Abstract
Elucidating the regulation of glucose-stimulated insulin secretion (GSIS) in pancreatic islet β cells is important for understanding and treating diabetes. MIN6 cells, a transformed β-cell line derived from a mouse insulinoma, retain GSIS and are a popular in vitro model for insulin secretion. However, in long-term culture, MIN6 cells' GSIS capacity is lost. We previously isolated a subclone, MIN6 clone 4, from the parental MIN6 cells, that shows well-regulated insulin secretion in response to glucose, glybenclamide, and KCl, even after prolonged culture. To investigate the molecular mechanisms responsible for preserving GSIS in this subclone, we compared four groups of MIN6 cells: Pr-LP (parental MIN6, low passage number), Pr-HP (parental MIN6, high passage number), C4-LP (MIN6 clone 4, low passage number), and C4-HP (MIN6 clone 4, high passage number). Based on their capacity for GSIS, we designated the Pr-LP, C4-LP, and C4-HP cells as “responder cells.” In a DNA microarray analysis, we identified a group of genes with high expression in responder cells (“responder genes”), but extremely low expression in the Pr-HP cells. Another group of genes (“non-responder genes”) was expressed at high levels in the Pr-HP cells, but at extremely low levels in the responder cells. Some of the responder genes were involved in secretory machinery or glucose metabolism, including Chrebp, Scgn, and Syt7. Among the non-responder genes were Car2, Maf, and Gcg, which are not normally expressed in islet β cells. Interestingly, we found a disproportionate number of known imprinted genes among the responder genes. Our findings suggest that the global expression profiling of GSIS-competent and GSIS-incompetent MIN6 cells will help delineate the gene regulatory networks for insulin secretion.
Introduction
MIN6 cells, which were isolated from an insulinoma of a transgenic mouse expressing the SV40 T antigen in pancreatic islet β cells, retain some features of differentiated pancreatic β cells [1]. However, long-term culture of MIN6 cells results in the loss of their insulin secretory capacity in response to glucose [2]–[4]. Several reports have described the isolation of glucose-responsive and -unresponsive MIN6 cell subclones, and these subclones have been used to identify genes associated with glucose-stimulated insulin secretion (GSIS) [5], [6]. However, these reports did not rule out the possibility that this differential gene expression was attributable simply to clonal variation. In addition, further analyses of the phenotypic stability of the glucose-responsive MIN6 subclones during long-term culture have not been performed.
Gene expression profile analysis has been performed to compare MIN6 cells after short-term and long-term culture [4]. In addition, comparative proteomic analyses of the cell lysates [7] and the conditioned media [8] were used to investigate the relative changes in the proteome that accompany the loss of GSIS over time in culture. Recently, impairment of glucose and lipid oxidation was suggested to be involved in the loss of GSIS in high passage MIN6 cells [9]. However, these studies did not verify that the differentially expressed genes and proteins were directly involved in the mechanisms for GSIS.
In the present study, we used a MIN6 subclone, designated MIN6 clone 4, which retains GSIS even after long-term culture. To identify the genes involved in the maintenance of GSIS capacity, we compared the gene expression profiles among parental MIN6 cells after short-term culture (Pr-LP) and long-term culture (Pr-HP) and of MIN6 clone 4 cells after short-term culture (C4-LP) and long-term culture (C4-HP). The results revealed one group of genes whose expression was high in well-regulated (i.e., low passage number) parental MIN6 cells and MIN6 clone 4 cells, but extremely low in the dysregulated (high passage number) parental MIN6 cells. Another group of genes was expressed at extremely low levels in the well-regulated MIN6 cells, but at high levels in the dysregulated ones. We discuss the roles of these differentially expressed genes in insulin secretion.
Materials and Methods
Culture of MIN6 parental cells and subclone
MIN6 cells, which we have previously established [1], were maintained in Dulbecco's modified Eagle's medium containing 25 mM glucose, 13% heat-inactivated fetal bovine serum, 0.1 mM 2-mercaptoethanol, 100 units/ml penicillin, and 0.05 mg/ml streptomycin in humidified 5% CO2 at 37°C [1]. MIN6 clone 4 cells are a subclone isolated from low-passage-number parental MIN6 cells by the limiting dilution method. This subclone was maintained in the same culture conditions as the parental cells, and retained good GSIS even after 6 months of continuous culture. For the low-passage-number parental MIN6 cells (Pr-LP), we used cells passaged 17–20 times; for the high-passage-number MIN6 cells (Pr-HP), we used cells passaged 35–40 times. Seventeen to 20 passages were also used for the low-passage-number MIN6 clone 4 cells (C4-LP), and the high-passage ones (C4-HP) were used after 40 to 50 passages. For all of the following methods sections, the parental MIN6 cells and their subclone were treated identically.
Measurement of insulin secretion and insulin content
MIN6 cells were cultured in 24-well plates for 4 days. Prior to the insulin secretion assay, the cells were starved in Krebs Ringer solution containing 0.1% bovine serum albumin (BSA) with 3 mM glucose for 30 min, and the wells were washed twice with the same buffer. The cells were then incubated in Krebs Ringer Solution with 3, 8, 15, or 25 mM glucose, 100 µM glybenclamide+3 mM glucose, or 30 mM KCl+3 mM glucose for 1 hr.
Insulin secreted into the medium and contained in the MIN6 cells was measured. For the secreted insulin, medium was collected and the insulin measured using an ELISA kit (Cat.#10-1250-01; Mercodia, Uppsala, Sweden). To normalize the amount of secreted insulin to the protein content of each well, the cells in each well were lysed with RIPA buffer, and the protein concentration of the cell lysates was measured by the Bradford method (Cat.#500-0006; Bio-Rad, Hercules, CA). To measure the insulin content of MIN6 cells, we lysed cells in a different set of wells with acid-ethanol, and centrifuged the cell extracts. The amount of insulin in the supernatants was assayed using the ELISA kit. Statistical analysis was performed by Student′s t-test.
DNA microarray analyses
Total RNA was extracted from MIN6 cells using Trizol reagent (Invitrogen, Carlsbad, CA) and subjected to double-strand cDNA synthesis using the Superscript Choice system (Cat.#18090-019; Invitrogen) and the T7-(dT)24 reverse transcription primer (Cat.#72-1591-01; Amersham Biosciences, Piscataway, NJ). Synthesis of biotin-labeled cRNA was carried out by in-vitro transcription using the Bio Array RNA Transcript Labeling Kit (Cat.#900182; Affymetrix, Santa Clara, CA) according to the manufacturer's instructions. The biotin-labeled cRNA was purified using RNeasy spin columns (Cat.#74106; Qiagen GmbH, Hilden, Germany) and fragmented in a reaction mixture. The biotin-labeled and fragmented cRNA was hybridized to the murine genome U74 version 2 GeneChip array (Affymetrix), incubated, and washed according to the manufacturer's instructions. The GeneChip arrays were then scanned with a Gene Array Scanner (Hewlett-Packard, Santa Clara, CA) and analyzed by GeneChip 5.1 software (Affymetrix). The microarray dataset has been deposited in NCBI's Gene Expression Omnibus and is accessible through GEO Series accession number GSE43774.
