Abstract
DNA methylome analysis in a variety of species has revealed elevated 5-methylcytosine at exons relative to introns. These associations raised the possibility that intragenic DNA methylation aids the spliceosome in the process of exon definition. Here, I highlight recent genome-wide associations and direct evidence linking DNA methylation to pre-mRNA splicing.
Keywords: DNA methylation, RNA polymerase II, alternative pre-mRNA splicing
The role of chromatin structure in pre-mRNA processing decisions has attracted significant attention over the last several years. At the root of this interest are a number of studies indicating that exonic DNA presents a distinct chromatin environment relative to intronic DNA. A variety of genome-wide profiling efforts have described elevated nucleosome occupancy, specific histone modifications and increased DNA methylation at exons relative to introns.1-8 These striking observations revealed a potential role for exonic chromatin structure in the process of pre-mRNA splicing. Spliceosome assembly occurs cotranscriptionally and is influenced by the rate of RNA polymerase II (RNAP II) elongation, such that a low rate favors inclusion of weak exons in spliced mRNAs.9 It was thus proposed that chromatin could promote exon definition by altering elongation kinetics or by acting as an adaptor for splicing factor recruitment.10,11 Evidence linking histone modifications to both these levels of regulation has accumulated over the last several years and is extensively reviewed elsewhere.10,12 In contrast, a potential role for nucleosomes and DNA methylation in splicing regulation has been relatively unexplored. Here, I present genome-wide studies as well as recent data from our laboratory that begin to reveal a direct role for DNA methylation in alternative splicing regulation.
While the overall extent of DNA methylation shows a wide-range of distribution in eukaryotes, certain trends are conserved. For example, the majority of DNA methylation occurs on cytosines found in a CpG context, and methylation is relatively enriched within gene bodies.13,14 Comparative analysis of several distinct eukaryotic genomes suggests that primitive DNA methylation was present in the last common ancestor of plants and animals and was preferentially targeted to gene bodies. In contrast, intergenic DNA methylation appears to have evolved independently in eukaryotic genomes as a defense against transposons.14 Although studies of DNA methylation have largely focused on its silencing activity in the human genome, the sum of these genome-wide comparisons suggest a conserved functional role for intragenic DNA methylation, which is explored in greater detail below.
In the human genome, with the exception of CpG islands (CGIs) at promoters, the bulk of CpGs are methylated.6 Hypermethylation of promoter CGIs has been associated with transcriptional silencing.15 In contrast, comparison of methylome and transcriptome data fails to identify a clear effect on gene expression, bringing the role of widespread intragenic DNA methylation into question.6 Potential insight may be gleaned from the distribution pattern of gene body methylation. Various techniques for methylome analysis in a variety of species have consistently come to the same conclusion: DNA methylation is enriched at exons relative to introns.6,13 This pattern is highlighted in the honeybee genome (Apis mellifera), wherein DNA methylation is notably absent in intergenic regions where it is abundantly found in humans, such as transposons and telomeres, and is instead concentrated at exonic DNA.13,16 While greater than 70% of CpGs are targeted for methylation in humans as compared with approximately 1% in honeybees,6,13 similar associations were yielded from human methylome analysis. In addition to a general enrichment of DNA methylation at exons,6,8 whole-genome bisulfite sequencing in several human tissues revealed abrupt transitions in 5-methyl-cytosine (5-mC) levels at exon-intron junctions.17 Specifically, DNA methylation showed a sharp spike at 5′ splice sites and sharp dip at 3′ splice sites. A downward methylation gradient was detected across exons, whereas an upward gradient was detected across introns.17 Additional evidence in support of a role for DNA methylation in exon definition came from a comprehensive reanalysis of several human methylome and RNA-sequencing (RNA-seq) studies.18 Exons identified as excluded through RNA-seq showed a lower level of DNA methylation than included or highly expressed exons. Similarly, non-coding untranslated regions (UTRs) showed a lower overall level of DNA methylation than all types of coding exons. A notable exception was 5′ coding exons, which showed no enrichment in DNA methylation relative to introns.18 Importantly, inclusion of this class of exons in spliced mRNA is not dependent on 5′ splice site recognition by the spliceosome, thereby bolstering the hypothesis that DNA methylation supports the spliceosome in the process of exon definition. While exonic DNA is intrinsically CpG-rich relative to intronic sequences,19 elevated DNA methylation at exons was maintained following correction for CpG content.18 Likewise, whereas DNA methyltransferases (DNMTs) are preferentially targeted to nucleosomes,20 analysis of nucleosome positioning and DNA methylation as a function of gene expression indicated a high-degree of independence of these variables.18 Altogether, these studies pointed to a conserved role for DNA methylation in exon definition, yet the mechanism remained unclear.
