Abstract
Bisulfite conversion of genomic DNA combined with next-generation sequencing (NGS) has become a very effective approach for mapping the whole-genome and sub-genome wide DNA methylation landscapes. However, whole methylome shotgun bisulfite sequencing is still expensive and not suitable for analyzing large numbers of human cancer specimens. Recent advances in the development of targeted bisulfite sequencing approaches offer several attractive alternatives. The characteristics and applications of these methods are discussed in this review article. In addition, the bioinformatic tools that can be used for sequence capture probe design as well as downstream sequence analyses are also addressed.
1. Introduction
DNA methylation occurs at the carbon-5 position of cytosine residue of the CpG dinucleotide in mammalian genomes and is one of the best-studied epigenetic modifications. DNA methylation is known to play important roles in several key physiological processes including regulation of gene expression, X-chromosome inactivation, imprinting, and maintenance of chromosomal stability. The majority of CpG dinucleotides (70–80%) are fully methylated in normal human cells [1]. However, 0.2–3kb stretches of GC-rich DNA, called CpG islands (CGIs), appear to be protected from modification in normal somatic cells [2]. Nearly half of all known human genes are associated with CGIs in their 5’-end regulatory regions. Patterns of DNA methylation are stably maintained through somatic cell division and can be inherited across generations. However, studies also show that DNA methylation is dynamically regulated during differentiation and aging [3,4]. A growing body of evidence has suggested that DNA methylation patterns can be modulated by environmental factors [5,6]. Twins whose separation resulted in exposure to variable environmental factors showed significant differences with respect to DNA methylation content [7]. Aberrant DNA methylation changes can cause a number of human diseases such as developmental diseases (ICF syndrome, Prader-Willi and Angelman syndromes etc), aging related diseases (i.e. Alzheimer’s disease), heart disease, diabetes, and autoimmune diseases [8,9,10,11,12]. Therefore, it is very important to understand how methylation patterns are established and maintained during normal development and under pathological conditions.
Accumulating evidence has indicated that epigenetic alterations are at least as common as, if not more frequent than, mutational events in the development of cancer [13]. Compared to normal cells, the malignant cells exhibit overall genomic hypomethylation (primarily in repeat elements and pericentromeric regions), but simultaneously show hypermethylation of normally protected CGIs [14,15]. Hypermethylation within promoters serves to turn off critical tumor suppressor genes that could otherwise suppress tumorigenesis [15]. Given their important functions in cancer initiation and progression, aberrant methylation patterns may be used as biomarkers for diagnosis and prognosis of cancer [16]. Unlike genetic mutations, epigenetic alterations are reversible, making them a therapeutic target. Treatment with inhibitors of DNA methylation and histone deacetylation can reactivate epigenetically silenced genes and has been shown to restore normal gene function [8,17]. The demethylating agent azacitidine and its deoxy derivative, decitabine, have been approved for treating hematological malignancies including MDS and AML [18].
2. Methods for DNA methylation analysis
The methylation status of DNA can be analyzed by many different methods which utilize three basic principles: 1) digest unmethylated or methylated DNA with methylation sensitive restriction enzymes; 2) use anti-methylcytosine antibodies or methyl-binding domain (MBD) proteins to enrich methylated DNA; 3) bisulfite treatment can convert unmethylated cytosine into uracil while leaving methylated cytosine unchanged. These principles have been integrated into high-throughput analytical applications such as microarray and next-generation sequencing (NGS) platforms. Several excellent reviews of the genome-wide DNA methylation analysis are available [19,20,21,22]. The representative microarray-based methods include DMH [23], HELP [24], MCA [25] (restriction enzyme based), MeDIP [26], and MIRA [27] (affinity enrichment based), which utilize promoter or CpG island tiling arrays to identify differentially methylated genes. Recently, some of these approaches have been moved to the NGS platforms (i.e. HELP-tagging [28], DREAM[29], MRE-seq [30], MeDIP-seq [30], and MIRA-seq [31]). One of the disadvantages of microarray-based analyses, particularly those using enrichment of methylated DNA, is that it can survey for the presence or absence of methylated DNA, but it gives little detail about the extent and pattern of CpG methylation within a given region. Often, extensive follow-up studies must be conducted on a single gene basis to confirm the results of such microarray experiments. The Illlumina Infinium beads array, which analyzes bisulfite-converted DNA, has become a popular platform for methylome analysis. The current version of Infinium 450K BeadChip is capable of measuring more than 450,000 CpG sites in promoters, CGIs, CGI shores, enhancer regions, and DNase I hypersensitive sites in the human genome curated from published literature. The chip has been proven to be highly accurate, reproducible and does not require a large amount of input DNA [32]. Most importantly, the data analysis is relative simple and straightforward making the Infinium array the best approach currently available for population-based epigenetic studies.
