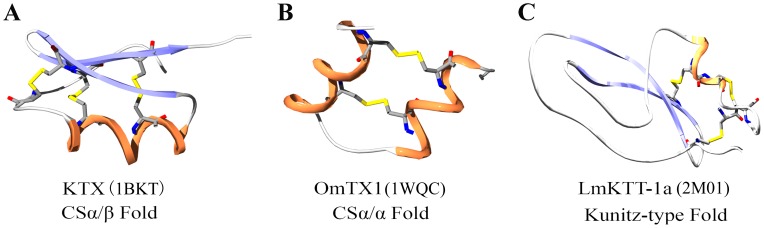
Figure 6. Diverse structural fold of scorpion potassium channel toxins.
(A) Ribbon presentation of the backbone of KTX from the α-KTX subfamily, which has a CSα/β fold. (B) Ribbon presentation of the backbone of OmTx1 from the κ-KTX subfamily, which has a CSα/α fold. (C) Ribbon presentation of the backbone of LmKTT-1a from a new subfamily with a Kunitz-type fold.
Diverse Genomic Organization of Scorpion Potassium Channel Toxins
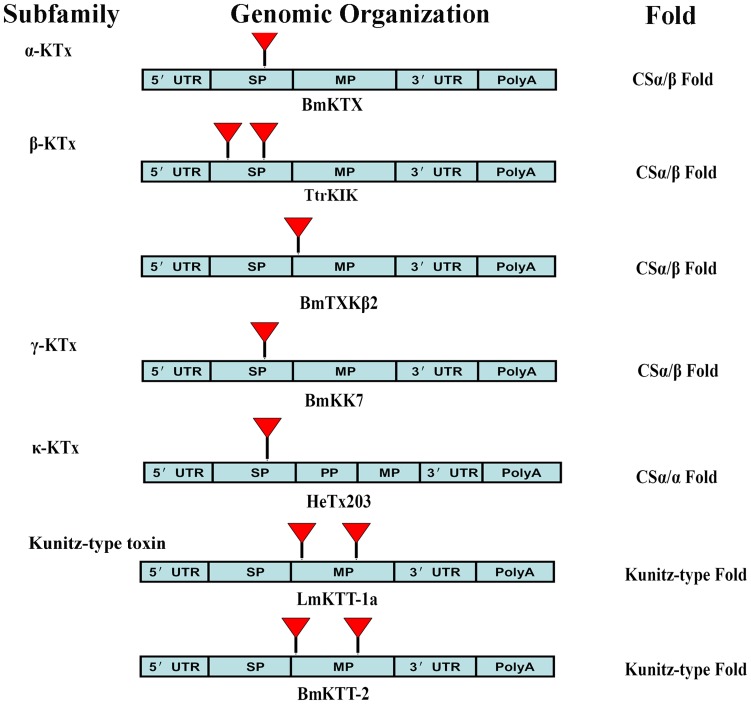
To compare the genomic organization of LmKTT-1a with known scorpion potassium channel toxins, we investigated the genomic organization of another scorpion Kunitz-type toxin, BmKTT-2, and representative scorpion potassium channel toxins from four known KTx subfamilies (Table 2 and Fig. 7). Our results showed that scorpion Kunitz-type potassium channel toxins have a unique genomic organization, which is different from that of scorpion toxins in the other four KTx subfamilies. The new genomic organization of scorpion Kunitz-type toxins further revealed the diversity of scorpion potassium channel toxins.
Table 2. Diversity of potassium channel toxins (KTxs) from scorpion venom.
| Classification | Gene Structure | 3-D Fold | Activity |
| α-KTx subfamily | One intron in the signal peptide | CSα/β | Kv1, hERG, SKCa, BKCa, IKCa effectors |
| β-KTx subfamily | One intron in the mature peptide or two introns in the signal peptide | CSα/β | Kv1.3 effectors, antimicrobial and anti-malaria |
| γ-KTx subfamily* | One intron in the signal peptide | CSα/β | hERG effectors |
| κ-KTx subfamily* | One big intron in the signal peptide | CSα/α | Kv1.3 effectors |
| δ-KTx subfamily* | Two introns in the mature peptide | Kunitz-type | Kv1.3 effectors and Trypsin inhibitors |
The genomic organization of this subfamily was reported firstly in this work.
