Abstract
The methylation of histones is a fundamental epigenetic process regulating gene expression programs in mammalian cells. Dysregulated patterns of histone methylation are directly implicated in malignant transformation. Here, we report the unexpected finding that the invasive extracellular matrix degrading endoglycosidase heparanase enters the nucleus of activated human T lymphocytes and regulates the transcription of a cohort of inducible immune response genes by controlling histone H3 methylation patterns. It was found that nuclear heparanase preferentially associates with euchromatin. Genome-wide ChIP-on-chip analyses showed that heparanase is recruited to both the promoter and transcribed regions of a distinct cohort of transcriptionally active genes. Knockdown and overexpression of the heparanase gene also showed that chromatin-bound heparanase is a prerequisite for the transcription of a subset of inducible immune response genes in activated T cells. Furthermore, the actions of heparanase seem to influence gene transcription by associating with the demethylase LSD1, preventing recruitment of the methylase MLL and thereby modifying histone H3 methylation patterns. These data indicate that heparanase belongs to an emerging class of proteins that play an important role in regulating transcription in addition to their well-recognized extra-nuclear functions.
Keywords: T lymphocytes, chromatin, heparanase, histone methylation, immune response genes, inducible genes, transcription
Introduction
Modulation of chromatin structure is a dynamic process that controls gene transcription programs in eukaryotic cells. The smallest repeating unit of chromatin is the nucleosome, which consists of 147 bp of DNA wrapped around a histone octamer.1,2 Chromatinized DNA can be remodeled via three main mechanisms: (1) the action of ATP-remodeling complexes, (2) the exchange of histone variants, and (3) post-translational modification (PTM) of the N-terminal tails. One of the best-characterized PTMs is the acetylation of lysine 9 on H3 histone tails (H3K9ac), which is typically associated with an open chromatin state conducive to active gene transcription.3-6 In contrast, methylation of lysine and/or arginine residues can be associated with either active or repressed gene states with histone H3K4 methylation primarily associated with active genes and H3K9 methylation correlating with gene repression.7 An additional layer of complexity in the histone code is that methylation can exist in mono-, di- or trimethylated forms, which have distinct transcriptional outcomes depending on the gene context.8 It is now emerging that the positioning of PTMs as well as the combination of histone marks govern gene transcription.9-11 Indeed, histone methylation is a highly dynamic process and the state of histone methylation at a particular gene locus appears to be finely balanced by the coordinated actions of histone methyltransferases and demethylases. Recent findings have elegantly exemplified that the dysregulation of the enzymes that catalyze histone methylation are associated with cancer progression.9,12-15 Although considerable advances have been made toward elucidating the landscape of histone methylation across gene loci, the molecular mechanisms underpinning the specific recruitment of chromatin modifying enzymes to targeted loci remain elusive.
Heparanase is an endo-β-glucuronidase that cleaves heparan sulfate and facilitates the passage of migrating cells through extracellular matrices (ECM), particularly basement membranes, as well as releasing heparan sulfate-bound growth factors from the ECM, whereby the released growth factors also aid wound healing and angiogenesis.16-18 Under normal physiological conditions heparanase aids the passage of leukocytes through sub-endothelial basement membranes and entry into sites of inflammation.19 It is also well established that heparanase, via a similar mechanism, plays a critical role in the metastatic spread of cancer.17,18,20 Recent studies have suggested, however, that heparanase has biological functions independent of its enzymatic activity. For example, extracellular heparanase can trigger the phosphorylation of specific protein kinases, such as p38 and Akt, and induce gene transcription, a process independent of heparanase endoglycosidic activity and mediated by the C-terminal domain of the molecule.21-24 Similarly, transfection of tumor cells with mutant heparanases or heparanase fragments that lack enzymatic activity can result in enhanced tumor development and metastasis.25,26 A number of studies have also reported the presence of heparanase in the nucleus of cells,27-30 with nuclear heparanase being associated with a favorable prognosis and cytoplasmic heparanase with poor survival in patients with lung, neck and gastric cancers.31 There is some evidence that nuclear heparanase expression is linked to cell differentiation28,30,32-34 but, despite such reports, the functional relevance of nuclear heparanase remains to be established.
In this study, we investigated whether nuclear heparanase can regulate transcription in resting and activated human T lymphocytes. This is an ideal model to investigate the functional role of nuclear heparanase, as T lymphocyte activation is a normal physiological process that involves the induction of a range of immune response-related genes, including heparanase. Also, we have previously shown T cells to be an ideal system to study heparanase gene regulation.35 Initial immunofluorescence studies revealed that nuclear heparanase is associated with euchromatin, and not heterochromatin, in both resting and activated human T cells. Subsequent single locus ChIP as well as genome wide ChIP-on-chip analysis identified a cohort of immune-related target genes that are bound by heparanase at their promoter and coding regions. Additional knockdown and overexpression experiments showed that chromatin-tethered heparanase is required for the inducible transcription of several of these genes in activated T cells. In fact, at the chromatin level, we found that heparanase regulates histone H3K4 and H3K9 methylation by binding to target gene control regions in association with the demethylase LSD1. We further showed that association of heparanase with the LSD1 complex is accompanied by lack of recruitment of the methylase, MLL, suggesting that heparanase plays a central role in mediating the on/off switch of inducible gene transcription. Taken together, these data suggest a hitherto unanticipated role for nuclear heparanase as a novel chromatin-associated regulator of transcription in mammalian cells.
Results
Heparanase associates with active chromatin and co-localizes with RNA polymerase II and active histone modifications within the nucleus of T cells
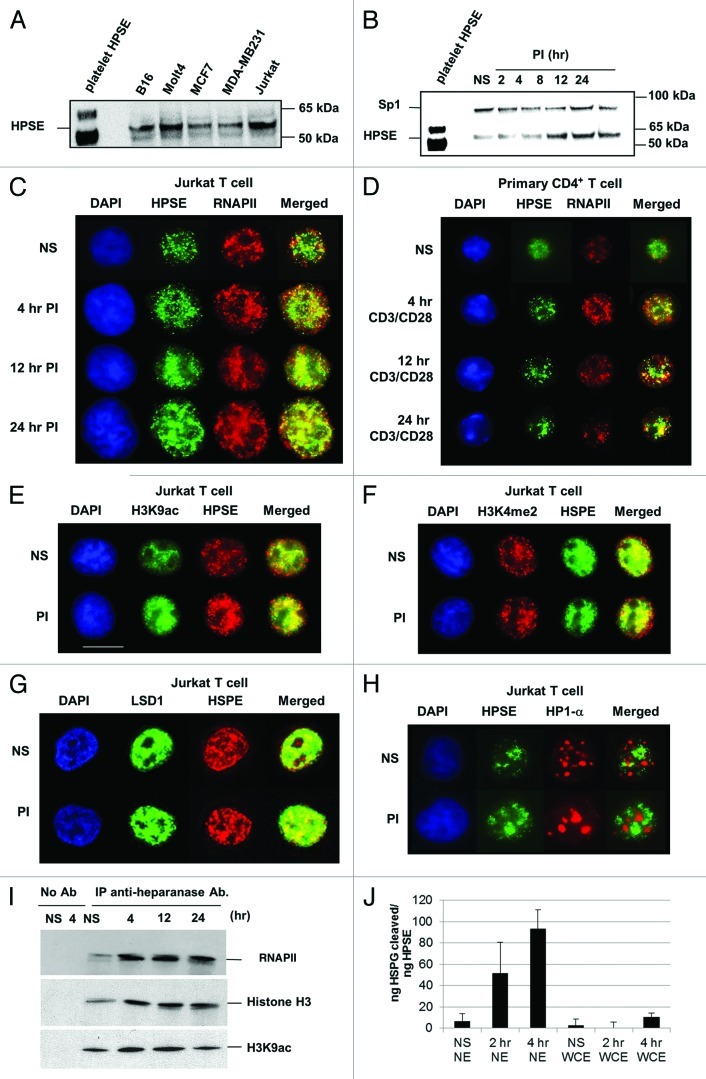
The human Jurkat T cell line has been widely used as a model for studying the regulation of transcription, a major advantage of this system being that it can be used to study the transcriptional regulation of immune response-related genes in resting and activated T cells. Initial experiments, involving immunoblot analysis of nuclear extracts, also revealed that Jurkat T cells, like many other cell types, contains nuclear heparanase (Fig. 1A). The polyclonal heparanase antibody used here from Insight recognizes both the enzymatically inactive 65 kDa pro-form and the active, proteolytically processed, 53 kDa form of the enzyme.36 These results were further confirmed using another heparanase antibody (Fig. S6). We found the ~53 kDa active form of heparanase to predominate in the Jurkat T cell nucleus, consistent with previous studies with other cell types.28,30 Immunoblot analysis of nuclear extracts from Jurkat T cells, which were either untreated (NS) or stimulated with phorbol 12-myristate 13-acetate/calcium ionomycin (PI) for the time periods indicated (2–24 h), also revealed that PI stimulation resulted in a substantial increase in nuclear heparanase, with the 53 kDa form of heparanase detected at all time points (Fig. 1B). Next, we monitored the nuclear distribution of heparanase in resting and activated Jurkat T cells using fluorescence microscopy. These experiments showed that nuclear heparanase was predominantly localized in the euchromatic compartment, which is stained weakly by DAPI, in both resting and activated Jurkat T cells (Fig. 1C and S4A). This was confirmed in primary CD4+ T cells, both before and after activation, with anti-CD3/CD28 antibodies, for the same time course (Figs. 1D and S4B). Consistent with the detection of heparanase in the euchromatin compartment, this enzyme co-resided with RNA polymerase II (denoted RNAP II herein), with a greater degree of co-localization in activated T cells (Figs. 1C and D). Additionally, we found that heparanase co-localizes with the active chromatin marks H3K9ac (acetylation of lysine-9 of histone H3), H3K4me2 (di-methylation of lysine-4 of histone H3) and the demethylase LSD1, but had a mutually exclusive staining profile for the repressive mark HP1-α (heterochromatin protein 1-α), both before and after 4 h of PI stimulation (Figs. 1E, 1F, 1G and 1H, S4C-F respectively). Collectively, these fluorescence microscopy data indicate that nuclear heparanase is associated with active chromatin. To confirm these observations, co-immunoprecipitation experiments were performed on nuclear extracts from Jurkat T cells using the anti-heparanase antibody followed by an immunoblot for RNAP II (using an antibody specific for the C-terminal domain repeat sequence YSPTSPS), histone H3 or H3K9ac. These experiments demonstrate that endogenous heparanase forms a complex with RNAP II, histone H3 (a key nucleosome component) and the H3K9ac activation mark, in resting and activated T cells (Fig. 1I). This was further confirmed by repeating the co-immunoprecipitation in reverse with an anti-RNAP II antibody, followed by an immunoblot for heparanase (Fig. S7). Heparanase failed to co-immunoprecipitate with the heterochromatin marker HP1-α. To verify the purity of the nuclear preparations, immunofluorescence staining was performed for the non-nuclear antigen, Integrin β1, which was excluded from the isolated T cell nuclei that were stained with DAPI (Fig. S5). Microscopy of Jurkat whole cells stained with heparanase and DAPI also show the specific distribution of heparanase in the cytoplasm and nucleus that is absent in cells treated with heparanase RNAi (Fig. S9). Using the time-resolved fluorescence energy transfer-based heparanase assay, heparanase activity was detected in nuclear extracts (NE) of Jurkat T cells, with a gradual increase in heparanase activity being detected at 2 and 4 h post PI stimulation. In contrast, minimal amount of heparanase activity was detected in whole cell extracts (WCE) of NS or PI stimulated Jurkat T cells (Fig. 1J). Although a consistent (and above control) level of heparanase activity was detected in the nucleus, the actual level of heparanase activity is low (only 5% substrate degradation for the 4 h sample). Taken together, these data indicate that active nuclear heparanase exists within the euchromatin compartment and is in close proximity to nucleosomes in resting and activated human T cells.