Due to the significantly increased throughput, a large number of differentially methylated genes can now be identified in a single experiment. Therefore, the traditional methods for validation experiments such as MSP, COBRA, and bisulfite clone sequencing are no longer sufficient to keep up with the increased demand. Pyrosequencing (PyroMark™, Qiagen) and MassArray (EpiTYPER™, Sequenom) have now largely replaced the more traditional validation methods. Bisulfite pyrosequencing is a quantitative method for DNA methylation analysis, and it can determine the CpG methylation status in a sequence up to 100-bp in length. Pyrosequencing is based on the sequence-by-synthesis principle; nucleotides are added in each pyrosequencing cycle and the amount of incorporated nucleotide results in a proportional emission of light. DNA methylation ratios are calculated from the levels of light emitted from each nucleotide incorporated at individual CpG positions in a strand-dependent manner. The methylation levels at each CpG position assayed are the average of all amplification products generated during a bisulfite-PCR reaction. DNA methylation analysis by MALDI-TOF mass spectrometry (MS) employs base-specific cleavage of single-stranded nucleic acids [33]. The idea is to generate a PCR product from bisulfite-treated DNA, which is then transcribed in vitro into a single-stranded RNA molecule; the synthesized RNA is cleaved in a base-specific manner by an endoribonuclease [33]. The base-specific cleavage products are analyzed using the MassArray instrument. Due to the conversion of unmethylated cytosine to thymine after bisulfite treatment and PCR, different cleavage products are generated for methylated and unmethylated DNA. The MassArray instrument is able to quantitatively determine the proportion of methylated vs. unmethylated DNA. However, due to the reduced sequence complexity of bisulfite-converted DNA, the often-occurring overlap of peaks is difficult to interpret. Therefore, not all CpG sites can be analyzed for a given bisulfite-PCR product. These two methods are capable of measuring 96 to 384 PCR products in a single reaction and provide a medium-throughput platform for candidate gene methylation analysis.
3. Whole genome bisulfite sequencing (WGBS-seq)
Whole genome shotgun bisulfite sequencing (WGBS-seq) is the best way of determining the landscape of the whole methylome. WGBS-seq has been successfully used to map the complete methylomes of several human embryonic stem (ES) cell lines [34,35], human peripheral mononuclear cells [36], and hematopoietic progenitor cells at the single methylcytosine resolution [37]. The single-base level analysis was instrumental in the discovery of non-CpG methylation in ES cells [35]. Recently, the WGBS-seq analyses of several cancer samples have been completed [38,39,40]. Again, WGBS-seq made a significant contribution to the discovery of large partially methylated domains in cancer cells [38,40]. However, despite its advantages, the WGBS-seq remains too expensive to be applied to a large number of samples. In order to achieve higher sensitivity capable of detecting methylation differences between samples, a greater sequencing depth is required ultimately leading to a significant increase in the cost of sequencing. Currently, most of the WGBS-seq studies were conducted using Illumina GAII or HiSeq 2000 sequencers, while only one study used the SOLiD sequencer [40]. Although we have previously used 454-sequencing for genome-wide bisulfite sequencing of methylation enriched DNA [31], it is cost-prohibitive to use 454-sequencing for WGBS-seq.
4. Targeted bisulfite sequencing (TBS-seq)
The development of the targeted sequencing approach was mainly driven by the desire to conduct whole exome sequencing that would allow for the identification of disease-causing genes at a low cost compared to whole genome sequencing. A number of methods for targeted genome sequencing have been developed [41,42,43,44]. These approaches have been gradually integrated with bisulfite sequencing over time. Table 1 summarizes the characteristics and some of the key performance parameters of these TBS-seq approaches. The following section will explore each of these approaches in more detail.
Table 1.
Comparison of the targeted bisulfite sequencing methods
| Method | BSPP | SHBS-seq | ACBS-seq | Bisulfite-Patch PCR |
Microdroplet PCR |
|||
|---|---|---|---|---|---|---|---|---|
| Reference | [47] | [48] | [49] | [51] | [52] | [58] | [60] | [62] |
| Capture probes/primer | ssDNA | ssDNA | ssDNA | Biotinylated RNA | Biotinylated RNA | ssDNA | Partial dsDNA | ssDNA |
| Probe/primer size | 150 bp | 150 bp | 150 bp | 160 bp | 120 bp | 60 bp | 40–50 bp | 20–30 bp |
| Number of probes/primers | 30,000 | 9,552 | 330,000 | 51,551 | n/a | 240,000 | 94 pairs | 3,500 pairs |