Quantitative RT-PCR
Total RNA was extracted from MIN6 cells by the acid guanidinium-phenol-chloroform (AGPC) method and subjected to cDNA synthesis using ReverTra Ace α (Cat.#FSK-101; Toyobo, Tokyo, Japan). Quantitative RT-PCR analysis was carried out using SYBR Premix Ex Taq (Cat.#RR041A; Takara, Otsu, Japan). The reaction was performed with 1 µl cDNA per 25 µl reaction in a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA) under the following thermal cycling conditions: 95°C for 10 sec followed by 40 cycles at 95°C for 5 sec and 60°C for 31 sec. The relative expression levels of the target genes were normalized to that of Rpl32. Statistical analysis was performed by Student′s t-test. Primer sequences are listed in Table S1.
Immunocytochemistry and FACS (fluorescence-activated cell sorting) analysis
For immunocytochemistry, the MIN6 cells were washed with PBS and fixed in 4% paraformaldehyde for 10 min, rinsed with PBS, incubated for 5 min in PBS with 1% Triton X-100, washed again with PBS, and incubated in blocking reagent (Blocking One, Cat.#03953-95; Nacalai Tesque, Kyoto, Japan). The samples were incubated with rat anti-CD24 antibody (Cat.#14-0241-81; eBioscience, San Diego, CA) for 1 hr at room temperature, washed with PBS, and then incubated with Alexa Fluor 488-conjugated anti-rat IgG (Cat.#A-11006; Molecular Probes, Eugene, OR) for 1 hr at room temperature. For FACS analysis, the cells were washed twice with PBS and then suspended in SM buffer (HEPES-buffered saline with 0.1% sodium azide and 1% BSA). CD24 staining was performed at 4°C for 30 min with rat anti-CD24 antibody, followed by incubation with Alexa Fluor 488-conjugated anti-rat IgG. FACS analyses were performed with a FACScan (Becton Dickinson, Franklin Lakes, NJ) using the CellQuest (Becton Dickinson) evaluation program.
In situ hybridization
Part of the mouse Hepacam2 cDNA (NM_178899; sequence position 58–699) was subcloned into the pGEMT-Easy vector (Cat.#A1360; Promega, Madison, WI) and used to generate sense and antisense RNA probes. Digoxigenin (DIG)-labeled RNA probes were prepared with the DIG RNA Labeling Mix (Cat.#1277073; Roche, Mannheim, Germany) according to the manufacturer's instructions. In situ hybridization was performed according to the protocol of Genostaff (Tokyo, Japan). In brief, a pancreas of a C57BL/6J mouse was dissected out after perfusion and fixation with Tissue Fixative (Genostaff, Tokyo, Japan), embedded in paraffin, and sectioned at 6 µm. The tissue sections were de-waxed and fixed with 4% paraformaldehyde in PBS. The sections were then hybridized with sense and antisense Hepacam2 probes. After treatment with blocking reagent in TTBS (10 mM Tris-HCl, pH 7.6, 137 mM NaCl, and 0.1% Tween 20) for 30 min, the sections were incubated with anti-DIG alkaline phosphatase (AP) conjugate (Cat.#1093274; Roche) diluted 1∶1000 with TTBS for 2 hr at room temperature, washed twice with TTBS, and then incubated in 100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 50 mM MgCl2, and 0.1% Tween 20 for 30 min. Coloring reactions were performed with NBT/BCIP solution (Cat.#B6404; Sigma, St. Louis, MO) overnight, and the sections were then washed with PBS and counterstained with Kernechtrot stain solution (Cat.#40872; Mutoh Chemical Co., Tokyo, Japan).
Bisulfite sequencing
Bisulfite treatment of the genomic DNA isolated from Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells was performed using the EpiTect Bisulfite Kit (Cat.#59104; Qiagen) according to the manufacturer's instructions. The CpG islands in the first intron of the Plagl1 gene (33 CpGs) and those in the DMR region of the Dlk1 gene (24 CpGs) were chosen for analysis. The primers for the Plagl1 gene were: forward, 5′-GGGTAGGTAAGTAGTGATAA-3′; reverse, 5′-CCTAAAACACCAAAATAACA-3′ [10]. The primers for the Dlk1 gene were: forward, 5′-GATTAGTGATTTATAATTTGTGTTTTGGTT-3′; reverse, 5′-AAACTCACCTAAATATACTAAAAACAAATA-3′ [11]. The PCR products were cloned into pBluescript and sequenced.
Statistical analysis
Results are presented as the mean ± SD. Statistical analyses were carried out by Student's t-test except for that of the distribution of the percentages of methylated CpGs in the Plagl1 locus, which was carried out by the F-test. A value of P<0.05 was considered statistically significant.
Results
Insulin secretion and insulin content
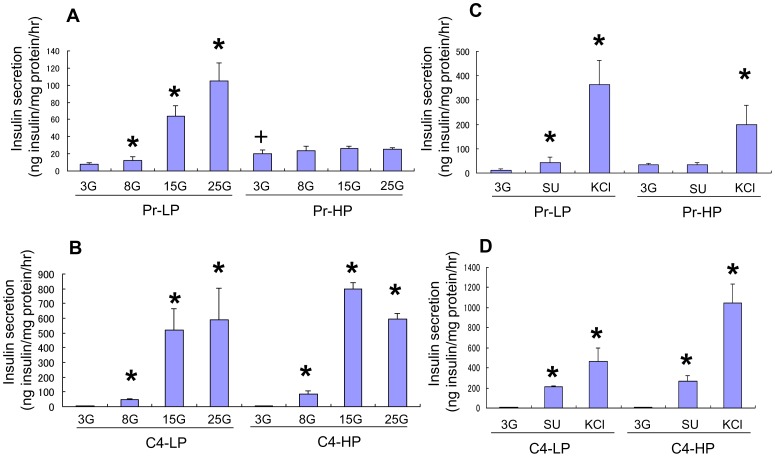
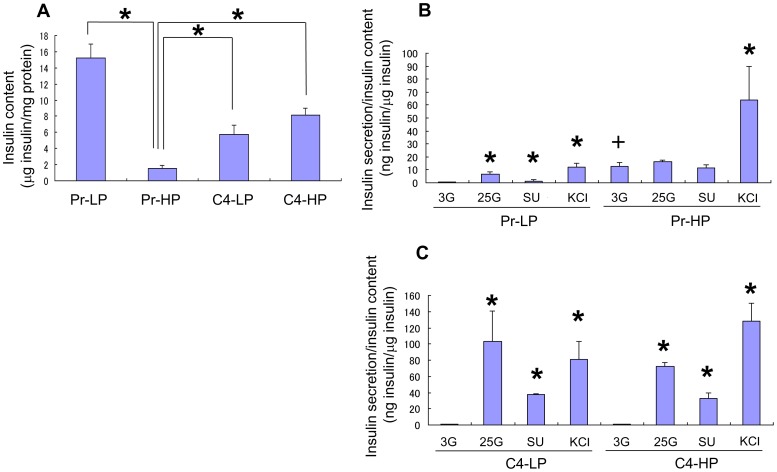
Insulin secretory capacity was compared by static incubation among parental MIN6 cells at 17–20 passages (Pr-LP) and 35–40 passages (Pr-HP) and MIN6 clone 4 cells at 17–20 passages (C4-LP) and 40–50 passages (C4-HP) ( Figure 1 and 2 ). Pr-HP cells showed a higher basal insulin secretion at 3 mM glucose than did the other groups, but the insulin secretion did not increase with higher glucose concentrations or the addition of glybenclamide (SU) or KCl, in contrast with the other three groups ( Figure 1 , 2B, and 2C ). In addition, the insulin content of the Pr-HP cells was much lower than that of the Pr-LP, C4-LP, or C4-HP cells ( Figure 2A ). Therefore, we designated the C4-LP, C4-HP, and Pr-LP cells as “responder” cells and the Pr-HP cells as “non-responder” cells.