Figure 1. Nuclear heparanase associates with discrete chromatin regions in T lymphocytes. (A) Immunoblot analysis of heparanase protein levels in nuclear extracts prepared from B16, Molt4, MCF7, MDA-MB231 and Jurkat tumor cell lines. Purified human platelet heparanase was used as a positive control (lane 1). (B) Immunoblot analysis of nuclear extracts from non-stimulated (NS) Jurkat T cells or cells stimulated with phorbol 12-myristate 13-acetate (20 ng/ml) and calcium ionomycin (1 μM) (PI) for the hours (hr) indicated using anti-heparanase (lower band) and anti-Sp1 (upper band) antibodies, the Sp1 being a loading control. Platelet heparanase was used as a positive control. (C) Fluorescence microscopy of fixed nuclei from Jurkat T cells (NS or PI-stimulated for the indicated time points) stained with DAPI and rabbit anti-heparanase or mouse anti-RNAP II antibodies. A rabbit isotype of equivalent concentration was used as a control and showed no fluorescence. (D) Fluorescence microscopy was performed as above using purified mouse primary CD4+ T cells that were NS or stimulated with anti-CD3 and anti-CD28 antibodies for the indicated time points. (E-H) Fluorescence microscopy was performed as in C, using rabbit polyclonal antibodies against H3K9ac (E), H3K4me2 (F), LSD1 (G) and a mouse monoclonal antibody against HP-1α (H). (I) Immunoprecipitation of heparanase from cross-linked sonicated nuclear lysates of either NS or PI-treated T cells followed by immunoblot analysis for RNAP II (upper panel), histone H3 (middle panel) and histone H3K9ac (lower panel). No antibody (no Ab) was used as a negative control. (J) HTRF heparanase activity assay on Jurkat nuclear and whole cell extracts. Data has been normalized to ng protein between samples after BCA assays. Data (n = 3) is representative of two independent experiments and includes errors as +/−SEM.
Nuclear heparanase controls transcription of a distinct cohort of T-cell inducible genes
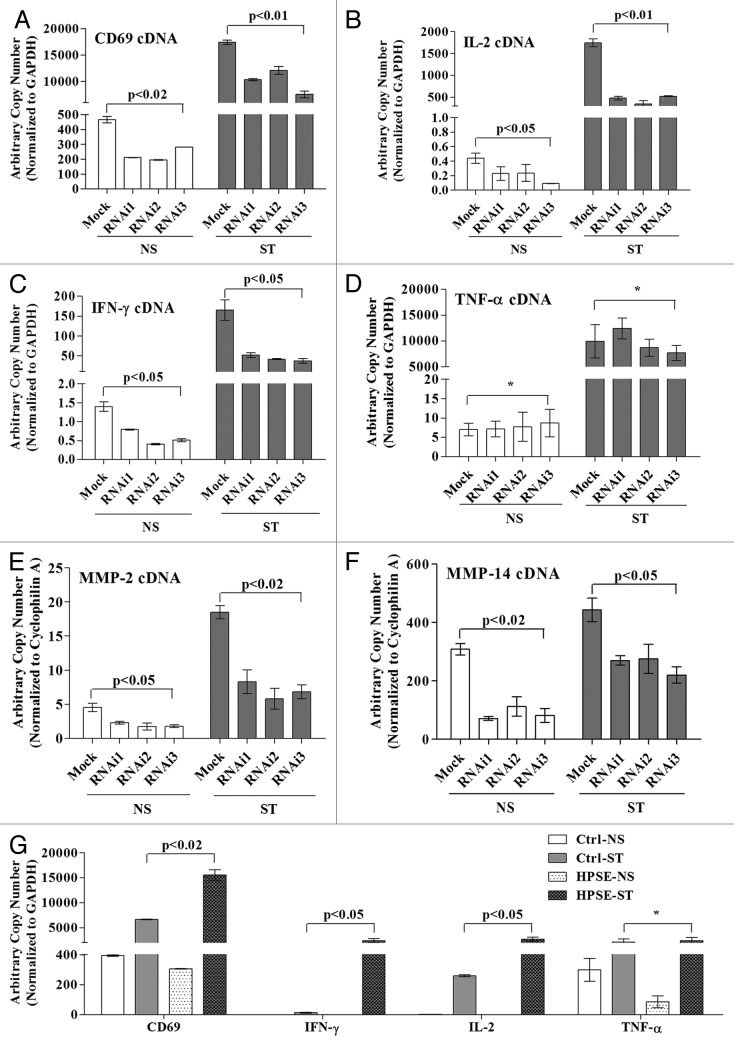
Since heparanase was found to associate with active chromatin marks and RNAP II, we examined whether this enzyme plays a direct role in activated gene transcription. We used a panel of three siRNAs, each targeting a distinct region of the heparanase transcript, to knock down heparanase levels in Jurkat T cells. All three siRNAs successfully reduced heparanase mRNA and protein production in Jurkat T cells, whereas a control siRNA did not (Figs. S1 and S2). To assess whether heparanase knockdown affected the expression of a panel of inducible T cell genes that are important for immune function, we compared the effects of each of the validated heparanase siRNAs on expression of CD69 (Fig. 2A), IL-2 (Fig. 2B), IFN-γ (Fig. 2C), TNF-α (Fig. 2D), MMP-2 (Fig. 2E) and MMP-14 (Fig. 2F). Strikingly, heparanase knockdown led to a significant reduction in the mRNA levels of five of the six genes tested (Figs. 2A-C and E-F), the level of inhibition ranging from 30–80% (Fig. S3), with the exception being TNF-α (Fig. 2D). Enforced expression of heparanase in Jurkat T cells by transfection with a heparanase expression plasmid resulted in increased expression of CD69, IL-2 and IFN-γ but not for the TNF-α gene in the PI stimulated T cells, confirming that TNF-α transcription appears to operate independently of heparanase (Fig. 2G). These data suggest that heparanase positively regulates the transcription of a specific cohort of inducible T-cell genes.
Figure 2. Heparanase is essential for inducible gene transcription in T cells. Jurkat T cells were transfected with validated heparanase siRNAs (RNAi1, RNAi2, RNAi3) or a negative control siRNA (Mock). At 48 h post-transfection, Jurkat T cells were either left untreated (NS) or stimulated with PI for 2 h (ST). Total RNA was extracted and TaqMan real-time PCR performed for CD69 (A), IL-2 (B), IFN-γ (C), TNF-α (D), MMP-2 (E) and MMP-14 (F). Results are plotted as arbitrary copies and normalized to GAPDH (A-D) or cyclophilin A (E-F). (G) Jurkat T cells were transfected with empty control vector (Ctrl) or a heparanase wild-type overexpression construct (HPSE) and at 48 h post-transfection cells were processed as above. Data represent the mean ± SE of three independent experiments and (*) indicates a non-significant change.