As described above, pre-mRNA processing occurs cotranscriptionally, raising the possibility that intragenic DNA methylation could directly promote exon definition through interaction with auxiliary proteins and/or through kinetic regulation of RNAP II elongation. By extension of this logic, perturbations to intragenic DNA methylation could thereby result in alternative pre-mRNA splicing. Several lines of indirect evidence support this premise. Returning to the honeybee, brain methylome analysis in honeybee castes demonstrated a correlative link between DNA methylation and alternative splicing.16 Queen bees and worker bees are identical at the DNA level, but show significant differences at the methylome level. RNAi-mediated depletion of the de novo methyltransferase (DNMT3) generated adult bees with Queen-like characteristics, suggesting a direct role for DNA methylation in shaping the downstream proteome.21 Indeed, combined RNA-seq and bisulfite sequencing in the brains of Queen and worker bees revealed a genome-wide association between DNA methylation and alternative pre-mRNA splicing.16 Additional correlative support linking DNA methylation and splicing in honeybees comes from the observation that 5-mC was detected at the exons of intron-containing but not intronless histone-encoding genes.16
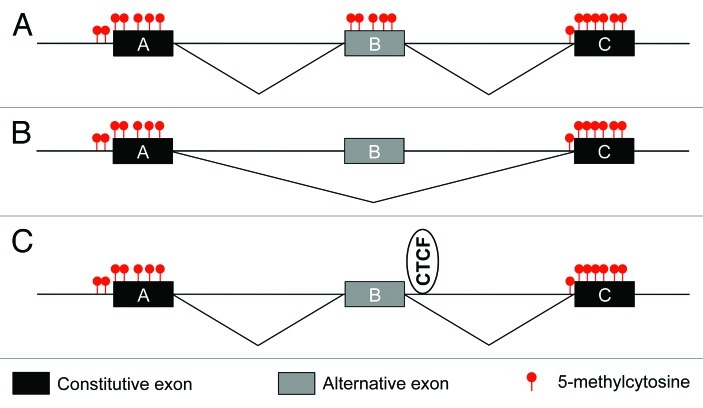
Recently, we demonstrated a direct mechanistic association between DNA methylation and alternative pre-mRNA splicing. In our studies of activation-induced alternative splicing of CD45 transcripts, we uncovered a role for the DNA binding protein, CCCTC-binding factor (CTCF) in splicing regulation.22 Alternative splicing of exons 4–6 of CD45 transcripts is tightly linked to the stages of lymphocyte development. In general, naïve peripheral lymphocytes include all three exons, whereas mature cells uniformly exclude the variable exons.23 We previously identified induction of heterogeneous ribonucleoprotein L-like as a critical switch for exons 4 and 6 exclusion from CD45 transcripts.24 However, neither our own efforts, nor those of others have succeeded in identifying a crucial RNA-binding regulator of exon 5 exclusion. Given the mounting evidence pointing to a role for epigenetics in splicing regulation, we examined the CD45 DNA. In so doing, we discovered that exon 5 of CD45 DNA is bound by CTCF and that binding is maintained in cells that express abundant CD45.22 CTCF has previously been shown to pose a barrier to RNAP II transcription,25 thereby positioning CTCF as a potential kinetic regulator of pre-mRNA splicing. Through utilizing lymphocytes with variable CTCF binding at CD45 exon 5 and RNAi-mediated depletion, we confirmed that CTCF promotes exon 5 inclusion in spliced mRNA through enforcing local RNAP II pausing.22 Importantly, CTCF interaction with DNA is inhibited by the presence of 5-mC.26 We showed that exclusion of exon 5 from CD45 transcripts was invariably associated with elevated methylation of exon 5 DNA with a reciprocal loss in CTCF and RNAP II pausing. Reversion of exon 5 DNA methylation through DNMT1 depletion, reestablished CTCF binding and RNAP II pausing, as well as exon 5 inclusion.22 Notably, earlier deletional minigene studies from the Lynch laboratory identified the region of CD45 exon 5 corresponding to the CTCF binding site to be important for exon 5 inclusion.27 Combined CTCF ChIP-seq and RNA-seq in CTCF-depleted cells vs. their relevant controls indicated that intragenically bound CTCF is a global regulator of alternative splicing. Specifically, intragenic binding of CTCF promotes inclusion of weak upstream exons, but not downstream exons, thereby supporting its role in the kinetic regulation of splicing.22 Overall, these data suggest that reciprocal variations in intragenic CTCF and DNA methylation during development or in pathological conditions, such as tumorigenesis, could have broad consequences on alternative pre-mRNA splicing (Fig. 1).
