Abstract
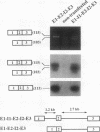
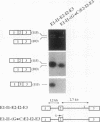
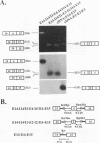
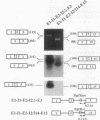
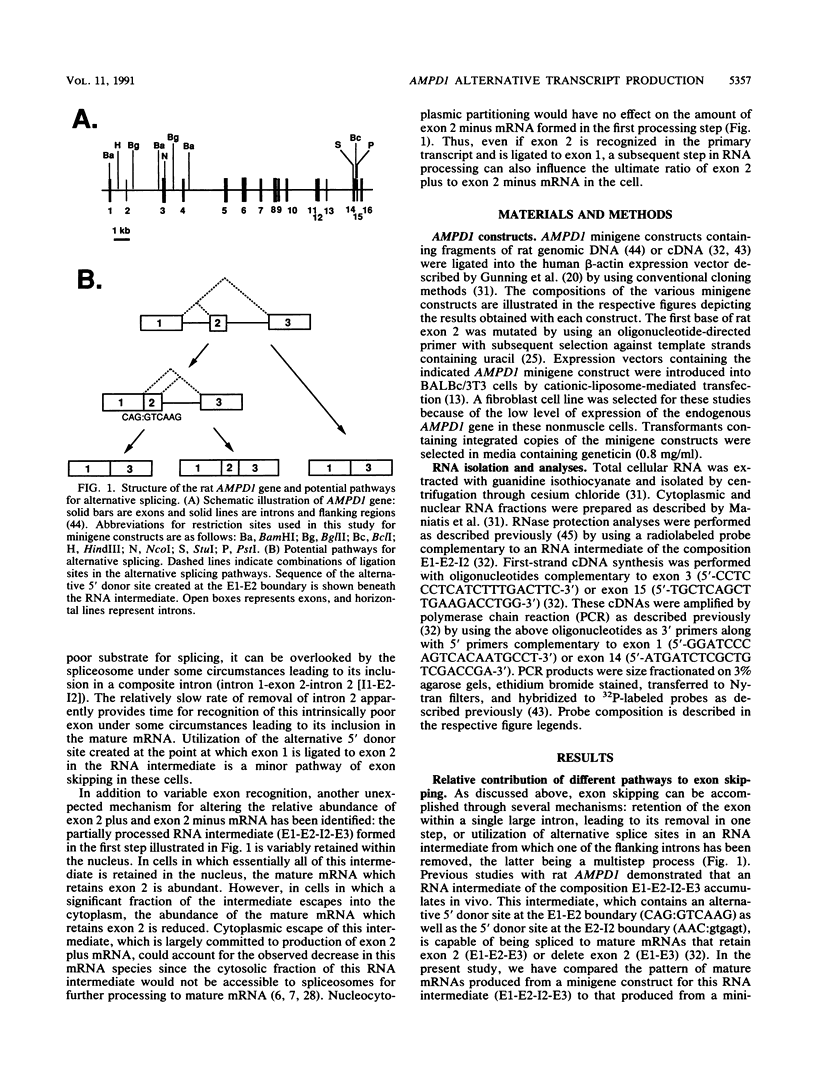
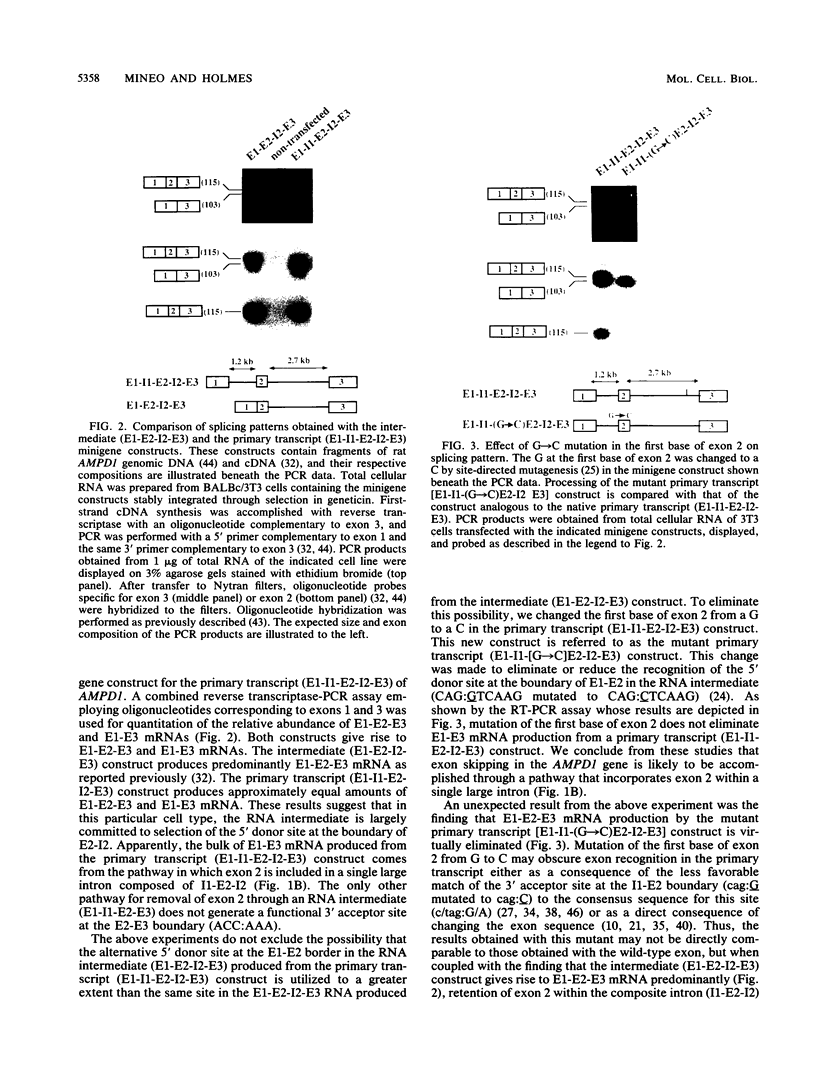
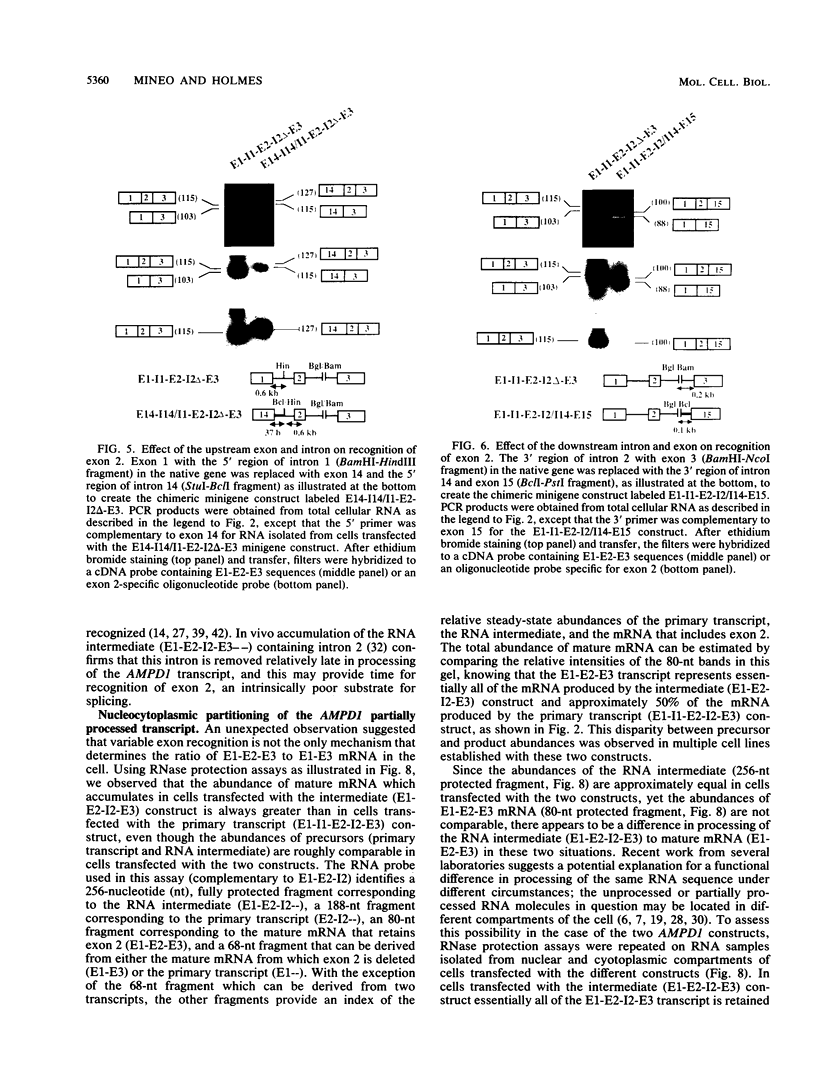
Two mature transcripts are produced from the rat AMP deaminase 1 (AMPD1) gene, one that retains exon 2 and one from which exon 2 has been removed. The ratio of these two transcripts is controlled by stage-specific and tissue-specific signals (I. Mineo, P. R. H. Clarke, R. L. Sabina, and E. W. Holmes, Mol. Cell. Biol. 10:5271-5278, 1990; R. L. Sabina, N. Ogasawara, and E. W. Holmes, Mol. Cell. Biol. 9:2244-2246, 1989). By using transfection studies with native, mutant, and chimeric minigene constructs, two steps in RNA processing that determine the ratio of these two transcripts have been identified. The first step is recognition of this exon in the primary transcript. The primary transcript is subject to alternative splicing in which exon 2 is either recognized and thereby included in the mature mRNA or is ignored and retained in a composite intron containing intron 1-exon 2-intron 2. The following properties of the primary transcript influence exon recognition. (i) Exon 2 is intrinsically difficult to recognize, possibly because of its small size (only 12 bases) and/or a suboptimal 5' donor site at the exon 2-intron 2 boundary. (ii) Intron 2 plays a permissive role in recognition of exon 2 because it is removed at a relatively slow rate, presumably because of the suboptimal polypyrimidine tract in the putative 3' branch site. The second step in RNA processing that influences the ratio of mature transcripts produced from the AMPD1 gene occurs subsequent to the ligation of exon 2 to exon 1. An RNA intermediate, composed of exon 1-exon 2-intron 2-exon 3, is produced in the first processing step, but it is variably retained in the nucleus. Retention of this intermediate in the nucleus is associated with accumulation of the mature mRNA containing exon 2, while cytoplasmic escape of this intermediate is reactions, exon recognition and nucleocytoplasmic partitioning, determine the relative abundance of alternative mRNAs derived from the AMPD1 gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adami G. R., Carmichael G. G. The length but not the sequence of the polyoma virus late leader exon is important for both late RNA splicing and stability. Nucleic Acids Res. 1987 Mar 25;15(6):2593–2610. doi: 10.1093/nar/15.