Nuclear heparanase is preferentially recruited to the regulatory regions of active genes but does not disrupt histone occupancy
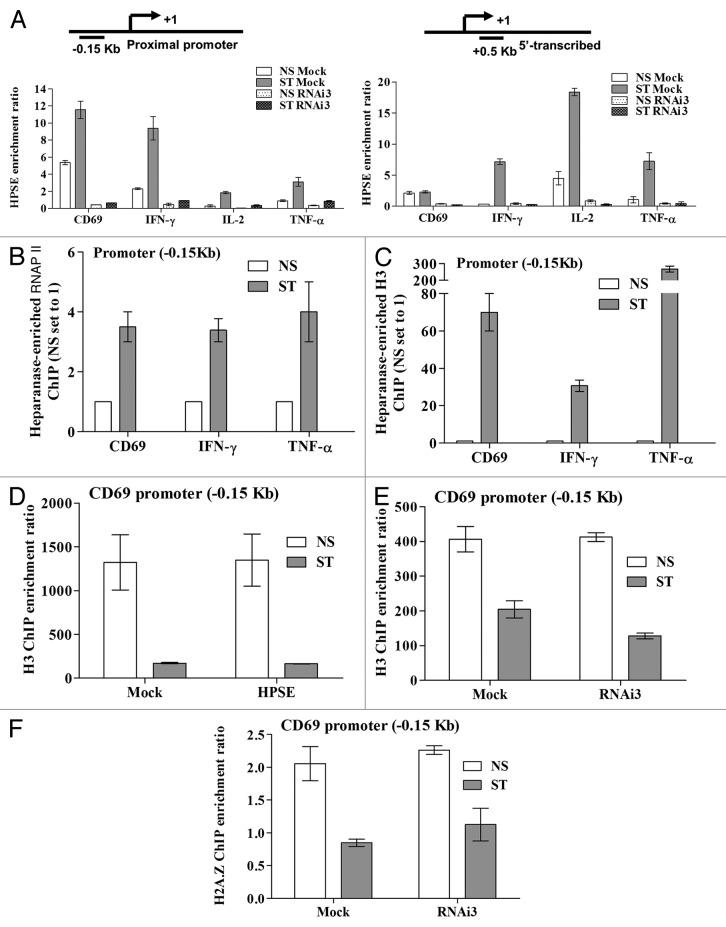
To determine the biochemical mechanism by which heparanase regulates inducible gene expression, ChIP experiments were performed on T cells depleted of heparanase by siRNA. These experiments showed that heparanase was specifically recruited to both the promoters and 5′ coding regions of the CD69, IFN-γ, IL-2 and TNF-α genes in stimulated T cells, and that recruitment of heparanase was abrogated in cells transfected with an siRNA against heparanase (RNAi3), but not in control (Mock) siRNA-transfected cells (Fig. 3A). RNAi3 was chosen as it had the best knockdown of heparanase at the protein level (Fig. S1) while there was no significant difference at the transcript level between the different RNAis (Fig. S2). Interestingly, although heparanase is recruited to the TNF-α gene, knockdown of heparanase did not inhibit transcription of this gene (Fig. 2D). To determine whether chromatin-bound heparanase was associated with RNAP II and nucleosomes at these genes, sequential ChIP analyses were performed. Sequential ChIP experiments using the anti-heparanase antibody followed by either anti-RNAP II or anti-histone H3 antibodies, revealed an increase in co-occupancy of heparanase with RNAP II and H3 following 4 h of PI stimulation (ST) (Figs. 3B and C, respectively).
Figure 3. Heparanase occupies regulatory regions of active genes in T cells. (A) Heparanase ChIP assays were performed on Jurkat T cells transfected with heparanase RNAi3 or a negative control siRNA (Mock). Samples were processed immediately (NS) or stimulated with PI for 2 h (ST). Primers covering both the -0.15 kb proximal promoter and +0.5 kb transcribed region of the CD69, IL-2, IFN-γ and TNF-α genes were tested (schematically illustrated above graphs). The ChIP enrichment ratio is calculated as the ratio of immunoprecipitated DNA relative to the no antibody control and normalized against the total input DNA. Sequential ChIP was performed on NS or 4 h PI stimulated Jurkat T cells (ST) initially with an anti-heparanase antibody and then with anti-RNAP II (B) or anti-histone H3 (C) antibodies. Real-time PCR analysis was performed on immunoprecipitated DNA using primers spanning the proximal promoters of the CD69, IFN-γ and TNF-α genes. NS ChIP enrichment was set to 1. (D) Histone H3 ChIP assays were performed using Jurkat T cells transfected with either empty vector (Mock) or a heparanase overexpression plasmid (HPSE). Samples were processed as in A. (E) H3 and (F) H2A.Z ChIP assays were performed on Jurkat T cells transfected with heparanase RNAi3 or a negative control siRNA as in (A) above. Real-time PCR analysis was performed on the proximal promoter of the CD69 gene (D-F). Data represent the mean ± SE of three independent experiments.
It is well established that chromatin remodeling accompanies inducible gene transcription and a key event in this process is the loss or exchange of histones.35,37-40 We have previously shown that histone exchange occurs across the CD69 proximal promoter region following T cell activation.35 Therefore, we examined whether heparanase could alter histone occupancy across the CD69 promoter. ChIP experiments with an antibody specific for the unmodified C-terminus of histone H3 showed that occupancy of this histone remained unchanged on the CD69 proximal promoter regardless of whether heparanase expression was enforced (Fig. 3D) or inhibited by siRNA (Fig. 3E). Since histone H3 is often partnered with H2A.Z,41 we assessed the occupancy profile of this variant and found that siRNA-mediated heparanase knockdown also failed to alter the level of H2A.Z associated with CD69 proximal regulatory elements (Fig. 3F). Collectively, these data suggest that while heparanase binds to the regulatory regions of transiently expressed genes following T cell activation, it does not alter H3 or H2A.Z histone occupancy.
Genome-wide ChIP-on-Chip analysis reveals that heparanase binds to the promoters of a cohort of transcriptionally active genes in T cells
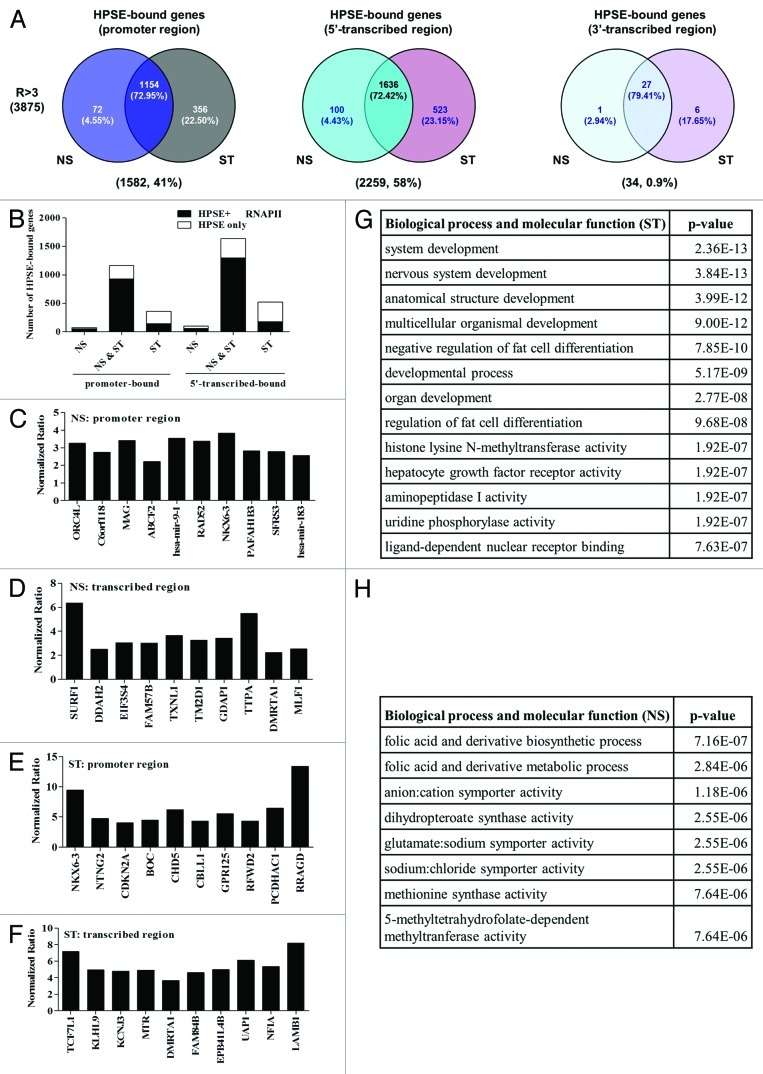
To determine the genome-wide binding pattern of heparanase, we performed ChIP coupled with genome-wide microarray analysis (ChIP-on-Chip) on resting vs. PI-activated T cells. ChIP DNA samples that were pooled from five independent ChIP experiments, were generated from resting (NS) and 12 h PI stimulated (ST) Jurkat T cells and hybridized onto Agilent human promoter microarrays that contained 17,000 well-characterized genes (-5.5 kb to +2.5 kb from TSS) from the human genome. Using a stringent analysis (p-value score < 0.05 and an enrichment ratio > 3), 3875 genes were identified that carried bound heparanase in both resting and activated T cells (Fig. 4A, Tables S1 and S2). The inducible immune response genes CD69, IFN-γ, IL-2 and TNF-α were among the subset of genes with bound heparanase, with enrichment ratios > 3 and p-value < 0.05. Analysis of the genomic distribution of heparanase showed enrichment for heparanase binding in the promoter (41%) and 5′ transcribed (58%) regions of genes, but only 0.9% in the 3′ transcribed region (Fig. 4A). As predicted from our earlier findings, RNAP II also occupied the promoter and 5′transcribed regions of the majority (~80%) of genes with bound heparanase in both resting and activated T cell subset (NS and ST) (Fig. 4B, Fig. S5). Interestingly, there was much less correlation between the heparanase-binding genes vs. the RNAP II-binding genes in the ST subset (Fig. 4B). The top 10 genes with the most significant heparanase binding (enrichment ratio > 3, p-value < 0.01) to either the promoter or transcribed regions of resting and activated T cells are depicted in Figures 4C-F. Interestingly, two key microRNA genes, miR-9 and miR-183, which have previously been implicated in cancer and EMT, were two of the top ten genes that carried bound heparanase on their promoters in NS T cells (Fig. 4C). Functional annotation of the top 100 ranked heparanase target genes in stimulated (ST) T cells (Fig. 4G) revealed that heparanase is recruited to genes linked to development and differentiation pathways. In contrast, functional annotation of heparanase target genes in non-stimulated (NS) T cells showed enrichment for several pathways, including metabolic and biosynthetic processes (Fig. 4H). The most significant transcription factors predicted to bind heparanase within promoter regions of heparanase-bound genes identified by ChIP-on-chip are graphed in Figure S8. Taken together, our genome-wide analysis suggests that nuclear heparanase is recruited to distinct cohorts of actively transcribed genes in resting and activated T cells.