Figure 1. Insulin secretion from MIN6 cells.
Insulin secretion from Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells stimulated with 3 mM (3G), 8 mM (8G), 15 mM (15G), or 25 mM (25G) glucose (A, B), 100 nM glybenclamide (SU), or 30 mM KCl (C, D). Pr-HP cells showed higher basal insulin secretion at 3 mM glucose compared with Pr-LP cells, but their insulin secretion did not increase further at higher glucose concentrations or with the addition of glybenclamide or KCl, whereas both C4-LP and C4-HP MIN6 cells showed a better insulin secretory response to glucose and glybenclamide than Pr-LP cells. Values are means ± SD and n = 5–6. *P<0.05 v.s. insulin secretion at 3 mM glucose.+P<0.05 v.s. insulin secretion of Pr-LP, C4-LP, and C4-HP cells at 3 mM glucose by Student's t-test.
Figure 2. Insulin content and secretion of MIN6 cells.
Insulin content of Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells (A). The insulin content of Pr-HP cells was lower than that of Pr-LP, C4-LP, or C4-HP cells. Values are means ± SD and n = 5–6. *P<0.05. Insulin secretion/insulin content from Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells stimulated with 3 mM (3G), 25 mM (25G) glucose, 100 nM glybenclamide (SU), or 30 mM KCl (B, C). Values are means ± SD and n = 5–6. *P<0.05 v.s. insulin secretion at 3 mM glucose.+P<0.05 v.s. insulin secretion of Pr-LP, C4-LP, and C4-HP cells at 3 mM glucose by Student's t-test.
Furthermore, both C4-LP and C4-HP cells showed a better insulin secretory response to glucose and glybenclamide than did the Pr-LP cells ( Figure 1 ), suggesting that the MIN6 clone 4 cells maintained stable regulatory mechanisms for insulin secretion. Interestingly, the insulin secretion induced with KCl was even higher in the C4-HP cells than in the C4-LP cells ( Figure 1D and 2C ).
DNA Microarray analysis
To identify genes involved in the regulation of the insulin secretory pathway, a comparative DNA microarray analysis was performed, using the Pr-LP, Pr-HP, C4-LP, and C4-HP cells. The results revealed one group of genes, which we call “responder genes,” that were highly expressed in the responder cells (Pr-LP, C4-LP, and C4-HP), but only weakly in the non-responder cells (Pr-HP). A different group of genes (“non-responder genes”) was highly expressed in the Pr-HP cells, but only weakly in the Pr-LP, C4-LP, and C4-HP cells. To compare the responder and non-responder results, we compared the mean value of the gene expression levels in the Pr-LP, C4-LP, and C4-HP cells with the expression level in the Pr-HP cells. Genes that showed a more than 5-fold difference in expression level are listed in Table 1 (40 responder genes) and Table 2 (41 non-responder genes). An extended list, including all the genes that showed a more than 3-fold difference, is provided in Table S2 (60 responder and 62 non-responder genes). Genes that were differentially expressed among Pr-LP, C4-LP, and C4-HP cells are also listed in Table S3, S4, S5.
Table 1. Genes preferentially expressed in responder MIN6 cells.
| Gene name | Mean value for Pr-LP, C4-LP, and C4-HP* | Pr-HP* | Fold change |
| Tmem59l | 3306.8 | 3.8 | 876.5 |
| Mlxipl | 750.0 | <1.0 | >750.0 |
| Scgn | 2686.6 | 10.3 | 261.6 |
| Tmed6 | 107.7 | <1.0 | >107.7 |
| Plagl1 | 4616.1 | 114.0 | 40.5 |
| Hepacam2 | 2767.3 | 73.5 | 37.6 |
| Dlk1 | 11245.0 | 323.7 | 34.7 |
| Rps6kb1 | 119.0 | 5.4 | 22.2 |
| Syt7 | 1327.9 | 65.9 | 20.1 |
| Slc29a4 | 1531.1 | 78.9 | 19.4 |
| Cd24a | 4806.0 | 282.5 | 17.0 |
| Rpgr | 119.3 | 7.3 | 16.4 |
| Meg3 | 2463.4 | 154.3 | 16.0 |
| Glul | 798.1 | 53.2 | 15.0 |
| 1700019D03Rik | 210.7 | 15.2 | 13.9 |
| Gm2115 | 607.8 | 44.8 | 13.6 |
| Nnat | 7113.3 | 637.7 | 11.2 |
| Ndn | 966.5 | 94.6 | 10.2 |
| Ly6e | 2512.2 | 250.9 | 10.0 |
| Gucy2c | 2118.9 | 217.6 | 9.7 |
| 3830403N18Rik | 345.9 | 38.0 | 9.1 |
| Kctd12 | 665.0 | 73.4 | 9.1 |
| Cd200 | 508.3 | 61.7 | 8.2 |
| Th | 3705.1 | 477.8 | 7.8 |
| Amdhd2 | 252.8 | 34.9 | 7.2 |
| Blnk | 2957.6 | 414.8 | 7.1 |
| Celsr2 | 221.2 | 31.6 | 7.0 |
| Cplx2 | 852.7 | 122.2 | 7.0 |
| Tmod2 | 530.2 | 77.1 | 6.9 |
| Cdhr1 | 1980.3 | 288.5 | 6.9 |
| Ppp1r3d | 495.8 | 73.8 | 6.7 |
| Pparg | 140.7 | 21.4 | 6.6 |
| Alcam | 278.1 | 42.5 | 6.6 |
| Unc80 | 579.0 | 93.3 | 6.2 |
| Epb4.1l4b | 1667.5 | 270.6 | 6.2 |
| Akr1c14 | 559.7 | 91.6 | 6.1 |
| A830039N20Rik | 285.8 | 48.3 | 5.9 |
| Mirg | 240.4 | 41.6 | 5.8 |
| Fgf12 | 414.6 | 73.3 | 5.7 |
| Slc44a1 | 218.2 | 39.1 | 5.6 |
Raw values of expression intensities measured by Affymetrix arrays.