Figure 1. Genome-wide associations support a role for 5-methylcytosine in pre-mRNA splicing regulation. (A and B) Constitutively included exons (black) show elevated DNA methylation relative to introns. Alternative exons (gray) with a high-degree of methylation show increased inclusion levels relative to alternative exons with reduced methylation. (C) Binding of CTCF to unmethylated binding sites promotes inclusion of weak upstream exons.
It is worth noting that in our model system, DNA methylation is associated with exon exclusion, which is opposite to the genome-wide trends discussed above. A similar relationship was seen for the GB18602 gene is honeybees, wherein decreased DNA methylation of an internal cassette exon in Queen bees relative to worker bees was associated with increased inclusion of the exon in mature transcripts.16 It is possible that interaction of methyl-binding proteins with 5-mC may pose similar barriers to RNAP II elongation as CTCF at CD45 exon 5, resulting in analogous effects on exon inclusion. Alternatively, we cannot exclude the possibility that methyl-sensitive proteins function as adaptors for factors that promote or inhibit spliceosome assembly. Indeed, it is conceivable that all these variables synergize to contribute to the exquisite regulation of pre-mRNA splicing in higher eukaryotes.
In ending, I will highlight an additional layer of complexity in the relationship between DNA methylation and pre-mRNA splicing. Several recent studies have identified a potential role for 5-hydroxymethyl-cytosine (5-hmC) in exon definition. Whole genome profiling of 5-hmC in human cells yielded a similar association to the 5-mC studies described above: 5-hmC is elevated in gene bodies with a substantial enrichment at exons relative to introns.28,29 Notably, 5-hmC overlaps with CTCF binding sites,29 hinting at a possible direct association. Moreover, these studies highlight an important caveat of the genome-wide methylome analyses described above: bisulfite sequencing does not distinguish 5-mC from 5-hmC. Pending the development of integrated data sets examining 5-hmC, 5-mC and transcript processing, we cannot conclude whether these DNA modifications mediate distinct effects on alternative pre-mRNA splicing. As the TET proteins that catalyze conversion of 5-mC to 5-hmC are variably expressed during development,30 it is tempting to speculate that plasticity between these modifications contributes to the developmental regulation of alternative splicing.
All in all, it is clear from the studies described herein that methylation of euchromatic DNA plays an important role in gene expression beyond the paradigm of promoter-associated silencing. Intragenic DNA methylation is emerging as a highly conserved, critical determinant of pre-mRNA processing. Whether decreased DNA methylation at the 5′ and 3′ UTRs functions in end processing of transcripts will be an interesting question for future studies. In addition, it will be important to determine the cellular signals that promote variations in intragenic DNA methylation that culminate in regulated alternative splicing decisions.
Acknowledgments
I apologize to colleagues whom I could not cite due to space constraints. This research was supported by the Intramural Research Program of the NIH, The National Cancer Institute, and The Center for Cancer Research.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/19816
References
- 1.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–41. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–81. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–54. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–5. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 6.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–89. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 10.Alló M, Schor IE, Muñoz MJ, de la Mata M, Agirre E, Valcárcel J, et al. Chromatin and alternative splicing. Cold Spring Harb Symp Quant Biol. 2010;75:103–11. doi: 10.1101/sqb.2010.75.023. [DOI] [PubMed] [Google Scholar]
- 11.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–94. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–9. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 15.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 16.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–31. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi JK. Contrasting chromatin organization of CpG islands and exons in the human genome. Genome Biol. 2010;11:R70. doi: 10.1186/gb-2010-11-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–92. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–76. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–30. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 22.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermiston ML, Xu Z, Weiss A. CD45: a critical regulator of signaling thresholds in immune cells. Annu Rev Immunol. 2003;21:107–37. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- 24.Oberdoerffer S, Moita LF, Neems D, Freitas RP, Hacohen N, Rao A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–91. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, et al. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci U S A. 2009;106:18357–61. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motta-Mena LB, Heyd F, Lynch KW. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol Cell. 2010;37:223–34. doi: 10.1016/j.molcel.2009.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–7. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 30.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]