Figure 4. Heparanase is recruited to the proximal regulatory regions of a distinct cohort of active genes in T cells. (A) Venn diagrams of genes that significantly bound heparanase (ratio > 3-fold, p < 0.05, n = 2), which were identified by ChIP-on-Chip microarray analysis to be only present in resting Jurkat T cells (NS), to be present in both resting and activated Jurkat T cells (NS and ST) or only present in activated Jurkat T cells (ST). The Venn diagrams illustrate the distribution of these gene categories within the promoter, coding and downstream regions of the heparanase binding genes. The number and percentage of genes present in a given subset are shown in brackets below each diagram. (B) Bar graphs showing the number of genes that bound (at their promoter and 5′-transcribed regions) heparanase alone or bound HPSE and RNAP II (HPSE and RNAP II) in the NS, NS and ST and ST subsets of the heparanase binding genes. (C-F) Graphical representation of the 10 most significant heparanase binding target genes (p-value < 0.01, n = 2) within: (C) NS, promoter regions; (D) NS, transcribed regions; (E) ST, promoter regions and (F) ST, transcribed regions. (G-H) Functional annotation of the heparanase binding genes in (G) stimulated (ST) and (H) non-stimulated (NS) T cells.
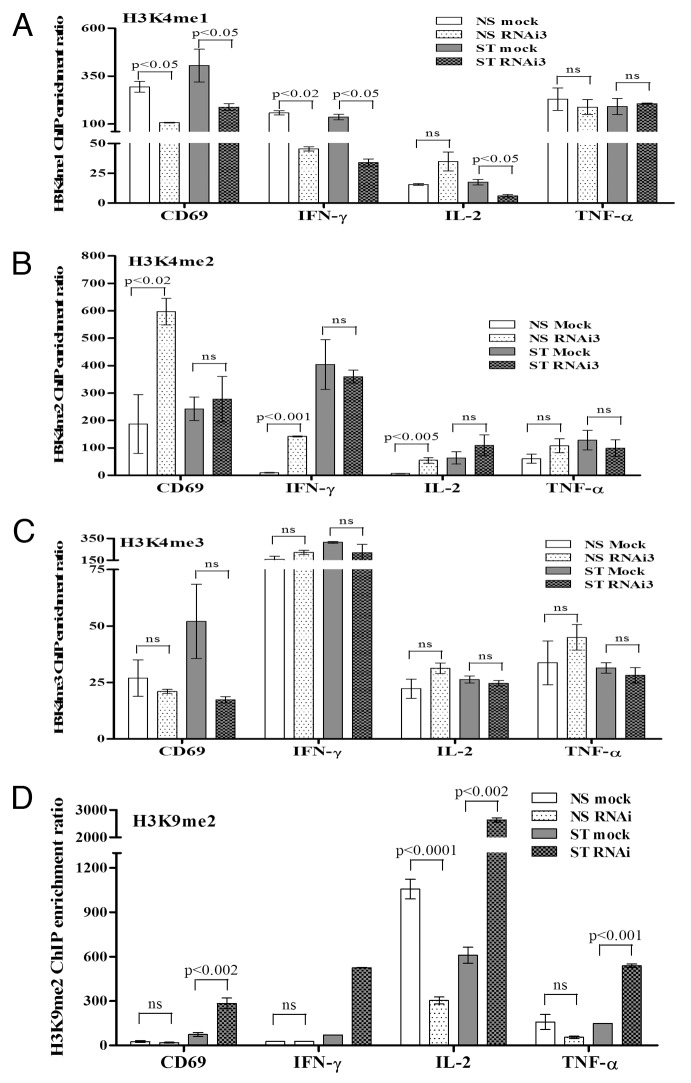
Heparanase knockdown reduces H3K4me1 formation and increases H3K9me2 deposition across the regulatory regions of inducible genes in T cells
Chromatin remodeling events at inducible genes are typically a consequence of changes in histone occupancy and composition, as well as histone PTMs. Since our findings above suggest that heparanase does not participate in modulating histone dynamics across inducible genes, we investigated whether this enzyme could impart its effects on gene expression via alteration of histone PTMs. Given that heparanase associates with RNAP II and active chromatin at activated target genes, we examined whether this protein could modify PTMs that are hallmarks of active gene transcription or repression. ChIP analysis was performed on resting or PI-stimulated Jurkat T cells, transfected with Mock or heparanase siRNA, for active histone H3 methylation marks (H3K4me1, H3K4me2 and H3K4me3) or the repressive mark H3K9me2 across the proximal promoter regions of the CD69, IFN-γ, IL-2 and TNF-α genes (Fig. 5). Remarkably, heparanase knockdown reduced H3K4me1 across three of the tested gene regulatory regions in both resting and activated T cells, with the main exceptions being IL-2 in resting T cells and the TNF-α gene, where the level of H3K4me1 remained unchanged (Fig. 5A). This result is consistent with the lack of effect of heparanase knockdown on TNF-α mRNA production (Fig. 2D). One possibility that could account for the decrease in H3K4me1 in heparanase-depleted T cells is that this residue is not converted as efficiently to the mono-methylated form in the absence of heparanase. Consistent with this observation, a corresponding accumulation in H3K4me2 was observed across all genes in the basal state in heparanase-depleted cells (Fig. 5B), coincident with the decrease in H3K4me1 (Fig. 5A). It is possible that no accumulation is observed between Mock and heparanase-depleted cells after stimulation due to the rapid loss of histones that occurs following activation. There was no common trend for H3K4me3 mark across the inducible gene promoters tested (Fig. 5C). In heparanase siRNA treated T cells, the repressive H3K9me2 mark decreased in resting T cells in the absence of heparanase and increased across all of the four inducible gene promoters examined following T cell activation (Fig. 5D), consistent with transcriptional repression of the CD69, IFN-γ and IL-2 genes and a concomitant decrease in H3K4me1 levels (Fig. 5A). In summary, heparanase appears to influence H3K4 and H3K9 methylation patterns across the regulatory regions of a subset of inducible genes in T cells.
Figure 5. Heparanase is required for H3K4/K9 methylation at the regulatory regions of inducible genes in T cells. ChIP assays for H3K4me1 (A), H3K4me2 (B), H3K4me3 (C) and H3K9me2 (D) on the proximal promoters of the CD69, IFN-γ, IL-2 and TNF-α genes were performed on Jurkat T cells transfected with heparanase RNAi3 or a negative siRNA control (Mock) and non-stimulated (NS) or stimulated with PI for 2 h (ST). Real-time PCR was performed using primers covering the -0.15 kb proximal promoter regions of these genes. Data represent the mean ± SE of three independent experiments.