Table 2. Genes preferentially expressed in non-responder MIN6 cells.
| Gene name | Pr-HP* | Mean value for Pr-LP, C4-LP, and C4-HP* | Fold change |
| Cd68 | 2507.1 | 79.8 | 31.4 |
| Car2 | 2841.2 | 93.0 | 30.6 |
| Stmn2 | 1994.6 | 65.5 | 30.5 |
| Maf | 536.5 | 22.0 | 24.4 |
| Il13ra1 | 237.9 | 11.1 | 21.4 |
| Perp | 998.0 | 52.7 | 19.0 |
| Sh3bgrl2 | 512.9 | 30.6 | 16.8 |
| Fgfr2 | 1035.3 | 66.6 | 15.6 |
| Pqlc3 | 2170.1 | 171.1 | 12.7 |
| Prr5l | 969.2 | 81.1 | 12.0 |
| Il4 | 217.4 | 19.9 | 10.9 |
| Ptgs2 | 195.8 | 18.2 | 10.7 |
| Scel | 316.3 | 30.5 | 10.4 |
| Lpl | 1474.6 | 160.6 | 9.2 |
| Rbfox1 | 491.0 | 54.5 | 9.0 |
| Lcp2 | 710.5 | 86.2 | 8.2 |
| Tbcel | 397.9 | 50.7 | 7.9 |
| Glyat | 204.5 | 26.9 | 7.6 |
| Tmcc3 | 879.6 | 117.5 | 7.5 |
| Basp1 | 153.9 | 21.5 | 7.2 |
| Pcdh7 | 357.1 | 52.0 | 6.9 |
| Ppap2a | 1835.0 | 271.5 | 6.8 |
| Edil3 | 1365.9 | 207.3 | 6.6 |
| S100a10 | 4357.1 | 661.4 | 6.6 |
| Sox11 | 1180.7 | 188.5 | 6.3 |
| Rpp25 | 346.5 | 55.8 | 6.2 |
| Enpep | 757.0 | 123.7 | 6.1 |
| Bach2 | 216.5 | 35.9 | 6.0 |
| 1810011O10Rik | 721.4 | 120.3 | 6.0 |
| Gcg | 8993.3 | 1553.4 | 5.8 |
| Eif2ak4 | 634.5 | 110.2 | 5.8 |
| Sash1 | 566.6 | 99.7 | 5.7 |
| Hspbp1 | 603.6 | 106.8 | 5.7 |
| Oat | 1753.3 | 327.6 | 5.4 |
| Ezr | 1299.8 | 243.9 | 5.3 |
| Gnai1 | 1080.2 | 202.5 | 5.3 |
| Gnptab | 1234.2 | 231.4 | 5.3 |
| Fam132a | 984.7 | 185.9 | 5.3 |
| Zfp185 | 669.7 | 126.7 | 5.3 |
| Ctsb | 199.4 | 39.5 | 5.1 |
| Hlf | 379.4 | 75.8 | 5.0 |
Raw values of expression intensities measured by Affymetrix arrays.
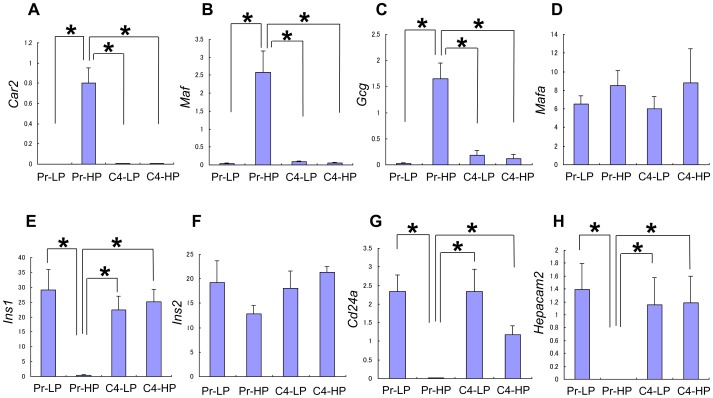
The expression pattern of some responder and non-responder genes was examined by quantitative RT-PCR ( Figure 3 ; Figure S1). The four responder genes chosen, Chrebp, Syt7, Cplx2, and Scgn ( Table 1 ), were highly expressed in the responder cells, but only weakly in Pr-HP cells (Figure S1A-D). These genes are known to be expressed in islet β cells and probably have some roles in the regulation of GSIS in β cells (see Discussion). The three non-responder genes chosen, Car2, Maf, and Gcg ( Table 2 ), were highly expressed in Pr-HP cells, but only weakly in the responder cells ( Figure 3A-C ). These genes are known to be expressed in the pancreas, but not in islet β cells. Thus, the Pr-HP cells can express genes that are not normally expressed in islet β cells.
Figure 3. Quantitative RT-PCR analysis.
Expression of the Car2 (A), Maf (B), Gcg (C), Mafa (D), Ins1 (E), Ins2 (F), Cd24a (G), and Hepacam2 (H) genes in Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells was examined by quantitative RT-PCR. Car2, Maf, and Gcg showed a non-responder gene expression pattern that was consistent with the DNA microarray analysis. Ins2 and Mafa did not show significant differences in expression among the four groups of MIN6 cells, whereas Ins1 was revealed to be a responder gene. Cd24a and Hepacam2 showed a responder gene expression pattern that was consistent with the DNA microarray analysis. Values are means ± SD and n = 4–5. *P<0.05.
Genes encoding β-cell-related transcription factors are listed in Table 3 . Pdx1, Nkx2-2, Nkx6-2, Foxa2, Pax4, and Pax6 were not differentially expressed among the four groups of MIN6 cells, but Neurod1 showed a 1.5-fold higher expression in the responder cells than in Pr-HP cells. Because the Mafa gene was not included in the U74 version 2 GeneChip array used in our analysis, we performed a quantitative RT-PCR analysis, which showed that Mafa gene expression did not differ significantly among the four MIN6 cell groups ( Figure 3D ).