| Probe/primer manufacturer | Agilent | Agilent | Agilent | Agilent | Agilent | Agilent | Sigma | RainDance |
| Capture method | Annealing, extension and circularization | Annealing, extension and circularization | Annealing, extension and circularization | Hybridization in solution | Hybridization in solution | Hybridization on array | Annealing, ligation, and PCR | Microdroplet PCR |
| Amount of input DNA | 200 ng bisulfite-treated | 1 µg bisulfite-treated | 200 ng bisulfite-treated | 20–30 µg native | 2–3 µg native | 20 µg bisulfite-treated and amplified | 250 ng bisulfite-treated | 2 µg bisulfite-treated |
| Bisulfite treatment | Pre-capture | Pre-capture | Pre-capture | Post-capture | Pre-capture | Pre-capture | Pre-capture | Pre-capture |
| Sequencing platform | Illumina | Illumina | Illumina | Illumina | Illumina | Illumina | 454 | Illumina |
| Target size | 2.1Mb | n/a | 34Mb | 8.2Mb | 38Mb | 258.9kb | 25.5kb | 1.35Mb |
| Number of targets | 10,582 | 9552 | 140,749 | 51,551 | n/a | 324 | 94 | 3,500 |
| Target features | CGI, ENCODE, (TSS±1000bp) | ENCODE regions | DMRs, CTCF, DNase I sites | CGI, TSS | Exon | CGI | Promoter | Promoter (TSS±1000bp) |
| Targets covered | n/a | 68% (>10 reads) | n/a | 85–88% (>10 reads) | 90% (>10 reads) | ~92% | 100% | 99% |
| On-target rate (%) | n/a | n/a | 96% | 77–84% | 71–75% | 6–12% | 90% | 90% |
| Number of CpG | 66,000 | 6,400 | 500,0000 | 0.9–1 million | n/a | 25,044 | n/a | 77,674 |
| Coverage per CpG | n/a | n/a | n/a | 86–146 (mean) | 58–73 | 95–105 (median) | 444 (median) | >100 (97% of CpGs) |
| Reproducibility (r) | 0.97 | 0.965 | 0.97–98 | 0.94 | n/a | n/a | 0.91 | n/a |
| Advantages | Highly specific and reproducible, require lower amount of input DNA | Flexible probe design; easy to scale up to target large regions | Use bisulfite-treated and amplified DNA | Require lower amount of DNA, use regular PCR primers | Highly multiplex PCR, low PCR bias | |||
| Disadvantages | Expensive probes, complex probe design | Capture small amount of DNA; challenges in bisulfite treatment | Low capture specificity, require specific instruments | Uncertainty about scale-up in throughput | Require specific instruments. | |||
4.1 Bisulfite padlock probes (BSPPs)
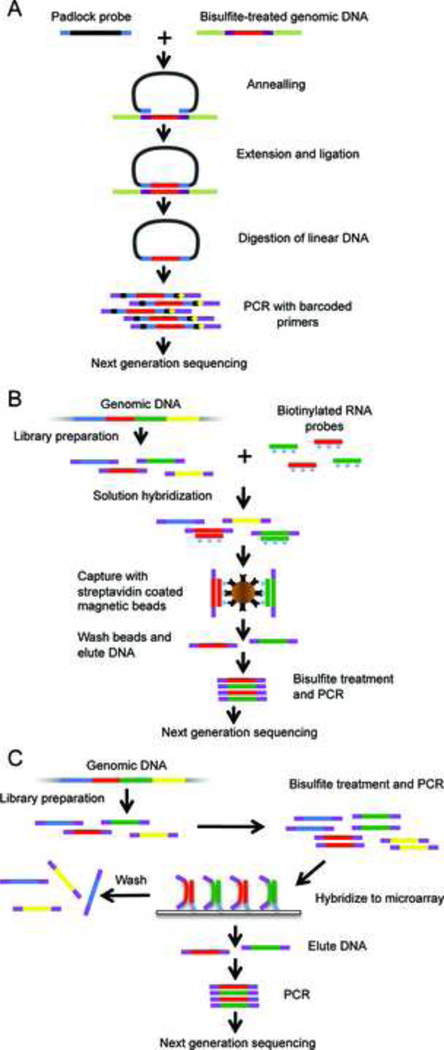
Padlock probes are single strand DNA fragments (100–150 nucleotide long) designed to hybridize to genomic DNA targets in a horseshoe manner (Fig. 1A) [45]. The targeted region for sequence capture is the gap between the two hybridized, locus-specific arms of a padlock probe. After the gap is filled in with a polymerase and ligated to form a circular strand of DNA, the target sequence is enriched by digestion of the linear DNA with nucleases. The target sequence in the circular DNA can be amplified using the common 'backbone' sequence that connects the two arms and is eventually converted into a sequencing library. This approach enables tens of thousands of probes to be used in a single reaction. Bisulfite padlock probes (BSPP) follow the same principle, but they are designed based on bisulfite-converted DNA sequences. Deng et al. used the BSPP approach to assess the methylation status of ~66,000 CpG sites in 2,020 CGI on human chromosomes 12 and 20 [46]. Ball and colleagues designed ~10,000 padlock probes to profile ~7,000 CpGs within the ENCODE pilot project regions [47]. Both studies demonstrated that the BSPP approach is highly specific and reproducible (Table 1). Initially, there was a significant degree of variability in the capture efficiency for each of the probes used. It was further demonstrated that the variability could be normalized using suppressor oligos. Recently, an improved BSPP protocol was developed [48] which included a design algorithm to generate efficient padlock probes and a library-free protocol that dramatically reduces sample-preparation cost and time. Using the new BSPP protocol, Diep and colleagues designed ~330,000 padlock probes that covered 140,749 non-overlapping regions with a total size of 34 megabases [48]. Their results showed that the improved BSPPs are highly specific (96% on-target) and more uniform than previous probes [48]. The new BSPPs allow for the accurate and reproducible measurement of the methylation status of 500,000 CpG sites from only 200 ng of bisulfite-converted DNA. The padlock probes can be synthesized by two commercial vendors (Agilent Technologies or LC biosciences); therefore the BSPP approach allows for a greater deal of flexibility. Users can design their own BSPPs for their unique targets of interest. The initial cost of BSPPs is high, although the cost per sample dramatically decreases with increased sample sizes [48]. Overall, the BSPP method allows for interrogation of the most informative loci across many samples quickly and cost-effectively.
Fig. 1. The targeted bisulfite sequencing methods.