Figure 7. Comparison of the gene structures of representative scorpion potassium channel toxins.
The gene structures of scorpion potassium channel toxins BmKTX from the α-KTX subfamily, which has a CSα/β fold, TtrKIK and BmTXKβ2 from the β-KTX subfamily, which has a CSα/β fold, BmKK7 from the γ-KTX subfamily, which has a CSα/β fold, HeTx203 from the κ-KTX subfamily, which has a CSα/α fold, and LmKTT-1a and BmKTT-2 from δ-KTX, the new subfamily of Kunitz-type fold toxins. The signal peptide (SP), propeptide (PP), mature peptide (MP), 5′-UTR, and 3′-UTR non-coding regions are shown. Introns are designated by triangles.
Structural Diversity of Scorpion Potassium Channel Toxins
The only structural folds that have been identified in scorpion potassium channel toxins are CS-α/β (cysteine-stabilized α-helix and β sheets) [35] and CS-α/α (cysteine-stabilized α-helix-loop-helix) [5]. Scorpion toxin LmKTT-1a possessed a unique cysteine framework and adopted a conserved Kunitz-type fold. To our knowledge, the LmKTT-1a structure is the third structural fold to be discovered for scorpion potassium channel toxins [15], demonstrating their structural diversity (Fig. 6).
A Unified Nomenclature for Scorpion Kunitz-type Toxins: δ-KTx Subfamily
Currently, the principle of classification and unified nomenclature for scorpion potassium channel toxins is based on their amino acid sequence similarities, type of structural folds, and genomic organization [5], [6]. Based on the structural, functional, and genomic comparison of scorpion Kunitz-type toxins with toxins from the four known subfamilies, we propose to classify scorpion Kunitz-type potassium channel toxins as a new KTx subfamily called δ-KTx. To our knowledge, this new KTx subfamily from scorpion contains seven members and can be classified into three groups with three different disulfide bridge patterns (Fig. 8).
Figure 8. Molecular diversity and classification of scorpion potassium channel toxins.
Representative potassium channel toxins from the α-KTxs, β-KTxs, γ-KTxs, κ-KTxs, and δ-KTxs subfamily are listed. All members from the δ-KTxs subfamily with a Kunitz-type fold are shown.
Evolutionary Diversity of Scorpion Potassium Channel Toxins
Given the selective pressure that occurs during the course of evolution, conservation between two proteins or peptides at the levels of genomic organization and 3-D structure provides strong evidence for a common evolutionary origin [36]. Previous research has shown that scorpion potassium channel toxins with a CSα/β fold share a common ancestor [34]. In this work, we provide the first report of the genomic organization of scorpion potassium channel toxins with Kunitz-type and CSα/α structural folds (Figs. 2, S5, and S6). Our results showed that scorpion potassium channel toxins with Kunitz-type fold and CSα/α fold adopted their unique genomic organization and structural folds and are completely different from the classical CSα/β toxins (Figs. 7 and S7). Combining the genomic organization, 3-D fold, and functional data together suggested that scorpion potassium channel toxins might have evolved from three different ancestors (Fig. 9). Our results highlight a new role for convergent evolution of animal toxins and demonstrate the evolutionary diversity of scorpion potassium channel toxins.
Figure 9. Schematic diagram of the evolution of scorpion potassium channel toxins.
Three putative ancestors were recruited from scorpion proteins to generate diverse potassium channel toxins from five different subfamilies (α-KTxs, β-KTxs, γ-KTxs, κ-KTxs, and δ-KTxs) with three different structural folds (CSα/β, CSα/β, and Kunitz-type).