Table 3. Expression levels of pancreas-related genes.
| Gene Name | Pr-LP* | Pr-HP* | C4-LP* | C4-HP* | Mean** | Fold change*** |
| Pdx1 | 701.4 | 519.5 | 591.7 | 570.0 | 621.0 | 0.84 |
| Nkx2-2 | 2408.6 | 1395.5 | 1612.7 | 1501.1 | 1840.8 | 0.76 |
| Nkx6-2 | 252.6 | 179.5 | 141.7 | 395.3 | 263.2 | 0.68 |
| Pax4 | 606.7 | 507.0 | 487.6 | 581.9 | 558.8 | 0.91 |
| Pax6 | 942.0 | 599.2 | 621.2 | 518.4 | 693.8 | 0.86 |
| Isl1 | 936.3 | 516.3 | 850.3 | 715.7 | 834.1 | 0.62 |
| Foxa2 | 1896.9 | 1349.5 | 1304.2 | 1318.7 | 1506.6 | 0.90 |
| Insm1 | 1358.5 | 488.6 | 986.8 | 1078.4 | 1141.3 | 0.43 |
| Neurod1 | 603.8 | 210.9 | 263.8 | 288.1 | 385.2 | 0.55 |
| Neurog3 | 176.9 | 192.3 | 192.0 | 203.6 | 190.8 | 1.01 |
| Pcsk2 | 4580.8 | 5429.3 | 5624.6 | 3924.2 | 4709.9 | 1.15 |
| Slc2a2 | 478.5 | 499.2 | 463.8 | 1754.5 | 898.9 | 0.56 |
| Gck | 803.2 | 550.6 | 788.4 | 716.6 | 769.4 | 0.72 |
| Abcc8 | 154.7 | 160.3 | 168.0 | 174.6 | 165.8 | 0.97 |
| Kcnj11 | 815.8 | 644.0 | 1171.2 | 933.1 | 973.4 | 0.66 |
| Ins1 | 28131.6 | 16852.5 | 27736.0 | 27214.7 | 27694.1 | 0.61 |
| Ins1 | 19939.3 | 8937.0 | 18275.3 | 20041.0 | 19418.5 | 0.46 |
| Ins2 | 14771.3 | 15525.9 | 15461.2 | 14919. 7 | 15050.7 | 1.03 |
| Sst | 206.1 | 206.4 | 211.3 | 191.4 | 203.0 | 1.02 |
| Ppy | 810.5 | 559.8 | 524.1 | 548.7 | 627.8 | 0.89 |
| Iapp | 8291.0 | 7443.6 | 9641.2 | 8110.5 | 8680.9 | 0.86 |
Raw values of expression intensities measured by Affymetrix arrays. **Mean value of C4-LP, C4-HP, and Pr-LP. ***Ratio of Pr-HP to mean value.
There was no significant difference in the expression of the insulin genes (Ins1 and Ins2) as assessed by the DNA microarray analysis. However, because the expression levels of the Ins1 and Ins2 genes were dramatically higher than those of the other genes, their signal intensities might have exceeded the linear range of detection. Therefore, we analyzed the expression of the Ins1 and Ins2 genes by quantitative RT-PCR. We found that the level of Ins2 gene expression was not significantly different among the four groups, but that of Ins1 was much lower in the Pr-HP cells than in the responder cells ( Figure 3E and 3F ). Thus, the Ins1 gene can be considered a responder gene.
Expression of a cell-surface protein, CD24
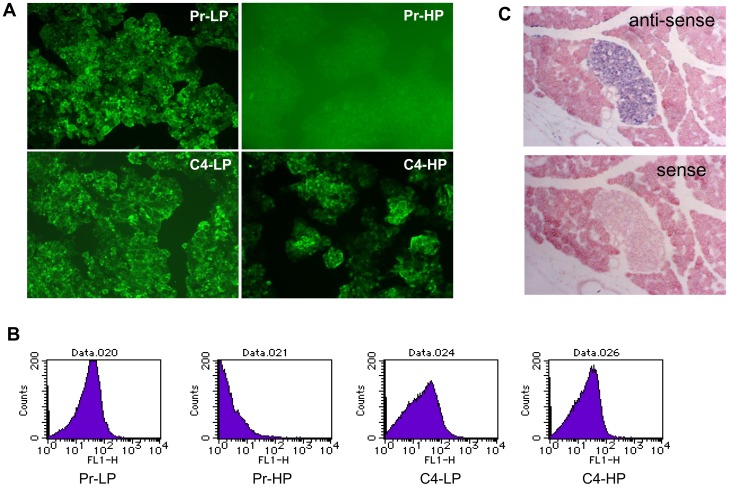
The DNA microarray data showed that Cd24a was expressed in the Pr-LP, C4-LP, and C4-HP cells, but not in the Pr-HP cells ( Figure 3G ). In addition, immunocytochemical analysis detected CD24 on the surface of Pr-LP, C4-LP, and C4-HP cells, but not on Pr-HP cells ( Figure 4A ). Flow-cytometric analysis also showed that CD24 was expressed similarly in Pr-LP, C4-LP, and C4-HP cells, but its expression was very low or lost in Pr-HP cells ( Figure 4B ). Interestingly, the Cd24a gene is expressed in the embryonic endocrine pancreas (E14.5), as shown by in situ hybridization (see http://www.genepaint.org/Frameset.html).
Figure 4. CD24 expression in MIN6 cells and analysis of Hepacam2 expression by in situ hybridization.
Immunocytochemical analysis (A) and flow cytometric analysis (B) of Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells with an anti-CD24 antibody. CD24 was not detected on the surface of Pr-HP cells. In situ hybridization analysis of Hepacam2 expression in pancreatic islets (C). Pancreatic sections were hybridized with DIG-labeled anti-sense and sense RNA probes for Hepacam2 transcripts (see Materials and Methods).
Analysis of the Hepacam2 gene
Hepacam2 was highly expressed in the responder MIN6 cells, but not in the non-responder ones ( Figure 3H ). It encodes a putative membrane-anchored protein. In situ hybridization analysis showed that Hepacam2 expression was restricted to the islets in the pancreas ( Figure 4C ). Hepacam2 is also expressed in the embryonic pancreas (E14.5) [12].