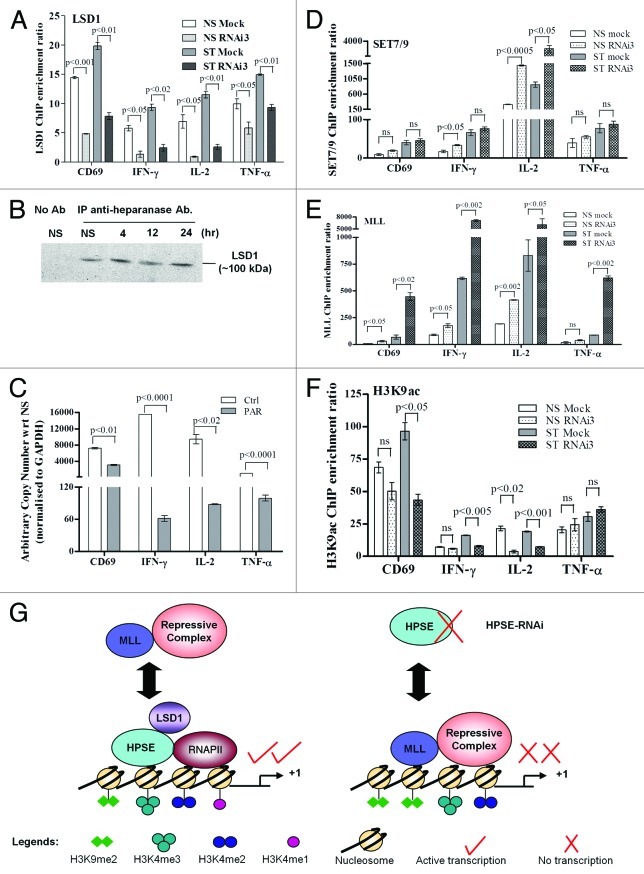
Nuclear heparanase controls recruitment of the demethylase LSD1 and the methyltransferase MLL to the promoters of transiently expressed genes in T cells
Our data thus far support the notion that in the absence of heparanase, there is less H3K4me2 converted to H3K4me1, and there is a corresponding accumulation of H3K9me2 in activated T cells. Since heparanase is not known to possess direct methylating activity, we used ChIP to investigate whether LSD1 (a key demethylase responsible for the demethylation of the di- to the monomethylated form of H3K4 and H3K9) was recruited to the promoter regions of the CD69, IFN-γ, IL-2 and TNF-α genes in the presence and absence of heparanase. These data showed that LSD1 recruitment to the regulatory regions of the four inducible genes examined is impaired by the inhibition of heparanase expression (Fig. 6A). To determine whether heparanase and LSD1 are associated within the same multi-protein complex, we performed co-immunoprecipitation experiments with T cell nuclear extracts. These experiments showed that heparanase and LSD1 interact in the nucleus of resting and activated T cells (Fig. 6B). Furthermore, treatment of cells with a LSD1 inhibitor, pargyline (PAR), significantly inhibited the transcription of all four genes examined, including TNF-α (Fig. 6C). These results were confirmed with LSD1 RNAi experiments that showed ~60% knockdown in LSD1 transcript was sufficient to reduce expression by ~60–80% for IL-2 and IFN-γ and to a lesser extent TNF-α (~20% inhibition), relative to the FAM control (Fig. S10). Together, these data suggest that heparanase modulates histone methylation via the histone demethylase LSD1. We next examined whether heparanase influenced the recruitment of two H3K4 methyltransferases, SET 7/9 and MLL. ChIP analysis revealed that heparanase depletion had little effect on the recruitment of SET7/9 to the CD69, IFN-γ and TNF-α promoters (Fig. 6D). However, there was a high level recruitment of SET7/9 to the IL-2 promoter and this was augmented in the absence of heparanase in both resting and activated T cells (Fig. 6D). This finding may explain the small increase of H3K4me3 that was observed for this gene in heparanase-depleted cells (Fig. 5C). In contrast, we found increased MLL recruitment to all four genes following stimulation, especially in heparanase-depleted T cells (Fig. 6E). Attempts to co-immunoprecipitate MLL and heparanase failed, further suggesting that these proteins do not co-exist within the same complex (data not shown). Since MLL has been shown to form a repressive complex with histone deacetylase 1 (HDAC1),42 we proposed that inhibition of heparanase would lead to increased occupancy of MLL/HDAC1 and thus lead to reduced levels of H3K9ac. Using ChIP analysis, we indeed showed that heparanase-depleted cells usually had reduced H3K9ac deposition at the proximal promoters of the CD69, IFN-γ and IL-2 genes (Fig. 6F), in both resting and stimulated cells. In contrast, heparanase depletion failed to inhibit H3K9ac deposition on TNF-α regulatory regions (Fig. 6F), further supporting the view that TNF-α transcription is regulated by a heparanase-independent mechanism. Taken together, we propose a model whereby heparanase exists in an active transcription complex with LSD1 and RNAP II that is conducive to active gene transcription with accompanying demethylation of H3K4me2 and H3K9me2 (Fig. 6G). Our findings also suggest that the presence of heparanase impedes the binding of an MLL-repressive complex to activated T-cell genes (Fig. 6G).
Figure 6. Nuclear heparanase may control H3 methylation profiles by recruiting LSD1 and excluding MLL from gene promoters in T cells. (A) LSD1 ChIP assays were performed on Jurkat T cells transfected with heparanase RNAi3 or a negative siRNA control (Mock). Real-time PCR was performed using primers covering the -0.15 kb proximal promoter of the CD69, IFN-γ, IL-2 and TNF-α genes. (B) Immunoprecipitation of heparanase from cross-linked sonicated nuclear lysates of either NS or PI-treated T cells followed by immunoblot analysis with an anti-LSD1 antibody. No antibody (no Ab) was used as a negative control. (C) Jurkat T cells untreated (Ctrl) or pre-treated with 3 mM pargyline (PAR) and non-stimulated or stimulated with PI for 2 h. Total RNA was extracted and TaqMan real-time PCR performed for CD69, IFN-γ, IL-2 and TNF-α transcripts. Results are plotted as arbitrary copies relative to the corresponding NS sample and normalized to GAPDH. (D) SET7/9, (E) MLL and (F) H3K9ac ChIP assays were performed as in (A). Data represent the mean ± SE of three independent experiments. (G) Proposed model of the interplay of nuclear heparanase with LSD1, RNAP II, MLL and histone methylation marks.
Discussion
Despite several reports documenting nuclear expression of the extracellular matrix endoglycosidase heparanase, its specific function within the nucleus has remained elusive. Here we provide, for the first time, a detailed molecular explanation for the functional relevance of nuclear heparanase in T lymphocytes showing that heparanase (1) associates with euchromatin and is closely associated with RNA polymerase II, (2) is specifically recruited to the promoters and 5′ transcribed region of a large cohort of genes, (3) is essential for the transcription of a subset of inducible, immune response target genes, (4) controls the acquisition by inducible gene promoters of specific H3 methylation marks associated with active transcription, and (5) appears to impart its effects by forming an active transcription complex with the demethylase LSD1, which counteracts the repressive actions of the methylase MLL (Fig. 6G). Collectively, these data imply that heparanase belongs to an emerging class of chromatin-associated proteins, which can directly modulate gene transcription in addition to their previously defined extra-nuclear roles in mammalian cells.
Non-traditional nuclear roles have now been described for an array of proteins with diverse biological functions, including hexokinase43 and Ca II.44 Furthermore, there is now emerging evidence of a novel class of signal transduction kinases that associate directly with chromatin to modulate transcription of their target genes, both in yeast and in mammalian cells.45-47 Similarly, the endoglycosidase heparanase was recently shown to play a non-conventional role as a signaling molecule at the cell surface.22 In this study, we have now shown that heparanase plays an important functional role in the nucleus as part of an active chromatin complex that regulates inducible gene transcription. This is the first time that a direct transcriptional role for heparanase has been described. Within the global context of the nucleus, heparanase appears to exist predominantly in the active chromatin fraction and co-localizes with RNAP II, both in a human T cell line and in primary CD4+ T cells. Furthermore, heparanase co-localizes with the active histone marks H3K9ac and H3K4me2, but is excluded from regions of chromatin carrying the repressor protein HP1α. We also found that the majority of nuclear heparanase exists in its enzymatically active (~50 kDa) form in a range of cancer cell lines, supporting previous observations.28,30 This suggests that the catalytic activity of heparanase may be important for its intra-nuclear function. Similarly, it has been shown for several yeast protein kinases that their stable association with chromatin in promoter complexes requires them to be in an enzymatically active state.46 It is likely that tethering of proteins like heparanase directly to chromatin may provide an additional level of control over the transcription of genes that are directly implicated in heparanase-mediated cell signaling and invasion processes. Indeed, we showed by knockdown and overexpression experiments, that heparanase was essential for inducible transcription of the CD69, IL-2, IFN-γ, MMP-2 and MMP-14 genes that are required for T cell effector function and migration. These findings are consistent with studies of heparanase knockout mice, which showed differential MMP-2 and MMP-14 expression in these animals.48 Interestingly, our investigations showed that heparanase is dispensable for TNF-α expression, suggesting that transcription of this gene occurs via a heparanase-independent mechanism requiring alternate intrinsic signaling, despite ChIP analyses showing that the regulatory regions of this gene carry heparanase.
In this study we demonstrate that nuclear heparanase is recruited in an activation-dependent manner to the proximal promoters of inducible T cell genes. This is akin to what has recently been observed for the signal transduction kinases tethered to chromatin,47,49 such as p38-α recruitment to pro-inflammatory gene promoters in macrophages.50 Heparanase was also found to associate with regulatory regions outside of gene promoters, namely the +0.5 kb transcribed regions of the CD69, IFN-γ and IL-2 genes. Using ChIP-on-chip, we showed that the recruitment of heparanase to the promoter and 5′ transcribed regions of active genes was a genome-wide phenomenon, which also coincided with recruitment of RNAP II. This finding is in accordance with previous studies that have shown that signal transduction kinases bind to the transcribed regions of target genes.47,49,51-56 Since heparanase is recruited to gene coding regions and associates with RNAP II, one may speculate that this enzyme may travel with the elongation complex through the coding regions of genes upon T cell activation. Indeed, such a mechanism has been proposed for yeast kinases that enter the coding regions of genes by “piggybacking” with RNAP II.45 Our findings suggest that heparanase may serve to increase mRNA production by directly associating with gene control regions in the nucleus.