Conclusions
In summary, we present the first characterization of the NMR solution structure and genomic organization of scorpion Kunitz-type potassium channel toxin. LmKTT-1a possessed a novel genomic organization and a conserved Kunitz-type structural fold with one α-helix and two β-sheets. Genomic and structural features were different from that of the classical scorpion potassium channel toxins, such as α-KTxs, β-KTxs, γ-KTxs, and κ-KTxs. Based on these analyses, we propose that a new subfamily of scorpion potassium channel toxins exists, which we have named δ-KTx. Our results highlight the structural, genomic, and evolutionary diversity of scorpion potassium channel toxins. These results may accelerate the design and development of diagnostic and therapeutic peptide agents for human potassium channelopathies.
Supporting Information
1H-15N HSQC spectrum of LmKTT-1a. The spectrum was labeled with all of the assignments including the expression tag (from residue tG1 to tM20) and the intact peptide (from residue K1 to C59). The amino acid sequence of rLmKTT-1a is GSSHHHHHHSSGLVPRGSHMKKKCQLPSDVGKGKASFTRYYYNEESGKCETFIYGGVGGNSNNFLTKEDCCRECAQGSC. The amide protons of Histidine residues (tH4–tH8) from the His-tag are severely overlapped and were not assigned. All assigned cross-peaks have been labeled with a one-letter amino acid code.
(TIF)
NOESY spectra of LmKTT-1a. Sequential assignment strips of the intact peptide are shown by 3D 15N-1H NOESY-HSQC.
(TIF)
1H-15N HSQC spectrum of LmKTT-1a after dissolving it in D2O for 8 hours.
(TIF)
NOE evidences for disulfide bonds C4–C54 (A), C29–C50 (B) and C51–C59 (C). The NOE connections were illustrated by 13C-edited NOESY-HSQC spectrum and a stick representation of the structure. The structure in the right panel was produced using the MOLMOL program. The sidechains of relevant residues were shown in red and indicated by residue name and number.
(TIF)
The HeTx203 gene sequence from the κ-KTx subfamily, which has a CSα/α fold, is shown. The signal peptide sequence predicted from the nucleotide sequence is underlined.
(TIF)
The BmKTT-2 gene sequence from the δ-KTx subfamily, which has a Kunitz-type fold, is shown. The signal peptide sequence predicted from the nucleotide sequence is underlined.
(TIF)
The BmKK7 gene sequence from the γ-KTx subfamily, which has a CSα/β fold, is shown. The signal peptide sequence predicted from the nucleotide sequence is underlined.
(TIF)
Funding Statement
This work was supported by grants from the National Basic Research Program of China (2010CB529800 and 2009CB918600), National High Technology Research and Development Program of China (2012AA020304), and National Natural Sciences Foundation of China (number 30530140, 31071942, 31200557 and 21173257). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rodriguez de la Vega RC, Schwartz EF, Possani LD (2010) Mining on scorpion venom biodiversity. Toxicon 56: 1155–1161. [DOI] [PubMed] [Google Scholar]
- 2. Ruiming Z, Yibao M, Yawen H, Zhiyong D, Yingliang W, et al. (2010) Comparative venom gland transcriptome analysis of the scorpion Lychas mucronatus reveals intraspecific toxic gene diversity and new venomous components. BMC Genomics 11: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. He QY, He QZ, Deng XC, Yao L, Meng E, et al. (2008) ATDB: a uni-database platform for animal toxins. Nucleic Acids Res 36: D293–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwartz EF, Diego-Garcia E, Rodriguez de la Vega RC, Possani LD (2007) Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones). BMC Genomics 8: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chagot B, Pimentel C, Dai L, Pil J, Tytgat J, et al. (2005) An unusual fold for potassium channel blockers: NMR structure of three toxins from the scorpion Opisthacanthus madagascariensis. Biochem J 388: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tytgat J, Chandy KG, Garcia ML, Gutman GA, Martin-Eauclaire MF, et al. (1999) A unified nomenclature for short-chain peptides isolated from scorpion venoms: alpha-KTx molecular subfamilies. Trends Pharmacol Sci 20: 444–447. [DOI] [PubMed] [Google Scholar]