Methylated genes
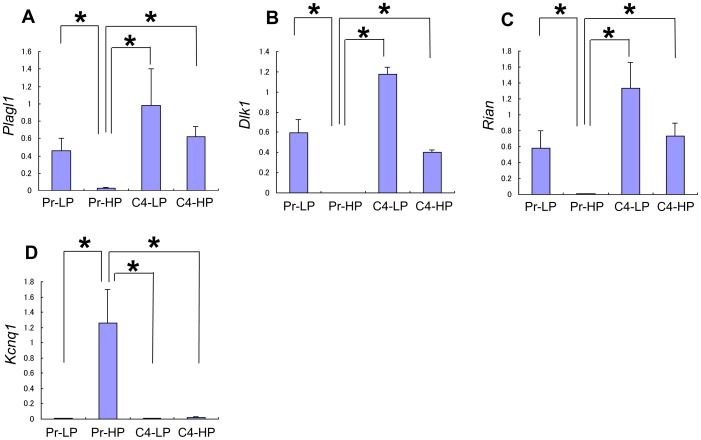
Interestingly, we found several imprinted genes among the differentially expressed genes. The responder genes included Plagl1 ( Figure 5A ), Dlk1 ( Figure 5B ), Meg3, Nnat, Ndn, Mirg, Peg3, and Th. Cdkn1c was a non-responder gene. Because only about 150 verified imprinted genes are known (http://igc.otago.ac.nz/1101Summary-table.pdf), the percentage of imprinted genes among the differentially expressed genes in our analysis seemed very high, especially for the responder genes. Therefore, we further investigated the expression of imprinted genes. Dlk1, Meg3, and Mirg are located in the well-known Dlk1-Gtl1 imprinted gene cluster [13], [14]. An analysis of the expression patterns of other imprinted genes in the Dlk1-Gtl1 locus indicated that Rian ( Figure 5C ) and Rtl1 (data not shown) are also responder genes.
Figure 5. Quantitative RT-PCR analysis of imprinted genes.
Expression of imprinted genes, Rian (A), Plagl1 (B), Dlk1 (C), and Kcnq1 (D) in Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells. Plagl1, Dlk1, and Rian were confirmed to be responder genes, whereas Kcnq1 was a non-responder gene. Values are means ± SD and n = 4–5. *P<0.05.
Recent reports have shown that variants in KCNQ1 are associated with susceptibility to human type 2 diabetes mellitus [15], [16]. Interestingly, Kcnq1 was reported to be an imprinted gene [13], [14]. Our DNA microarray analysis showed that the Kcnq1 expression in Pr-HP cells was 2.2-fold higher than in responder cells. Therefore, we analyzed the expression of Kcnq1 by quantitative RT-PCR. As shown in Figure 5D , Kcnq1 was a typical non-responder gene.
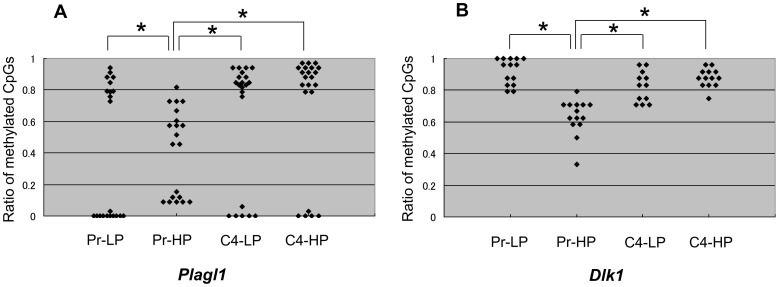
Genomic imprinting is an epigenetic form of gene regulation that involves differential DNA methylation of the paternal and maternal alleles of a gene. Such methylation is inherited in a parent-of-origin-specific manner. We investigated the methylation status of the known regulatory regions of the Plagl1 and Dlk1 genes, which were highly expressed in the responder MIN6 cells, but hardly at all in the non-responder cells ( Figure 5B and 5C ). Bisulfite methylation analysis revealed that in the Plagl1 gene locus, the pattern of methylation of CpG islands was quantitatively different between the responder MIN6 cells (Pr-LP, C4-LP, and C4-HP) and the non-responder cells (Pr-HP) ( Figure 6A ). In the responder cells, one allele was almost completely unmethylated and the other almost completely methylated, whereas in the Pr-HP cells, both alleles appeared to be methylated at random. In contrast, the Dlk1 locus was significantly less methylated in Pr-HP cells than in the responder cells ( Figure 6B ). Thus, the methylation levels of the CpG islands of imprinted genes differed between the responder and non-responder MIN6 cells, but the pattern of methylation differed according to the gene locus.
Figure 6. Analysis of CpG methylation of Plagl1 and Dlk1.
The ratio of methylated CpGs of each allele from Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells, in the Plagl1 gene region (A) and the Dlk1 gene region (B), by bisulfite sequencing. In the Plagl1 gene locus, the pattern of methylation of CpG islands was quantitatively different between the responder MIN6 cells (Pr-LP, C4-LP, and C4-HP) and the non-responder cells (Pr-HP). In responder cells, one allele was either almost completely unmethylated and the other was almost completely methylated, whereas in the Pr-HP cells, both alleles were randomly methylated. In contrast, the Dlk1 locus was significantly less methylated in Pr-HP cells than in the responder MIN6 cells. Values are means ± SD and n = 8-10. *P<0.
Discussion
Although MIN6 cells are widely used as a model of pancreatic β cells, the GSIS is gradually lost with long-term culture. We previously isolated a subclone, MIN6 clone 4, from the parental MIN6 cells. This subclone showed stable GSIS even after long-term culture. To look for candidate genes for the maintenance of GSIS, we decided to analyze the gene expression patterns of Pr-LP, Pr-HP, C4-LP, and C4-HP cells. Our DNA microarray analysis extracted 60 differentially expressed responder genes (with consistently higher expression in Pr-LP, C4-LP, and C4-HP cells) and 62 non-responder genes (higher expression in the Pr-HP cells) (Table S2). Genes differentially expressed among Pr-LP, C4-LP, and C4-HP cells are also listed (Table S3, S4, S5). Among these genes, Tmem59l is also one of the responder genes (Table 1). As shown in Figure S1E, Tmem59l was highly expressed in the responder cells, but not in the non-responder cells. Its expression was higher in C4-LP and C4-HP cells than in Pr-LP cells. Further functional analysis is needed to know the roles of these differentially expressed genes in β cells.
Some of the responder genes have been implicated in the function or development of pancreatic β cells. One responder gene, Chrebp (official name: Mlxipl), encodes a transcription factor called carbohydrate-responsive element binding protein (ChREBP), and its expression increases in response to extracellular glucose in β cells [17]. It is also involved in the differentiation [18] and proliferation of β cells [19]. Chrebp was also reported to be expressed in the embryonic pancreas and to be a target of FOXA1 and FOXA2, which are crucial for the mature β-cell phenotype [20]. Our data suggested that Chrebp is also important for the physiological function of β cells.