It is well established that chromatin regulatory mechanisms play a fundamental role in transcriptional regulation of inducible genes.57,58 Here, we found that heparanase operates independently of the histone H3 loss that occurs at this gene locus following T cell activation.35 These observations are supported in primary CD4+ and CD8+ T cells, where acquisition of distinct PTMs, and not histone loss, was found to drive gene transcription.59 To decipher chromatin regulatory signatures associated with the depletion of heparanase and the abolished expression of heparanase target genes, we performed a detailed ChIP analysis of H3 methylation PTMs before and after T cell stimulation. Our data suggest that the combination of H3K4me1, H3K4me2, H3K4me3 across heparanase tethered inducible gene loci is important for gene transcription (Fig. 6G), which may provide a histone code for the recruitment of essential chromatin modifying proteins.9,60 Loss of heparanase led to a decrease of H3K4me1 with a concomitant accumulation of H3K9me2 in activated T cells. This could be explained by several scenarios: (1) H3K4me2 cannot be converted to H3K4me1, (2) rapid conversion of H3K4me1 to H3K4me0, and (3) loss of H3K4me2-containing nucleosomes in activated T cells. Since we do not observe a substantial accumulation of H3K4me2 and as LSD1 is not recruited in the absence of heparanase to demethylate H3K4me2 or H3K4me1, we favor the third scenario, which is supported by the notion that histone loss accompanies inducible gene transcription.35,38
In line with these changes in H3 methylation, our data also shows that association of heparanase with its chromatin template is essential for the recruitment of the histone modifier LSD1 (Fig. 6G), an enzyme responsible for converting the dimethylated forms of H3K4 and H3K9 to their monomethylated configurations.61-63 Inhibition of LSD1 enzymatic activity or removal of heparanase abolished transcription of the inducible genes tested, suggesting that these proteins may coexist as an active transcription complex. Thus, we propose that heparanase may function as a structural adaptor by forming an integral component of a chromatin modifying complex, akin to that previously shown for the yeast Hog1 and mammalian p38 kinase.45 This notion is also supported by a recent study showing a direct interaction between LSD1 and the signaling molecule PKC-β, which has traditionally been defined as a “non-nuclear” kinase.64 Our discovery of heparanase in association with LSD1 comes at an opportune time, as little is known regarding the molecular switches that allow LSD1 to activate some genes, but repress others. Previous studies have shown that LSD1 cannot erase H3K4 and H3K9 methylation marks simultaneously on histone peptides.65,66 Our findings in vivo for the first time suggest that the heparanase/LSD1 complex can simultaneously mediate H3K4 and H3K9 methylation marks on activated genes. It is plausible that the tethering of heparanase to LSD1 may induce conformational changes to facilitate the binding of LSD1 and demethylation of both H3K4 and H3K9 residues. However, we acknowledge the possibility that one of the caveats of ChIP assays is that we may be monitoring H3K4 and H3K9 methylation on separate nucleosomes or, alternatively, on different H3 variants that contain these residues.67
A previous study has shown that LSD1 associates with MLL as part of an active transcription complex.68 In contrast, our model (Fig. 6G) depicting the interplay between LSD1 and MLL on inducible genes is analogous to the previously described “on/off switch” on the interferon-β gene mediated by the exchange of repressor (HDAC1) and activator (p300) proteins.69 One fascinating finding from our study is that in the absence of heparanase, MLL recruitment is increased, particularly at the IL-2 and IFN-γ target genes. This suggests that heparanase and MLL are mutually exclusive on the promoters of inducible heparanase target genes (Fig. 6G), which we further confirmed by co-immunoprecipitation studies that failed to show the association of these proteins in the nucleus (data not shown). However, since we also showed inducible recruitment of heparanase and MLL to target genes following T cell stimulation, it is plausible that MLL could function to dampen the transcriptional response as part of a negative feedback system to prevent aberrant cytokine production. It is possible that reduced binding of heparanase and MLL occurs in the basal state due to the compact nature of chromatin in resting T cells, which may preclude binding to DNA. Taken together, the recruitment of the MLL repressive complex, the accumulation of H3K9me2 and concomitant decline in H3K4me1 may provide a molecular explanation for the reduced transcription of inducible genes in situations where heparanase is essential (Fig. 6G). Since H3K4me3 levels are unaltered when MLL is recruited, we speculate that the histone methyltransferase activity of this protein is likely to be redundant on the majority of heparanase-target genes. Rather we propose that the DNA binding domain of MLL recruits a repressive complex involving histone deacetylases,42,70 which complements our data with the HDAC inhibitor TSA.
Here we have uncovered a novel molecular function for nuclear heparanase, whereby it is recruited as part of an active transcription complex to a cohort of inducible genes in human T cells. We show for the first time that heparanase is a key regulator of histone H3 methylation via the specific recruitment of LSD1 and displacement of MLL. Ultimately, future studies will help unravel the precise function(s) of heparanase in controlling distinct H3 methylation patterns on target genes. Since dysregulation of nuclear heparanase, LSD1 and MLL are intimately linked with cancer metastasis, it may be important to consider these findings for the design of future therapeutic strategies in the treatment of cancer.
Materials and Methods
Cell culture
Human Jurkat T cells and the acute T lymphocytic cell line, Molt4, were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FCS, 2 mM L-glutamine, 9 mM HEPES and antibiotics. The human mammary adenocarcinoma lines, MDA-MB231 and MCF-7, and the mouse melanoma cell line B16 were maintained as above but in low glucose DMEM (Gibco). Jurkat T cells at a density of 5x105/ml were stimulated with 20 ng/ml phorbol 12-myristate 13-acetate and 1 µM Ca 2+ ionophore A23187 (PI) for the times indicated. For pargyline treatment, cells were treated with 3 mM of inhibitor for 18 h prior to stimulation.
Primary T-cell preparation and stimulation conditions
Mouse spleens were isolated from wild type C57BL/6 mice (4 - 5 weeks old) and CD4+ T cells were purified using MACS CD4+ microbeads according to the manufacturer’s guidelines (Miltenyi Biotec). Purified CD4+ T cell populations were stained and analyzed by flow cytometry and shown to be greater than 90% pure with CD4-, CD8-, B220 (B cell)-, and CD11b (macrophage)-specific antibodies (BD Biosciences). T cells were stimulated with anti-human CD3 antibody immobilised on six-well plates and soluble CD28 antibody (BD Biosciences) as previously described.59 All animal work was approved by the JCSMR Ethics Committee (ANU).
HPSE activity assay
A homogeneous time-resolved fluorescence-based (HTRF) assay was used to measure the heparan-sulfate (HS) degrading activity of samples. HS conjugated with biotin and europium (Eu3+) cryptate was purchased from Cisbio (Bedford, MA). Nine μl of cell lysate (whole cell or nuclear) was combined with an equal volume of sample dilution buffer (20 mM TRIS-HCl pH 5.5, 0.15 M NaCl, 0.1% CHAPS) and incubated with 9 μL of 0.7 μg/ml Biotin-HS-Eu(K) (0.2 M NaCH3 pH 5.5) at 37°C for 2 h. The reaction was stopped by the addition of 18 μl of 1 μg/ml Streptavidin-XL665 (Cisbio, Bedford, MA, diluted in 0.1 M NaPO4 pH 7.5, 1.2 M KF, 0.1% BSA, 2 mg/ml heparin) and incubated at room temperature in the dark overnight to allow conjugation of Streptavidin-XL665 to the remaining substrate. Samples were added to a 96 well white plate and fluorescent readings (excitation at 314 nm and emissions at 620 and 668 nm) were measured using a M5e plate reader (Molecular Devices, Sunnyville, CA) utilizing SoftMaxPro 5.2 software. Positive control samples that exhibited maximal FRET had only lysis buffer added in place of the cell lysate. Negative control samples that exhibited no FRET contained sample dilution buffer as for the positive control, but only the Streptavidin dilution buffer was added at the second step. Protein concentration of samples was determined using a BCA assay and the HS cleavage activity of samples was normalized to amount of protein. The resulting data was analyzed using the following equations:
Delta F% = ((sample ratio 688/620/blank ratio 668/620)/blank ratio 668/620)*100
% substrate degradation = 100*(1-(delta F%(sample)/delta F%(max FRET-positive control)
Amount of HS cleaved (pg) = % substrate degradation x 6300 pg
Immunofluorescence
Nuclei were isolated from Jurkat T cells as described previously71 and fixed for 10 min at room temperature with 2% paraformaldehyde (pH 7–8) (UNIVAR). Nuclei were permeabilized with 1% Triton X-100 (Sigma-Aldrich) and incubated with 1% BSA (Sigma-Aldrich) in PBS containing 0.1% Tween 20 (blocking solution) for 45 min at room temperature. Primary antibodies used for staining were: rabbit polyclonal anti-heparanase (Insight, Ins-AB-04002), mouse monoclonal anti-heparanase (Insight, Ins-AB-04001), anti-H3K4me2 (Abcam, ab7766) and anti-H3K9ac (Millipore, 06–942) and mouse monoclonal antibodies anti-HP1α (Millipore, 05–689), anti-RNAP II (Abcam, ab817) and anti-LSD1 (Millipore, 05–939). Next, donkey anti-rabbit IgG-FITC (1:100) or donkey anti-mouse IgG-Cy3 conjugated secondary antibody (1:200) were diluted in blocking solution and incubated with the nuclei at 37°C for 30 min in a hybridization chamber. Samples were then stained with 100 ng/ml of DAPI (Sigma-Aldrich) for 5 min. Cells were viewed under oil immersion at x100 magnification using an Olympus fluorescence IX71 microscope (Olympus) and images were captured using DPController software (Olympus) and analyzed using Photoshop 7.0 (Adobe Systems). Scans were taken with a 10 μm scale bar.