(A) Bisulfite padlock probe (BSPP): 150bp single stranded DNA probes are prepared and hybridized to bisulfite-converted genomic DNA. Capture occurs by filling in sequences between the probe-targeting arms with polymerase followed by ligation to form circularized DNA. After removing linear DNA with nucleases, the remaining circularized captured targets are amplified by PCR prior to next-generation sequencing. (B) Solution hybrid selection bisulfite sequencing (SHBS-seq): A sequencing library is prepared from native genomic DNA. The library is then hybridized to biotinylated RNA probes in solution and recovered with streptavidin beads. Eluted products are treated with bisulfite and amplified by PCR prior to sequencing. (C) Array-capture bisulfite sequencing (ACBS-seq): A sequencing library is prepared from genomic DNA, treated with bisulfite and amplified by PCR. The amplified library is hybridized to an oligonucleotide capture array that is designed based on the bisulfite-converted DNA sequences. Eluted products are amplified prior to sequencing. Note that in (B) and (C), the adaptors used in the sequencing library construction contain methyl-cytosine so that the adaptor sequences will not change during bisulfite treatment.
4.2 Solution hybrid selection bisulfite sequencing (SHBS-seq)
Similar to BSPP, the solution hybrid selection (SHS) method was initially developed for exome sequencing [49], and it has been commercialized by Agilent Technologies (SureSelect Target Enrichment System). Lee et al. first demonstrated the successful application of this technology to targeted methylation analysis [50]. The most critical component of this technology is the synthesis of thousands of biotinylated RNA capture probes. This was achieved by using Agilent Technologies’ proprietary technology to synthesize tens of thousands of unique, long oligonucleotides (up to 190-mer) in fmol amounts on parallel on microarrays. The final product is delivered as a pool of single-stranded DNA oligonucleotides in a single test tube. Each oligonucleotide consisted of a target-specific 160-mer sequence flanked by 15 bases of a universal primer sequence on each end to allow PCR amplification. After an initial round of PCR, a T7 promoter was added to the 5’-end by a second PCR reaction. The biotin-labeled RNA probes were then synthesized by in vitro transcription using T7 RNA polymerase. In the study published by Lee et al., we designed 51,551 highly specific capture probes to target a total of 8.2 Mb that covered 23,441 CpG islands (~83% of the 28,226 CGIs in the human genome) and the transcription start sites (TSS) of 19,369 RefSeq genes [50]. The workflow of the solution hybrid selection combined with bisulfite sequencing is illustrated in Figure 1B. The biotinylated RNA probes were first hybridized to a pre-made sequencing library. The RNA:DNA hybrids were captured with streptavidin-coated magnetic beads. Finally, the captured DNA was treated with sodium bisulfite, amplified by PCR, and subsequently sequenced using the Illumina Genome Analyzer (Fig. 1B). Using several cancer cell lines, we demonstrated that SHBS-seq is very specific (average on-target rate of ~80%) [50], which is comparable with exome sequencing [49]. Up to 1 million CpG sites can be analyzed quantitatively using the SHBS-seq approach. Extensive validation experiments demonstrated that this method is both reproducible and accurate [50]. One of the bottlenecks limiting the successful application of SHBS-seq is that the amount of captured DNA is very low simply because the targeted regions only represent 0.2% of human genome. Since a substantial amount of DNA is expected to be lost during the harsh process of bisulfite conversion, we pooled DNA from 5–6 capture reactions in order to maintain low cycle numbers in the post-bisulfite PCR amplifications. As a result, 20–30 µg of input DNA was required for successful sequencing. However, Wang et al. applied the Agilent SureSelect Human All Exon Kit to enrich all human exons, which totaled approximately 38 Mb, for TBS-seq of whole exons [51], and reported that only 2–3 µg of genomic DNA was required. Recently, Agilent launched the SureSelect Human Methyl-seq kit that is based on the SHBS-seq approach. This commercial kit allows researchers to analyze over 3.7 million individual CpG sites for their methylation status. The capture probes are designed to target 84 Mb of genomic sequences which covers a comprehensive collection of features including CGIs, CGI shores/shelves, cancer- and tissue-specific DMRs, GENCODE promoters [52], enhancers, conserved undermethylated regions, and DNase I hypersensitive sites in the human genome. The Agilent protocol requires 2–3 µg of genomic DNA and can achieve an on-target-rate of 82%, which is similar to previous reports [50]. Overall, although it requires more input DNA than the BSPP method, SHBS-seq has the advantage that it can be easily scaled up to accommodate larger genomic regions. Furthermore, because the capture probes are designed based on native genomic DNA, it does not suffer from the limitations of the BSPP approach that are caused by the reduced sequence complexity after bisulfite treatment. Theoretically, biotin-labeled DNA probes can also be used for SHBS-seq. Nautiyal and colleagues developed a unique method for capturing target DNA via oligo-guided ligation before bisulfite conversion [53]. They demonstrated that their capture probes could be used to determine the methylation status of more than 145,000 CpGs from 5472 promoters. One of the challenges of their methodology is the construction of the capture probe panels, which were prepared by single-plex PCR reactions [53].