Genes known to be involved in exocytosis were also found among the responder genes. Syt7 is a member of the synaptotagmin family, which is crucial for the exocytosis of vesicles [21]. Its protein product, Synaptotagmin VII, participates in the calcium-dependent insulin release from pancreatic β cells [22], [23], and the ablation of Syt7 impairs GSIS [24], [25]. Complexin II, encoded by Cplx2, is a cytoplasmic pre-synaptic protein believed to regulate neurotransmitter release from pre-synaptic terminals [26]. In our results, the Complexin II level appeared to be high in pancreatic β cells. Because Cplx2 is a homologue of Cplx1, which is implicated in insulin secretion [27], [28], Cplx2 may also play a role in insulin secretion. Scgn, which encodes a calcium-binding protein, is highly expressed in the pancreatic islets [29] and is also involved in the regulation of insulin secretion [30].
The Pr-HP MIN6 cells expressed some pancreas-related genes that are not normally expressed in mature β cells. For example, Car2, which encodes carbonic anhydrase 2, is expressed in duct cells and pancreatic α cells [31]. Gcg, encoding preproglucagon, is expressed in pancreatic α cells. cMaf (official name: Maf) encodes a transcription factor that belongs to the large Maf family, and the cMaf mRNA is detected in the embryonic pancreas [32], but it is not expressed in adult pancreatic β cells. cMaf was also reported to be expressed in pancreatic α cells and to have a role in their differentiation and maintenance [33]–[35].
The Ins2 gene was expressed at high levels in all four MIN6 cell groups, whereas the Ins1 gene expression was high in responder cells, but extremely low in Pr-HP cells ( Figure 3E ). Ins1 and Ins2 are regulated differently [36]–[39] and are under the control of different enhancer-promoter regions. The human insulin gene (INS) and the mouse Ins2 gene have two introns, but the mouse Ins1 gene lacks the second intron. Thus, the mouse Ins2 gene appears more like the human insulin gene. Compensatory responses were reported in mice carrying a null mutation for Ins1 or Ins2 [40]. Therefore, the downregulation of Ins1 might not account for the dysregulation of GSIS in Pr-HP cells. In pancreatic β-cell-specific Neurod1 knockout mice, the Ins1 gene expression is almost absent, whereas the Ins2 gene expression is not much affected [41]. Neurod1 expression in Pr-HP cells was approximately half that in the responder cells ( Table 3 ), which may partly explain the low level of Ins1 expression in the Pr-HP cells.
Our DNA microarray analysis identified Cd24a as a responder gene ( Figure 3G ). Immunohistochemical analysis confirmed that CD24 was expressed on the surface of Pr-LP cells and MIN6 clone 4 cells, but was low or absent in Pr-HP cells. Similarly, Cram et al. showed that the Cd24 gene is expressed at higher levels in RIN-A12 insulinoma cells, which produce a larger amount of insulin, than in RIN-5AH insulinoma cells, which are low insulin producers [42]. Our present results may suggest that CD24 is a useful marker for β-cell lines that retain their differentiated phenotypes.
The Hepacam2 gene was identified as a responder gene ( Figure 3H ). Hepacacm2 encodes a putative membrane-anchored protein and was reported to be expressed in pancreatic endocrine cells, including β cells [43]. Our in situ hybridization analysis confirmed this ( Figure 4C ). It should be interesting to examine the possible roles of Hepacam2 in the regulation of GSIS in β cells.
The present study also found a set of imprinted genes among the responder genes. The majority of known imprinted genes in mammals have roles in the control of embryonic growth and development, including development of the placenta [44]. Others are involved in post-natal development, with roles in suckling behavior and metabolism [44]. The imprinted responder genes Plagl1, Dlk1, and Nnat have been implicated in pancreatic β-cell function [45]–[51]. Plagl1, also called Zac1, was first isolated as a candidate gene for transient neonatal diabetes (TNDM) [45], which is a rare inherited diabetic syndrome apparent in the first weeks of human life and again during early adulthood. Plagl1 encodes a proapoptotic zinc finger protein. To investigate its pancreatic function, Ma et al. developed a high-copy transgenic mouse line, TNDM29, carrying the human TNDM locus, including the Plagl1 gene [46]. TNDM29 neonates display hyperglycemia, and older adults have impaired glucose tolerance. Plagl1 overexpression in β-cell lines impairs insulin secretion [47]. Combined with our data, these findings suggest that β-cell function requires the careful regulation of Plagl1 expression levels.
Dlk1 is expressed in the embryonic pancreas and is gradually restricted to β cells during development [48]. Its protein, Dlk1, is secreted from islets and a β-cell line, and Dlk1 overexpression in RINm5F cells attenuates their proliferation [49].
Nnat expression was first discovered in the brain and is regulated by Neurod1; its suppression by siRNA represses the GSIS by β cells [50]. A recent report also revealed that Nnat is involved in the insulin secretory mechanism and in apoptosis [51]. Thus, the low expression of Nnat may partly account for the impairment of insulin secretion in Pr-HP cells.
Two other imprinted genes, Meg3 (Gtl2) and Mirg, were identified as responder genes. These genes are located in the Dlk1-Gtl2 gene cluster. Upon examination, we found that other genes in this locus, Rtl1 and Rian, were also responder genes. The human DLK1-MEG3 (GTL2) gene region was recently reported to affect susceptibility for type 1 diabetes [52]. Thus, this locus may contribute to the regulation of β-cell function.
In contrast to the above imprinted genes, Kcnq1 was expressed at very low levels in the responder cells, but at high levels in the non-responder, Pr-HP cells. Recently Kcnq1 was reported to be a candidate gene for type 2 diabetes susceptibility [13], [14]. Kcnq1-overexpressing MIN6 cells show a defect in insulin secretion in response to various secretagogues [53]. Thus, it is possible that high levels of Kcnq1 expression lead to poor β-cell function.
DNA methylation is necessary for the proper expression of imprinted genes [13], [14] and is believed to represent the primary imprinting mark. Imprinted genes can be divided into two classes based on whether they are activated or repressed by the loss of DNA methylation. To clarify the mechanism for the regulation of imprinted genes in MIN6 cells, we analyzed the methylation status of the known regulatory loci of the Plagl1 and Dlk1 genes. One allele of the Plagl1 gene locus was unmethylated and the other was highly methylated in responder cells, whereas both alleles were randomly methylated at various levels in Pr-HP cells ( Figure 6A ). Plagl1 expression is reported to be downregulated by methylation [54]. Therefore, the random methylation of the Plagl1 locus may account for its reduced expression in Pr-HP cells. On the other hand, both alleles of the Dlk1 locus were highly methylated in responder cells, but were less methylated in non-responder cells ( Figure 6B ). An analysis of Dnmt −/− embryos indicated that Dlk1 is positively regulated by DNA methylation [55]. Therefore, the reduced methylation of the Dlk1 locus may account for the lower Dlk1 gene expression in the Pr-HP cells.