Heparanase overexpression in Jurkat T cells
The full-length human heparanase wild-type cDNA, kindly donated by Dr. Mark Hulett (Latrobe University, Melbourne, Australia), was cloned into a pcDNA3.1 expression vector containing an IRES/eGFP insertion, using PCR products amplified to generate the pcDNA3.1-heparanase-IRES/eGFP expression construct. Jurkat T cells (5x105 cells in 300 µl of warmed RPMI complete medium supplemented with 20% FCS) were electroporated in 0.4 cm gap Gene pulser Cuvettes (BioRad) with a BioRad Gene Pulser at 270 V and a capacitance of 975 µF. With Jurkat T cell transfections, 15 µg of the pCDNA3.1-IRES/eGFP or pCDNA3.1-heparanase- IRES/eGFP plasmids was used. Jurkat T cells were left to recover for approximately 24 to 30 h at 5x105 cells/ml in complete medium prior to PI stimulation. Transfected and untransfected Jurkat T cells were harvested and sorted by FACS.
Transfection of heparanase siRNA
Jurkat T cells were suspended at 1x105 cells/well in 24-well plates and transfected with synthetic heparanase siRNAs at a final concentration of 50 nM (4390824 HSPE Silencer® select siRNA 1 to 3 (s21304, s21305, s21306), Ambion) or a negative control (Silencer® FAMTM labeled negative control siRNA #1 (AM4620), Ambion) using Lipofectamine 2000 transfection reagent (11668–027, Invitrogen) according to the manufacture’s guidelines. Cells were transfected for 48 h and either processed immediately or stimulated with PI for 2 h.
RNA extraction and real-time PCR analysis
Total RNA was extracted from Jurkat T cells with TRIzol reagent (Sigma-Aldrich) as described previously.72 RNA (1 µg) was subsequently reverse transcribed with the SuperScript III RNase H¯ reverse transcriptase kit (Invitrogen). TaqMan Gene Expression assays and SYBR Green real-time PCR was performed on an ABI PRISM 7900 sequence detector (PerkinElmer/PE Applied Biosystems). The real-time PCR was performed in a total volume of 10 µl, as detailed in the manufacturer’s guidelines (Applied Biosystems). Human TaqMan primer sets used for gene specific transcript analysis were: CD69 (Hs00156399), IFN-γ (Hs00156399), IL-2 (Hs00174114), TNF-α (Hs00156399), Heparanase (Hs00180737), MMP-2 (Hs00156399) and MMP-14 (Hs00156399). Threshold cycle values from the PCR amplification plots were converted to arbitrary copy number using the Equation 105/2(Ct-17) and normalized to either the housekeeping gene GAPDH (Hs99999905) or Cyclophilin A (Hs99999904) (Applied Biosystems).
ChIP assays
ChIP assays were performed as previously detailed.35 The soluble chromatin fraction was incubated overnight at 4°C with 5 to 10 µg of one of the following antibodies: anti-heparanase (Insight, Ins-AB-04002), anti-histone H3 (Abcam, ab1791), anti-H3K9ac (Millipore, 06–942), anti-H3K4me3 (Abcam, ab8580), anti-H3K4me2 (Millipore, 07–030), anti-H3K4me1 (Abcam, ab8895), anti-LSD1 (Millipore, 05–939), anti-MLL (Millipore, 05–764) and anti-RNAP II CTD repeat YSPTSPS (8WG16; Abcam, ab817) antibodies or without antibody as a control. Standard curves were generated for each primer set to correct for differences in primer efficiency. Ct values from PCR amplification plots were converted to arbitrary copy number using the Equation 105/2(Ct-17). ChIP enrichment ratios were calculated and sequential ChIP assays were performed as described previously.35 ChIP data was also calculated using the ∆∆Ct method and presented data as % total input for the K27 methylation ChIP data set (Fig. S11) for comparison to show that all three methods generate similar profiles. Detail on how each data set was calculated is provided in the figure legend.
Co-immunoprecipitation and immunoblot analysis
The half-way ChIP procedure was performed as specified by Upstate Biotechnology. Briefly, following immunoprecipitation of the cross-linked chromatin fraction, the salmon sperm DNA/Protein A agarose beads were resuspended in 100 μl of PBS before the addition of 100 μl of 2 x SDS Loading Buffer. Samples were boiled in the reducing SDS Loading Buffer at 95°C for 5 min to denature proteins and briefly centrifuged for 2 min at room temperature to remove the beads from the supernatant. Next, 40 μl of the supernatant was resolved by SDS-PAGE and subjected to western blot analysis. Immunoblot analysis was performed as previously described,72 with anti-heparanase (Insight, Ins-AB-04002), anti-RNAP II (8WG16) (Abcam, ab817), anti-histone H3 (Abcam, ab1791) and anti-LSD1 (Millipore, 05–939) specific antibodies or without an antibody as a control.
ChIP-on-Chip
Heparanase and RNAP II ChIP DNA was generated from resting and 12 h PI treated Jurkat T cells pooled from five independent ChIP assays. ChIP samples were subsequently amplified based on one round of the whole genome amplification method using the WGA2 kit (Sigma-Aldrich), as described previously.73-75 Labeling, hybridization and scanning were performed as described in the Mammalian ChIP-on-chip protocol (version 9.1; Agilent Technologies). Briefly, 2 μg of the control ChIP DNA was labeled with cyanine-3 and 2 μg of heparanase or RNAP II ChIP DNA was labeled with cyanine-5 (Invitrogen BioPrime CGH labeling kit). Then 5 μg of cy3-labeled control ChIP DNA was hybridized on each microarray slide competitively with 5 μg of cy5-labeled heparanase or RNAP II ChIP DNA for 40 h at 65°C and 20 rpm. Agilent human promoter microarrays were utilized comprising of two slides per set defined to cover ~17,000 promoters of human transcripts from -5.5 to +2.5 kb relative to the transcriptional start site (part G4489A with design ID 014706 and 014707). The microarrays were scanned on an Agilent DNA Microarray Scanner (G2565BA) at 100% PMT gain. Two replicates (biological) of each ChIP-on-chip experiment were performed.
ChIP-on-chip data analysis
Microarray images were processed with Agilent Feature Extraction software (version 9.5.3) using the default protocol for ChIP-on-chip data. The software generates a median pixel intensity calculated per feature and this was then imported into Agilent ChIP Analytics software (version 1.3) to normalize the intensity in the cyanine-3 and the cyanine-5 channels. Lowess normalization to correct for intensity-dependent dye-bias was applied for the given replicates of heparanase or RNAP II, using a subset of common control probes on the array. This normalizes intensities throughout the entire signal range and reduces the occurrence of false high-fold enrichment results for low intensity measurements. The fold enrichment was calculated based on a weighted average of the replicate log ratios of the normalized intensity in the ChIP enriched channel compared with the normalized intensity in the control genomic DNA channel for each probe. Identification of bound probes and genes was performed using the Whitehead Per-array Neighborhood model76-79 within the ChIP Analytics software. In the neighborhood model, if a candidate bound probe set P-value of a weighted average of the replicate log ratios was less than 0.05, the central probe was marked as potentially bound and the probe sets were required to pass one of two additional □lters: the center probe in the probe set has a single-point P-value less than 0.1 or one of the 〉anking probes has a single-point P-value less than 0.1. These two □lters cover situations where binding events of broadly acting transcription factors occur midway between two probes. Genes most likely to be bound were identified by ranking results of the ‘gene report’ by P-value; highest likelihood corresponds to lowest P-value generated from the neighborhood model. The GO biological processes and molecular functions enriched among the heparanase target genes were determined using Agilent GeneSpring GX software (version 7.3).
Data analysis
Data was analyzed and graphs were generated using Prism (version 5.0, GraphPad software).