4.3 Array capture bisulfite sequencing (ACBS-seq)
Oligonucleotide microarrays have also been used as substrates for hybrid selection of targeted regions from complex genomes [54,55]. Several commercial microarray platforms such as Nimblegen and Agilents are available for this application. Hodges and colleagues first adapted the array capture approach for targeted bisulfite sequencing (Fig. 1C). Similar to the SHBS-seq approach described above, the region of interest is first captured from native genomic DNA followed by bisulfite treatment and sequencing. In practice, however, this strategy has two major shortcomings: first, the array hybridization requires large amounts of native, unamplified DNA[54,55], which is not readily available in many cases. Second, only very small amounts of material are generally eluted from the capture arrays. Since a substantial amount of DNA will be further lost during the harsh process of bisulfite conversion, bisulfite conversion post-capture significantly restricts the number of amplifiable molecules available for cluster generation and sequencing. Therefore, Hodges and colleagues chose to design a customized oligonucleotide tiling array to capture bisulfite-converted DNA. For each target sequence, they designed two sets of capture probes: one that assumed full methylation of all CpG sites and one that assumed full conversion of CpGs to TpGs. The rationale for this design is based on the results from previous studies which suggest that up to six distributed mismatches are tolerated without a substantial impact on hybridization efficiency for the 60-mer oligonucleotide microarray [56]. Therefore, even if the methylation statuses of CpG sites are heterogeneous, the target DNA can still be efficiently captured by the array. By performing pre-capture bisulfite-treatment, the sequencing library can be amplified before hybridizing to the capture array. In this way, although 20 µg of DNA is still required for array hybridization, the amount of native input DNA can be significantly reduced. Coupled with next-generation sequencing, Hodges and colleagues demonstrated that their approach allowed for the capture of 25,044 CpGs in 324 CGIs spanning 300 kb of sequence [57]. Confirmation studies showed that even partially methylated states could be successfully evaluated. However, one of most significant drawbacks of the ACBS-seq approach is that the specificity is dramatically lower than the BSPP and solution hybrid selection methods, only 6–12% [57] as compared to 96% for BSPP [48] and 77–83% for SHBS-seq [50]. The array capture procedures also require specific equipment for DNA hybridization and elution of target DNA.
4.4 Bisulfite-Patch PCR
Before the sequence capture techniques were developed, PCR was used to enrich the target genes for methylation analyses using next-generation sequencing [58]. However, this laborious approach does not efficiently utilize the high-throughput capacity of the next-generation sequencing. To address this problem, Varley and Mitra developed an approach called Bisulfite-Patch PCR [59], which enables highly multiplexed PCR amplification of DNA for 94 amplicons across a number of clinical samples. By incorporating next-generation sequencing, they have successfully sequenced 94 targeted gene promoters simultaneously in the same reaction. The method requires small amounts of starting DNA (250 ng), does not require a shotgun library construction, and is highly specific (90% on-target rate). Bisulfite-Patch PCR first uses a restriction enzyme to digest human genomic DNA and defines the ends of the fragments that will be selected. Targeted loci are then selected from the genomic restriction fragments by annealing patch oligos to the ends of the targeted genomic fragments [59]. These patch oligos serve two functions: 1) guide the selection of target loci; 2) help to ligate the universal primers. After ligation, unselected genomic DNA is eliminated with exonucleases. Since the boundaries of target loci are defined by restriction enzyme used, the number of loci that can be targeted in a single reaction is constrained by availability of restriction sites. However, this can be overcome by using multiple different enzymes [59].
4.5 Microdroplet PCR
One significant advance in multiplex PCR was the microdroplet PCR amplification system developed by RainDance Technologies [60]. This system allows the user to set up 1.5 million parallel PCR amplifications in a single reaction in under an hour. Single nucleic acids and PCR reagents are put into the picoliter-sized droplets one droplet at a time effectively creating a single-plex PCR reaction inside of each droplet. Up to 10,000 droplets per second can be generated using one of RainDance’s commercial instrument systems. PCR amplification bias is significantly decreased due to the nature of microdroplet emulsion PCR reactions [60]. Komori and colleagues first demonstrated targeted methylation analysis of nearly 3500 amplicons covering 77,674 CpGs in 2127 genes in primary CD4 T cells using the RainDance technology [61]. This approach generated high-quality bisulfite sequencing data for 97% of the targeted CpG sites and 99% of the targeted amplicons. If it is combined with 454-sequencing’s long read ability, microdroplet PCR bisulfite sequencing could reveal single molecule methylation patterns better than the short-read sequencing platforms.
5. Primer and capture probe design
The successful application of TBS-seq largely depends on the optimal design of capture probes and primers. Diep et al. developed a program called ppDesigner (http://genome-tech.ucsd.edu/public/Gen2_BSPP/ppDesigner/ppDesigner.php) for designing optimal BSPPs [48]. Whole genome sequences or a user's list of arbitrary targets can be uploaded into ppDesigner, and the program can automatically perform in silico bisulfite-conversion of the input sequences and generate padlock probes to cover the chosen targets while avoiding CpGs on the capturing arms that could be methylated. ppDesigner is extremely flexible and has been successfully used to design a variety of genomic and bisulfite probes [48]. For array-based and solution-based sequence capture, several commercial vendors such as Agilent, Illumina, and Nimblegen have developed websites that allow users to upload their target sequences and capture probes or arrays are designed for free (i.e. https://earray.chem.agilent.com/earray/). RainDance will also design the bisulfite PCR primers for customers who provide their own genes of interest.
PRIMEGENSw3 (http://primegens.org) is a set of web-based utilities for high-throughput primer/probe design with a focus on the epigenetic application. These utilities allow users to select genomic regions and to design thousands of primer pairs/probes to cover the targeted regions in an automatic fashion. The results can be visualized online in a tabular format or can be downloaded. A stand-alone version of the software, PRIMEGENSv2, is also available for download at the website. We have previously used this program to design bisulfite PCR primers with great success [62]. PRIMEGENSv2 was also crucial for designing the capture probes used in the SHBS-seq work [50]. One of the key features of this program is that it vigorously checks cross-hybridization across the genome to ensure the uniqueness of output primers and probes.