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho S., Tate V., Boedtker H. Location of the 11 bp exon in the chicken pro alpha 2(I) collagen gene. Nucleic Acids Res. 1984 Aug 10;12(15):6117–6125. doi: 10.1093/nar/12.15.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A. S., Jr, Kittler E. L., Emerson C. P., Jr Structure, evolution, and regulation of a fast skeletal muscle troponin I gene. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8080–8084. doi: 10.1073/pnas.82.23.8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 1991 Mar;5(3):389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Messenger RNA transport and HIV rev regulation. Science. 1990 Aug 10;249(4969):614–615. doi: 10.1126/science.2143313. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989 Dec 1;59(5):789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. A gene chimaera of SV40 and mouse beta-globin is transcribed and properly spliced. Nature. 1981 Jan 29;289(5796):378–382. doi: 10.1038/289378a0. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. A single cardiac troponin T gene generates embryonic and adult isoforms via developmentally regulated alternate splicing. J Biol Chem. 1985 Sep 15;260(20):11140–11148. [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Feener C. A., Koenig M., Kunkel L. M. Alternative splicing of human dystrophin mRNA generates isoforms at the carboxy terminus. Nature. 1989 Apr 6;338(6215):509–511. doi: 10.1038/338509a0. [DOI] [PubMed] [Google Scholar]
- Felgner P. L., Gadek T. R., Holm M., Roman R., Chan H. W., Wenz M., Northrop J. P., Ringold G. M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Ge H., Manley J. L. The role of the polypyrimidine stretch at the SV40 early pre-mRNA 3' splice site in alternative splicing. EMBO J. 1988 Mar;7(3):809–817. doi: 10.1002/j.1460-2075.1988.tb02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M. E., Nadal-Ginard B. Myosin light-chain 1/3 gene alternative splicing: cis regulation is based upon a hierarchical compatibility between splice sites. Mol Cell Biol. 1990 May;10(5):2133–2144. doi: 10.1128/mcb.10.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Manley J. L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990 Jul 13;62(1):25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Green M. R., Zapp M. L. Human immunodeficiency virus. Revving up gene expression. Nature. 1989 Mar 16;338(6212):200–201. doi: 10.1038/338200a0. [DOI] [PubMed] [Google Scholar]