- 7. Wulff H, Castle NA, Pardo LA (2009) Voltage-gated potassium channels as therapeutic targets. Nat Rev Drug Discov 8: 982–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gimenez-Gallego G, Navia MA, Reuben JP, Katz GM, Kaczorowski GJ, et al. (1988) Purification, sequence, and model structure of charybdotoxin, a potent selective inhibitor of calcium-activated potassium channels. Proc Natl Acad Sci U S A 85: 3329–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu Y, Cao Z, Yi H, Jiang D, Mao X, et al. (2004) Simulation of the interaction between ScyTx and small conductance calcium-activated potassium channel by docking and MM-PBSA. Biophys J 87: 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kharrat R, Mansuelle P, Sampieri F, Crest M, Oughideni R, et al. (1997) Maurotoxin, a four disulfide bridge toxin from Scorpio maurus venom: purification, structure and action on potassium channels. FEBS Lett 406: 284–290. [DOI] [PubMed] [Google Scholar]
- 11. Zhang M, Korolkova YV, Liu J, Jiang M, Grishin EV, et al. (2003) BeKm-1 is a HERG-specific toxin that shares the structure with ChTx but the mechanism of action with ErgTx1. Biophys J 84: 3022–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mouhat S, Jouirou B, Mosbah A, De Waard M, Sabatier JM (2004) Diversity of folds in animal toxins acting on ion channels. Biochem J 378: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen ZY, Hu YT, Yang WS, He YW, Feng J, et al. (2012) Hg1, novel peptide inhibitor specific for Kv1.3 channels from first scorpion Kunitz-type potassium channel toxin family. J Biol Chem 287: 13813–13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao R, Dai H, Qiu S, Li T, He Y, et al. (2011) SdPI, The First Functionally Characterized Kunitz-Type Trypsin Inhibitor from Scorpion Venom. PLoS ONE 6: e27548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith JJ, Hill JM, Little MJ, Nicholson GM, King GF, et al. (2011) Unique scorpion toxin with a putative ancestral fold provides insight into evolution of the inhibitor cystine knot motif. Proc Natl Acad Sci U S A 108: 10478–10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harvey AL (2001) Twenty years of dendrotoxins. Toxicon 39: 15–26. [DOI] [PubMed] [Google Scholar]
- 17. Han S, Yi H, Yin SJ, Chen ZY, Liu H, et al. (2008) Structural basis of a potent peptide inhibitor designed for Kv1.3 channel, a therapeutic target of autoimmune disease. J Biol Chem 283: 19058–19065. [DOI] [PubMed] [Google Scholar]
- 18. Zhao R, Dai H, Qiu S, Li T, He Y, et al. (2011) SdPI, the first functionally characterized Kunitz-type trypsin inhibitor from scorpion venom. PLoS One 6: e27548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng YC, Yan FJ, Chang LS (2005) Taiwan cobra chymotrypsin inhibitor: cloning, functional expression and gene organization. Biochim Biophys Acta 1747: 213–220. [DOI] [PubMed] [Google Scholar]
- 20. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, et al. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- 21. Masse JE, Keller R (2005) AutoLink: automated sequential resonance assignment of biopolymers from NMR data by relative-hypothesis-prioritization-based simulated logic. J Magn Reson 174: 133–151. [DOI] [PubMed] [Google Scholar]
- 22. Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- 23. Guntert P, Mumenthaler C, Wuthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273: 283–298. [DOI] [PubMed] [Google Scholar]
- 24. Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486. [DOI] [PubMed] [Google Scholar]