6. Sequencing data analysis
Tens of millions of bisulfite sequencing reads can be generated for each sample using the TBS-seq approaches discussed above. Analyzing the bisulfite sequencing data is quite challenging because the sequence reads do not exactly match the reference genome, and the complexity of sequences is reduced due to bisulfite-conversion. The bisulfite treatment and subsequent PCR amplification results in the conversion of unmethylated cytosines to thymine without changing methylated cytosines. Therefore, Ts in bisulfite sequencing reads are ambiguous during the sequence alignment process because they can be either original Ts or unmethylated Cs converted by bisulfite-treatment. Two basic strategies have been developed to map the bisulfite sequences to the reference genome. The first strategy considers both cytosine and thymine as potential matches to a cytosine in the reference sequences. The second strategy, which is unbiased, converts all cytosines in the bisulfite sequencing reads and the reference genome into thymines before the alignment is performed. Both strategies have advantages and disadvantages [63]. The first method has higher mapping efficiency because it considers all the information available in the read; however, it may result in overestimation of methylation levels due to the greater alignment efficiency of reads containing methylated cytosines. The second approach is unbiased because the alignment of any read sequence is not affected by its methylation status; however, this approach generally has a slightly lower mapping efficiency due to the significant reduction in sequence complexity. Another significant factor that influences the alignment rate is read length. Bisulfite sequencing reads that were generated with early versions of sequencers with read lengths of less than 40 bp were frequently poorly aligned. However, most of the recent bisulfite sequencing data was generated using longer reads (50–100bp) and the alignment rate increased to above 70% for most of the cases.
A number of software tools have been developed for mapping bisulfite sequencing reads such as BSMAP [64], BS-Seeker [65], BisMark [66], BRAT[67], RMAP-BS [68], MethylCoder [69], SOCS-B [69], and B-SOLANA [70]. Most of these programs use the existing short-read aligner (i.e. SOAP [71] for BS-MAP and Bowtie [72] for Bismark, BS-Seeker, and B-SOLANA). B-SOLANA and SOCS-B can handle the SOLiD sequencing data in color space. An excellent review on the advantages and drawbacks of these software packages can be found elsewhere [63]. The workflow for analyzing the bisulfite sequencing data also includes various quality control reporting, adaptive trimming based on quality score, adaptor trimming etc [63]. Apparently, some of these tasks have to be handled separately when using specific software other than the programs mentioned above. In addition, most of these programs only output the DNA methylation level of each CpG site, but do not provide downstream analysis, such as annotation, statistical comparison, and multiple sample comparison. Therefore, more integrated pipelines for bisulfite sequencing analysis are still highly desirable for the end users of TBS-seq approaches.
7. Conclusions and future directions
Currently, there are a number of methods available for TBS-seq. One of them, the SHBS-seq is commercially available (Agilent SureSelect Methyl-seq kit). Compared to WGBS-seq, the TBS-seq methods offer a series of less expensive alternatives. However, most of these methods require an initial investment to generate capture probes, and the cost per sample is highly dependent on the number of samples analyzed. Further development of commercial products based on these methods will be needed in order to drive down the cost. Nevertheless, these methods can be used for generating single-base resolution DNA methylation maps in a large number of cancer specimens. Each method has its own advantages and disadvantages. For instance, the BSPP approach can provide highly specific and reproducible DNA methylation measurements and can tolerate a small amount of input DNA; however, it requires the synthesis of a large number of padlock probes. In addition, the requirement that the capturing arms be CpG-free might exclude some highly GC rich sequences. The SHBS-seq approach provides an unbiased capture of target DNA and can cover a large size of DNA; however, due to the extremely small amount of DNA captured in each reaction, and the loss of DNA during post capture bisulfite-treatment, this method is probably sensitive to technical variations. Selection of target loci from bisulfite treated DNA probably will not work well with either solution or array-based sequence capture due to significant cross-hybridization. Bisulfite-Patch PCR is a simple and effective method for studies that investigate a relatively small number of genes in a large number of patient samples. This method does not require special equipment and is well suited for use in smaller laboratories. If the access to RainDance instruments is available, the Microdroplet PCR bisulfite sequencing is recommended for medium to large-scale studies involving thousands of target genes. Software tools have been developed to assist in the design of primers and capture probes. A number of programs for bisulfite sequence analysis are now available although further development of more integrative pipelines is still needed. The bioinformatic analysis of amplicon sequencing should be easier than that of shotgun sequencing.
With the improvement of next-generation sequencing technologies, the cost of whole genome sequencing will continue to decrease. It is expected that we could soon read the entire DNA methylome for under $1,000. Once this happens, TBS-seq may lose its cost advantage over WGBS-seq. However, TBS-seq can still be used as a validation tool as it can provide more quantitative measurements of DNA methylation differences due to its ultra-deep sequencing depth. Furthermore, single-molecule and nanopore sequencing approaches are likely to usher in the next revolution in high-throughput DNA methylation analysis. These technologies will be able to read the 5-methylcytosine in the native state of DNA without using the bisulfite-treatment strategy. The single molecule real-time (SMRT) technology developed by Pacific Biosciences can already detect 5-methylcytosine directly without bisulfite conversion [73]. These novel NGS platforms also have many other advantages over the current platforms such as less bias during template preparation, possibly longer read length, lower cost, higher speed, and better accuracy. The integration of sequence-capture methods with these new NGS platforms will likely make targeted methylation sequencing more effective. One of the remaining obstacles in DNA methylation analysis is to measure the genome-wide DNA methylation patterns in a small amount of tissues or cells. Due to cellular heterogeneity in DNA methylation patterns, it has become increasingly important to analyze methylation in single cells or tissues isolated by laser capture microdissection. However, most of the TBS-seq approaches discussed in this review require a significant amount of input DNA (more than 100ng of DNA or equivalent of 1.7×105 cells), and therefore cannot be used for single cell analysis. Further works on integrating microfluidics into the single cell methylation analysis may ultimately resolve this difficulty. In summary, although there is still much to improve, the application of various TBS-seq approaches can significantly reduce the sequencing cost, improve the sensitivity and accuracy of methylation detection, and enable population-based methylation studies in cancer research.