- Gunning P., Leavitt J., Muscat G., Ng S. Y., Kedes L. A human beta-actin expression vector system directs high-level accumulation of antisense transcripts. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson R. K., La Follette L., Rottman F. M. Alternative processing of bovine growth hormone mRNA is influenced by downstream exon sequences. Mol Cell Biol. 1989 Apr;9(4):1604–1610. doi: 10.1128/mcb.9.4.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989 Jul 25;17(14):5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990 Jul 13;62(1):35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Kuivaniemi H., Kontusaari S., Tromp G., Zhao M. J., Sabol C., Prockop D. J. Identical G+1 to A mutations in three different introns of the type III procollagen gene (COL3A1) produce different patterns of RNA splicing in three variants of Ehlers-Danlos syndrome. IV. An explanation for exon skipping some mutations and not others. J Biol Chem. 1990 Jul 15;265(20):12067–12074. [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Lang K. M., Keller W. Sequence requirements in different steps of the pre-mRNA splicing reaction: analysis by the RNA modification-exclusion technique. Mol Cell Biol. 1990 Sep;10(9):4942–4947. doi: 10.1128/mcb.10.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain P., Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989 May 19;57(4):573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- Malim M. H., Hauber J., Le S. Y., Maizel J. V., Cullen B. R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989 Mar 16;338(6212):254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Mineo I., Clarke P. R., Sabina R. L., Holmes E. W. A novel pathway for alternative splicing: identification of an RNA intermediate that generates an alternative 5' splice donor site not present in the primary transcript of AMPD1. Mol Cell Biol. 1990 Oct;10(10):5271–5278. doi: 10.1128/mcb.10.10.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H., Fukamizu A., Hirose S., Hayashi T., Hori H., Ohkubo H., Nakanishi S., Murakami K. Structure of the human renin gene. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5999–6003. doi: 10.1073/pnas.81.19.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi R. N., Baker B. S. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990 Jan;4(1):89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Pan Z. Q., Prives C., Manley J. L. Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell. 1987 Jul 17;50(2):227–236. doi: 10.1016/0092-8674(87)90218-2. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Sabina R. L., Marquetant R., Desai N. M., Kaletha K., Holmes E. W. Cloning and sequence of rat myoadenylate deaminase cDNA. Evidence for tissue-specific and developmental regulation. J Biol Chem. 1987 Sep 15;262(26):12397–12400. [PubMed] [Google Scholar]
- Sabina R. L., Morisaki T., Clarke P., Eddy R., Shows T. B., Morton C. C., Holmes E. W. Characterization of the human and rat myoadenylate deaminase genes. J Biol Chem. 1990 Jun 5;265(16):9423–9433. [PubMed] [Google Scholar]
- Sabina R. L., Ogasawara N., Holmes E. W. Expression of three stage-specific transcripts of AMP deaminase during myogenesis. Mol Cell Biol. 1989 May;9(5):2244–2246. doi: 10.1128/mcb.9.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski B. A., Belote J. M., McKeown M. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell. 1989 Aug 11;58(3):449–459. doi: 10.1016/0092-8674(89)90426-1. [DOI] [PubMed] [Google Scholar]
- Ulfendahl P. J., Kreivi J. P., Akusjärvi G. Role of the branch site/3'-splice site region in adenovirus-2 E1A pre-mRNA alternative splicing: evidence for 5'- and 3'-splice site co-operation. Nucleic Acids Res. 1989 Feb 11;17(3):925–938. doi: 10.1093/nar/17.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaud M., Gattoni R., Stevenin J., Vidaud D., Amselem S., Chibani J., Rosa J., Goossens M. A 5' splice-region G----C mutation in exon 1 of the human beta-globin gene inhibits pre-mRNA splicing: a mechanism for beta+-thalassemia. Proc Natl Acad Sci U S A. 1989 Feb;86(3):1041–1045. doi: 10.1073/pnas.86.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Leung H., Weiner A. M. The natural 5' splice site of simian virus 40 large T antigen can be improved by increasing the base complementarity to U1 RNA. Mol Cell Biol. 1987 Aug;7(8):3018–3020. doi: 10.1128/mcb.7.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]