- 25.Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- 26. Yuan CH, He QY, Peng K, Diao JB, Jiang LP, et al. (2008) Discovery of a distinct superfamily of Kunitz-type toxin (KTT) from tarantulas. PLoS One 3: e3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peigneur S, Billen B, Derua R, Waelkens E, Debaveye S, et al. (2011) A bifunctional sea anemone peptide with Kunitz type protease and potassium channel inhibiting properties. Biochem Pharmacol 82: 81–90. [DOI] [PubMed] [Google Scholar]
- 28. Smith LA, Lafaye PJ, LaPenotiere HF, Spain T, Dolly JO (1993) Cloning and functional expression of dendrotoxin K from black mamba, a K+ channel blocker. Biochemistry 32: 5692–5697. [DOI] [PubMed] [Google Scholar]
- 29. Bayrhuber M, Vijayan V, Ferber M, Graf R, Korukottu J, et al. (2005) Conkunitzin-S1 is the first member of a new Kunitz-type neurotoxin family. Structural and functional characterization. J Biol Chem 280: 23766–23770. [DOI] [PubMed] [Google Scholar]
- 30. Hansen D, Macedo-Ribeiro S, Verissimo P, Yoo Im S, Sampaio MU, et al. (2007) Crystal structure of a novel cysteinless plant Kunitz-type protease inhibitor. Biochem Biophys Res Commun 360: 735–740. [DOI] [PubMed] [Google Scholar]
- 31. Chakrabarty A, Green JA, Roberts RM (2006) Origin and evolution of the TKDP gene family. Gene 373: 35–43. [DOI] [PubMed] [Google Scholar]
- 32. Szopa J (1974) Interaction with DNA of the acetylated and non-acetylated polyvalent basic trypsin inhibitor of the Kunitz type. Acta Biochim Pol 21: 151–157. [PubMed] [Google Scholar]
- 33.Zhu S, Peigneur S, Gao B, Luo L, Jin D, et al.. (2011) Molecular diversity and functional evolution of scorpion potassium channel toxins. Mol Cell Proteomics 10: M110 002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu S, Huys I, Dyason K, Verdonck F, Tytgat J (2004) Evolutionary trace analysis of scorpion toxins specific for K-channels. Proteins 54: 361–370. [DOI] [PubMed] [Google Scholar]
- 35. Rodriguez de la Vega RC, Possani LD (2004) Current views on scorpion toxins specific for K+-channels. Toxicon 43: 865–875. [DOI] [PubMed] [Google Scholar]
- 36. Zhu S, Darbon H, Dyason K, Verdonck F, Tytgat J (2003) Evolutionary origin of inhibitor cystine knot peptides. Faseb J 17: 1765–1767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1H-15N HSQC spectrum of LmKTT-1a. The spectrum was labeled with all of the assignments including the expression tag (from residue tG1 to tM20) and the intact peptide (from residue K1 to C59). The amino acid sequence of rLmKTT-1a is GSSHHHHHHSSGLVPRGSHMKKKCQLPSDVGKGKASFTRYYYNEESGKCETFIYGGVGGNSNNFLTKEDCCRECAQGSC. The amide protons of Histidine residues (tH4–tH8) from the His-tag are severely overlapped and were not assigned. All assigned cross-peaks have been labeled with a one-letter amino acid code.
(TIF)
NOESY spectra of LmKTT-1a. Sequential assignment strips of the intact peptide are shown by 3D 15N-1H NOESY-HSQC.
(TIF)
1H-15N HSQC spectrum of LmKTT-1a after dissolving it in D2O for 8 hours.
(TIF)
NOE evidences for disulfide bonds C4–C54 (A), C29–C50 (B) and C51–C59 (C). The NOE connections were illustrated by 13C-edited NOESY-HSQC spectrum and a stick representation of the structure. The structure in the right panel was produced using the MOLMOL program. The sidechains of relevant residues were shown in red and indicated by residue name and number.
(TIF)
The HeTx203 gene sequence from the κ-KTx subfamily, which has a CSα/α fold, is shown. The signal peptide sequence predicted from the nucleotide sequence is underlined.
(TIF)
The BmKTT-2 gene sequence from the δ-KTx subfamily, which has a Kunitz-type fold, is shown. The signal peptide sequence predicted from the nucleotide sequence is underlined.
(TIF)
The BmKK7 gene sequence from the γ-KTx subfamily, which has a CSα/β fold, is shown. The signal peptide sequence predicted from the nucleotide sequence is underlined.
(TIF)