Acknowledgement
The author would like to thank Dr. Justin Choi for helpful discussion throughout this work. Huidong Shi is a Georgia Cancer Coalition Distinguished Cancer Scientist. This work was supported in part by National Institute of Health Grants CA134304, DA025779.
Glossary
Abbreviations
- ACBS-seq
array capture bisulfite sequencing
- AML
acute myeloid leukemia
- BSPP
bisulfite padlock probes
- CGI
CpG island
- COBRA
combined bisulfite and restriction analysis
- DMH
differential methylation hybridization
- DREAM
digital restriction enzyme analysis of methylation
- ES
embryonic stem
- HELP
HpaII tiny fragment enrichment by ligation-mediated PCR
- ICF
immunodeficiency, Centromere instability and Facial anomalies
- MALDI-TOF MS
Matrix-assisted laser desorption/ionization-time of flight mass spectrometry
- MBD
methyl-binding domain
- MCA
methylated CpG island amplification
- MDS
myelodysplastic syndromes
- MeDIP-seq
methylated DNA immunoprecipitation sequencing
- MeDIP
methylated DNA immunoprecipitation
- MIRA-seq
methylated CpG island recovery assay sequencing
- MIRA
methylated CpG island recovery assay
- MRE-seq
methylation sensitive restriction enzyme sequencing
- MSP
methylation specific PCR
- NGS
Next-generation sequencing
- PCR
polymerase chain reaction
- SHBS-seq
solution hybrid selection bisulfite sequencing
- SHS
solution hybrid selection
- TBS-seq
targeted bisulfite sequencing
- WGBS-seq
whole genome shotgun bisulfite sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 2.Craig JM, Bickmore WA. The distribution of CpG islands in mammalian chromosomes. Nat Genet. 1994;7:376–382. doi: 10.1038/ng0794-376. [DOI] [PubMed] [Google Scholar]
- 3.Winnefeld M, Lyko F. The aging epigenome: DNA methylation from the cradle to the grave. Genome Biol. 2012;13:165. doi: 10.1186/gb-2012-13-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock C, Beerman I, Lien W-H, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA Methylation Dynamics during In-Vivo Differentiation of Blood and Skin Stem Cells. Molecular Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147:S43–S49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 6.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 9.Irier HA, Jin P. Dynamics of DNA Methylation in Aging and Alzheimer's Disease. DNA Cell Biol. 2012 doi: 10.1089/dna.2011.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayaraman S. Epigenetics of autoimmune diabetes. Epigenomics. 2011;3:639–648. doi: 10.2217/epi.11.78. [DOI] [PubMed] [Google Scholar]
- 11.Movassagh M, Vujic A, Foo R. Genome-wide DNA methylation in human heart failure. Epigenomics. 2011;3:103–109. doi: 10.2217/epi.10.70. [DOI] [PubMed] [Google Scholar]
- 12.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 13.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 16.Shi H, Wang MX, Caldwell CW. CpG islands: their potential as biomarkers for cancer. Expert Rev Mol Diagn. 2007;7:519–531. doi: 10.1586/14737159.7.5.519. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Lay F, Han H, Jones PA. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol Sci. 2010;31:536–546. doi: 10.1016/j.tips.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Issa J-PJ. DNA Methylation as a Therapeutic Target in Cancer. Clinical Cancer Research. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 19.Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum Mol Genet. 2006;15:R95–R101. doi: 10.1093/hmg/ddl095. [DOI] [PubMed] [Google Scholar]
- 20.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 21.Lister R, Ecker JR. Finding the fifth base: Genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- 23.Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- 24.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, Hatchwell E, Selzer RR, Richmond TA, Green RD, Melnick A, Greally JM. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Estecio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, Issa JP. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 27.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-Assisted Microarray Analysis, a New Technology for the Determination of DNA Methylation Patterns, Identifies Frequent Methylation of Homeodomain-Containing Genes in Lung Cancer Cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki M, Jing Q, Lia D, Pascual M, McLellan A, Greally J. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol. 2010;11:R36. doi: 10.1186/gb-2010-11-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa J-PJ, Godley LA, Li W, Goodell MA. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O'Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi JH, Li Y, Guo J, Pei L, Rauch TA, Kramer RS, Macmil SL, Wiley GB, Bennett LB, Schnabel JL, Taylor KH, Kim S, Xu D, Sreekumar A, Pfeifer GP, Roe BA, Caldwell CW, Bhalla KN, Shi H. Genome-wide DNA methylation maps in follicular lymphoma cells determined by methylation-enriched bisulfite sequencing. PLoS One. 2010;5:e13020. doi: 10.1371/journal.pone.0013020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 33.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, Wei CL. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zheng H, Yu J, Wu H, Sun J, Zhang H, Chen Q, Luo R, Chen M, He Y, Jin X, Zhang Q, Yu C, Zhou G, Huang Y, Cao H, Zhou X, Guo S, Hu X, Li X, Kristiansen K, Bolund L, Xu J, Wang W, Yang H, Wang J, Li R, Beck S, Zhang X. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000533. e1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodges E, Molaro A, Dos-Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, Smith AD, Hannon GJ. Directional DNA Methylation Changes and Complex Intermediate States Accompany Lineage Specificity in the Adult Hematopoietic Compartment. Molecular Cell. 2011;44:17–28. doi: 10.1016/j.molcel.