Supplementary Material
Acknowledgments
This work was supported by the Australian National Health and Medical Research Council (NHMRC) project and program grants awarded to S.R. and C.R.P., respectively. We acknowledge the extensive bioinformatics analysis performed by Andrew Pinson. We are grateful to Vijay Randev for support that allowed smooth progression of the ChIP-on-chip experiments.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/19998
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Wolffe AP. Transcription: in tune with the histones. Cell. 1994;77:13–6. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–27. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–91. doi: 10.1016/S0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 6.Weissmann F, Lyko F. Cooperative interactions between epigenetic modifications and their function in the regulation of chromosome architecture. Bioessays. 2003;25:792–7. doi: 10.1002/bies.10314. [DOI] [PubMed] [Google Scholar]
- 7.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 8.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 9.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–72. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 13.Biancotto C, Frigè G, Minucci S. Histone modification therapy of cancer. Adv Genet. 2010;70:341–86. doi: 10.1016/B978-0-12-380866-0.60013-7. [DOI] [PubMed] [Google Scholar]
- 14.Black JC, Whetstine JR. Chromatin landscape: Methylation beyond transcription. Epigenetics; 6. [DOI] [PMC free article] [PubMed]
- 15.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–20. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 16.Cohen E, Doweck I, Naroditsky I, Ben-Izhak O, Kremer R, Best LA, et al. Heparanase is overexpressed in lung cancer and correlates inversely with patient survival. Cancer. 2008;113:1004–11. doi: 10.1002/cncr.23680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 18.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–7. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parish CR. The role of heparan sulphate in inflammation. Nat Rev Immunol. 2006;6:633–43. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 20.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–30. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Zaken O, Gingis-Velitski S, Vlodavsky I, Ilan N. Heparanase induces Akt phosphorylation via a lipid raft receptor. Biochem Biophys Res Commun. 2007;361:829–34. doi: 10.1016/j.bbrc.2007.06.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fux L, Feibish N, Cohen-Kaplan V, Gingis-Velitski S, Feld S, Geffen C, et al. Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009;69:1758–67. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J Biol Chem. 2004;279:23536–41. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- 24.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66:1455–63. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 25.Barash U, Cohen-Kaplan V, Arvatz G, Gingis-Velitski S, Levy-Adam F, Nativ O, et al. A novel human heparanase splice variant, T5, endowed with protumorigenic characteristics. FASEB J. 2010;24:1239–48. doi: 10.1096/fj.09-147074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen-Kaplan V, Doweck I, Naroditsky I, Vlodavsky I, Ilan N. Heparanase augments epidermal growth factor receptor phosphorylation: correlation with head and neck tumor progression. Cancer Res. 2008;68:10077–85. doi: 10.1158/0008-5472.CAN-08-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Sanderson RD. Heparanase regulates levels of syndecan-1 in the nucleus. PLoS One. 2009;4:e4947. doi: 10.1371/journal.pone.0004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi M, Naomoto Y, Nobuhisa T, Okawa T, Takaoka M, Shirakawa Y, et al. Heparanase regulates esophageal keratinocyte differentiation through nuclear translocation and heparan sulfate cleavage. Differentiation. 2006;74:235–43. doi: 10.1111/j.1432-0436.2006.00072.x. [DOI] [PubMed] [Google Scholar]
- 29.Nobuhisa T, Naomoto Y, Takaoka M, Tabuchi Y, Ookawa K, Kitamoto D, et al. Emergence of nuclear heparanase induces differentiation of human mammary cancer cells. Biochem Biophys Res Commun. 2005;331:175–80. doi: 10.1016/j.bbrc.2005.03.129. [DOI] [PubMed] [Google Scholar]
- 30.Schubert SY, Ilan N, Shushy M, Ben-Izhak O, Vlodavsky I, Goldshmidt O. Human heparanase nuclear localization and enzymatic activity. Lab Invest. 2004;84:535–44. doi: 10.1038/labinvest.3700084. [DOI] [PubMed] [Google Scholar]
- 31.Doweck I, Kaplan-Cohen V, Naroditsky I, Sabo E, Ilan N, Vlodavsky I. Heparanase localization and expression by head and neck cancer: correlation with tumor progression and patient survival. Neoplasia. 2006;8:1055–61. doi: 10.1593/neo.06577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobuhisa T, Naomoto Y, Ohkawa T, Takaoka M, Ono R, Murata T, et al. Heparanase expression correlates with malignant potential in human colon cancer. J Cancer Res Clin Oncol. 2005;131:229–37. doi: 10.1007/s00432-004-0644-x. [DOI] [PubMed] [Google Scholar]
- 33.Nobuhisa T, Naomoto Y, Okawa T, Takaoka M, Gunduz M, Motoki T, et al. Translocation of heparanase into nucleus results in cell differentiation. Cancer Sci. 2007;98:535–40. doi: 10.1111/j.1349-7006.2007.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohkawa T, Naomoto Y, Takaoka M, Nobuhisa T, Noma K, Motoki T, et al. Localization of heparanase in esophageal cancer cells: respective roles in prognosis and differentiation. Lab Invest. 2004;84:1289–304. doi: 10.1038/labinvest.3700159. [DOI] [PubMed] [Google Scholar]
- 35.Sutcliffe EL, Parish IA, He YQ, Juelich T, Tierney ML, Rangasamy D, et al. Dynamic histone variant exchange accompanies gene induction in T cells. Mol Cell Biol. 2009;29:1972–86. doi: 10.1128/MCB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahtouk K, Hose D, Raynaud P, Hundemer M, Jourdan M, Jourdan E, et al. Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma. Blood. 2007;109:4914–23. doi: 10.1182/blood-2006-08-043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brettingham-Moore KH, Sprod OR, Chen X, Oakford P, Shannon MF, Holloway AF. Determinants of a transcriptionally competent environment at the GM-CSF promoter. Nucleic Acids Res. 2008;36:2639–53. doi: 10.1093/nar/gkn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, Wang J, Woltring D, Gerondakis S, Shannon MF. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol Cell Biol. 2005;25:3209–19. doi: 10.1128/MCB.25.8.3209-3219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinke H, Hörz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–607. doi: 10.1016/S1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 40.Svaren J, Hörz W. Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem Sci. 1997;22:93–7. doi: 10.1016/S0968-0004(97)01001-3. [DOI] [PubMed] [Google Scholar]
- 41.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–29. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci U S A. 2003;100:8342–7. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–89. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 44.Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edmunds JW, Mahadevan LC. Cell signaling. Protein kinases seek close encounters with active genes. Science. 2006;313:449–51. doi: 10.1126/science.1131158. [DOI] [PubMed] [Google Scholar]
- 46.Lawrence MC, McGlynn K, Shao C, Duan L, Naziruddin B, Levy MF, et al. Chromatin-bound mitogen-activated protein kinases transmit dynamic signals in transcription complexes in beta-cells. Proc Natl Acad Sci U S A. 2008;105:13315–20. doi: 10.1073/pnas.0806465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–6. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 48.Zcharia E, Jia J, Zhang X, Baraz L, Lindahl U, Peretz T, et al. Newly generated heparanase knock-out mice unravel co-regulation of heparanase and matrix metalloproteinases. PLoS One. 2009;4:e5181. doi: 10.1371/journal.pone.0005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pascual-Ahuir A, Struhl K, Proft M. Genome-wide location analysis of the stress-activated MAP kinase Hog1 in yeast. Methods. 2006;40:272–8. doi: 10.1016/j.ymeth.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 50.Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- 51.Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell. 2001;7:767–77. doi: 10.1016/S1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- 52.Chow CW, Davis RJ. Proteins kinases: chromatin-associated enzymes? Cell. 2006;127:887–90. doi: 10.1016/j.cell.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 53.Edmunds JW, Mahadevan LC. MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J Cell Sci. 2004;117:3715–23. doi: 10.1242/jcs.01346. [DOI] [PubMed] [Google Scholar]
- 54.Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, et al. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23:241–50. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 55.Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–17. doi: 10.1016/S1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 56.Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr Opin Cell Biol. 2004;16:263–71. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 57.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–78. doi: 10.1016/S0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 58.Smale ST, Fisher AG. Chromatin structure and gene regulation in the immune system. Annu Rev Immunol. 2002;20:427–62. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- 59.Juelich T, Sutcliffe EL, Denton A, He Y, Doherty PC, Parish CR, et al. Interplay between chromatin remodeling and epigenetic changes during lineage-specific commitment to granzyme B expression. J Immunol. 2009;183:7063–72. doi: 10.4049/jimmunol.0901522. [DOI] [PubMed] [Google Scholar]
- 60.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 61.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–7. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 62.Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 63.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 64.Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792–6. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- 65.Forneris F, Battaglioli E, Mattevi A, Binda C. New roles of flavoproteins in molecular cell biology: histone demethylase LSD1 and chromatin. FEBS J. 2009;276:4304–12. doi: 10.1111/j.1742-4658.2009.07142.x. [DOI] [PubMed] [Google Scholar]
- 66.Forneris F, Binda C, Adamo A, Battaglioli E, Mattevi A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J Biol Chem. 2007;282:20070–4. doi: 10.1074/jbc.C700100200. [DOI] [PubMed] [Google Scholar]
- 67.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15:1312–7. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–36. doi: 10.1016/S1097-2765(02)00477-X. [DOI] [PubMed] [Google Scholar]
- 70.Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS One. 2008;3:e3255. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 2001;167:4494–503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 72.Rao S, Gerondakis S, Woltring D, Shannon MF. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J Immunol. 2003;170:3724–31. doi: 10.4049/jimmunol.170.7.3724. [DOI] [PubMed] [Google Scholar]
- 73.Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res. 2009;19:1374–83. doi: 10.1101/gr.089185.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–94. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–80. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, et al. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci U S A. 2008;105:15837–42. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 78.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–77. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.