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, Van Den Berg D, Laird PW. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, Camargo AA, Stevenson BJ, Ecker JR, Bafna V, Strausberg RL, Simpson AJ, Ren B. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, Briem E, Zhang K, Irizarry RA, Feinberg AP. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, Gao Y, Church GM, Shendure J. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 42.Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 43.Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, Weinstock GM, Gibbs RA. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 44.Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 45.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, Gao Y, Church GM, Shendure J. Multiplex amplification of large sets of human exons. Nat Meth. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 46.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, Daley GQ, Eggan K, Hochedlinger K, Thomson J, Wang W, Gao Y, Zhang K. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ball MP, Li JB, Gao Y, Lee J-H, LeProust EM, Park I-H, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotech. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diep D, Plongthongkum N, Gore A, Fung HL, Shoemaker R, Zhang K. Library-free methylation sequencing with bisulfite padlock probes. Nat Methods. 2012;9:270–272. doi: 10.1038/nmeth.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee EJ, Pei L, Srivastava G, Joshi T, Kushwaha G, Choi JH, Robertson KD, Wang X, Colbourne JK, Zhang L, Schroth GP, Xu D, Zhang K, Shi H. Targeted bisulfite sequencing by solution hybrid selection and massively parallel sequencing. Nucleic Acids Res. 2011;39:e127. doi: 10.1093/nar/gkr598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Jiang H, Ji G, Gao F, Wu M, Sun J, Luo H, Wu J, Wu R, Zhang X. High resolution profiling of human exon methylation by liquid hybridization capture-based bisulfite sequencing. BMC Genomics. 2011;12:597. doi: 10.1186/1471-2164-12-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrow J, Denoeud F, Frankish A, Reymond A, Chen C-K, Chrast J, Lagarde J, Gilbert J, Storey R, Swarbreck D, Rossier C, Ucla C, Hubbard T, Antonarakis S, Guigo R. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7:S4. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nautiyal S, Carlton VE, Lu Y, Ireland JS, Flaucher D, Moorhead M, Gray JW, Spellman P, Mindrinos M, Berg P, Faham M. High-throughput method for analyzing methylation of CpGs in targeted genomic regions. Proc Natl Acad Sci U S A. 2010;107:12587–12592. doi: 10.1073/pnas.1005173107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, Weinstock GM, Gibbs RA. Direct selection of human genomic loci by microarray hybridization. Nat Meth. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 55.Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Meth. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 56.Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 57.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, McCombie WR, Wigler M, Hannon GJ, Hicks JB. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor KH, Kramer RS, Davis JW, Guo J, Duff DJ, Xu D, Caldwell CW, Shi H. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 59.Varley KE, Mitra RD. Bisulfite Patch PCR enables multiplexed sequencing of promoter methylation across cancer samples. Genome Res. 2010;20:1279–1287. doi: 10.1101/gr.101212.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH, Kotsopoulos SK, Samuels ML, Hutchison JB, Larson JW, Topol EJ, Weiner MP, Harismendy O, Olson J, Link DR, Frazer KA. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotech. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Komori HK, LaMere SA, Torkamani A, Hart GT, Kotsopoulos S, Warner J, Samuels ML, Olson J, Head SR, Ordoukhanian P, Lee PL, Link DR, Salomon DR. Application of microdroplet PCR for large-scale targeted bisulfite sequencing. Genome Res. 2011;21:1738–1745. doi: 10.1101/gr.116863.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srivastava GP, Guo J, Shi H, Xu D. PRIMEGENS-v2: genome-wide primer design for analyzing DNA methylation patterns of CpG islands. Bioinformatics. 2008;24:1837–1842. doi: 10.1093/bioinformatics/btn320. [DOI] [PubMed] [Google Scholar]
- 63.Krueger F, Kreck B, Franke A, Andrews SR. DNA methylome analysis using short bisulfite sequencing data. Nat Methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- 64.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harris EY, Ponts N, Levchuk A, Roch KL, Lonardi S. BRAT: bisulfite-treated reads analysis tool. Bioinformatics. 2010;26:572–573. doi: 10.1093/bioinformatics/btp706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith AD, Chung WY, Hodges E, Kendall J, Hannon G, Hicks J, Xuan Z, Zhang MQ. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25:2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pedersen B, Hsieh TF, Ibarra C, Fischer RL. MethylCoder: software pipeline for bisulfite-treated sequences. Bioinformatics. 2011;27:2435–2436. doi: 10.1093/bioinformatics/btr394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kreck B, Marnellos G, Richter J, Krueger F, Siebert R, Franke A. B-SOLANA: an approach for the analysis of two-base encoding bisulfite sequencing data. Bioinformatics. 2012;28:428–429. doi: 10.1093/bioinformatics/btr660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 72.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]