Recently, an epigenetic mechanism was reported to be involved in the dysfunction of pancreatic β cells in vivo, resulting in the development of diabetes mellitus [56]–[60]. The imprinted genes found in the responder and non-responder genes may be important for the maintenance of β-cell functions. Further analysis of the genes identified by the global expression profiling of well-regulated and dysregulated MIN6 cells will help us to clarify the regulatory networks that control insulin secretion.
Supporting Information
Quantitative RT-PCR analysis. Expression of the genes of interest, Chrebp (A), Syt7 (B), Cplx2 (C), Scgn (D), and Tmem59l (E) in Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells was examined by quantitative RT-PCR. These genes were confirmed to be responder genes. n = 4–5. Values are means ± SD. *P<0.05.
(TIF)
PCR primers used in the present study.
(PDF)
Genes differentially expressed between responder and non-responder MIN6 cells.
(PDF)
Genes differentially expressed between Pr-LP and clone 4 MIN6 cells.
(PDF)
Genes differentially expressed between Pr-LP and C4-LP MIN6 cells.
(PDF)
Genes differentially expressed between C4-LP and C4-HP MIN6 cells.
(PDF)
Acknowledgments
The authors are grateful to Ms. Mayu Yamamoto and Mr. Masafumi Ashida for technical assistance. We acknowledge the editorial assistance of Drs. Leslie A. Miglietta and Grace E. Gray.
Funding Statement
This work was supported by a Grant-in-Aid (No. 23617011) from the Japanese Society for the Promotion of Science (http://www.jsps.go.jp/english/index.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, et al. (1990) Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127: 126–132. [DOI] [PubMed] [Google Scholar]
- 2. Kayo T, Sawada Y, Suzuki Y, Suda M, Tanaka S, et al. (1996) Proprotein-processing endoprotease furin decreases regulated secretory pathway-specific proteins in the pancreatic beta cell line MIN6. J Biol Chem 271: 10731–10737. [DOI] [PubMed] [Google Scholar]
- 3. Sawada Y, Zhang B, Okajima F, Izumi T, Takeuchi T (2001) PTHrP increases pancreatic beta-cell-specific functions in well-differentiated cells. Mol Cell Endocrinol 182: 265–275. [DOI] [PubMed] [Google Scholar]
- 4. O'Driscoll L, Gammell P, McKiernan E, Ryan E, Jeppesen PB, et al. (2006) Phenotypic and global gene expression profile changes between low passage and high passage MIN-6 cells. J Endocrinol 191: 665–676. [DOI] [PubMed] [Google Scholar]
- 5. Minami K, Yano H, Miki T, Nagashima K, Wang CZ, et al. (2000) Insulin secretion and differential gene expression in glucose-responsive and -unresponsive MIN6 sublines. Am J Physiol Endocrinol Metab 279: E773–781. [DOI] [PubMed] [Google Scholar]
- 6. Lilla V, Webb G, Rickenbach K, Maturana A, Steiner DF, et al. (2003) Differential gene expression in well-regulated and dysregulated pancreatic beta-cell (MIN6) sublines. Endocrinology 144: 1368–1379. [DOI] [PubMed] [Google Scholar]
- 7. Dowling P, O'Driscoll L, O'Sullivan F, Dowd A, Henry M, et al. (2006) Proteomic screening of glucose-responsive and glucose non-responsive MIN-6 beta cells reveals differential expression of proteins involved in protein folding, secretion and oxidative stress. Proteomics 6: 6578–6587. [DOI] [PubMed] [Google Scholar]
- 8. Dowling P, Shields W, Rani S, Meleady P, Henry M, et al. (2008) Proteomic analysis of conditioned media from glucose responsive and glucose non-responsive phenotypes reveals a panel of secreted proteins associated with beta cell dysfunction. Electrophoresis 29: 4141–4149. [DOI] [PubMed] [Google Scholar]
- 9. Cheng K, Delghingaro-Augusto V, Nolan CJ, Turner N, Hallahan N, et al. (2012) High passage MIN6 cells have impaired insulin secretion with impaired glucose and lipid oxidation. PLoS One 7: e40868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hiura H, Obata Y, Komiyama J, Shirai M, Kono T (2006) Oocyte growth-dependent progression of maternal imprinting in mice. Genes to Cells 11: 353–361. [DOI] [PubMed] [Google Scholar]
- 11. Takada S, Paulsen M, Tevendale M, Tsai CE, Kelsey G, et al. (2002) Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: implications for imprinting control from comparison with Igf2-H19. Hum Mol Genet 11: 77–86. [DOI] [PubMed] [Google Scholar]
- 12. Hoffman BG, Zavaglia B, Witzsche J, Ruiz de Algara T, Beach M, et al. (2008) Identification of transcripts with enriched expression in the developing and adult pancreas. Genome Biol 9: R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morison IM, Ramsay JP, Spencer HG (2005) A census of mammalian imprinting. Trends Genet 21: 457–465. [DOI] [PubMed] [Google Scholar]
- 14. Edwards CA, Ferguson-Smith AC (2007) Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol 19: 281–289. [DOI] [PubMed] [Google Scholar]
- 15. Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, et al. (2008) Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 40: 1092–1097. [DOI] [PubMed] [Google Scholar]
- 16. Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, et al. (2008) SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 40: 1098–1102. [DOI] [PubMed] [Google Scholar]
- 17. Leclerc I, Rutter GA, Meur G, Noordeen N (2012) Roles of Ca2+ions in the control of ChREBP nuclear translocation. J Endocrinol 213: 115–122. [DOI] [PubMed] [Google Scholar]
- 18. Metukuri MR, Zhang P, Basantani MK, Chin C, Stamateris RE, et al. (2012) ChREBP mediates glucose-stimulated pancreatic β-cell proliferation. Diabetes 61: 2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soggia A, Flosseau K, Ravassard P, Szinnai G, Scharfmann R, et al. (2012) Activation of the transcription factor carbohydrate-responsive element-binding protein by glucose leads to increased pancreatic beta cell differentiation in rats. Diabetologia 55: 2713–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao N, Le Lay J, Qin W, Doliba N, Schug J, et al. (2010) Foxa1 and Foxa2 maintain the metabolic and secretory features of the mature beta-cell. Mol Endocrinol 24: 1594–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gauthier BR, Wollheim CB (2008) Synaptotagmins bind calcium to release insulin. Am J Physiol Endocrinol Metab 295: E1279–1286. [DOI] [PubMed] [Google Scholar]
- 22. Gao Z, Reavey-Cantwell J, Young RA, Jegier P, Wolf BA (2000) Synaptotagmin III/VII isoforms mediate Ca2+-induced insulin secretion in pancreatic islet beta-cells. J Biol Chem 275: 36079–36085. [DOI] [PubMed] [Google Scholar]
- 23. Gauthier BR, Duhamel DL, Iezzi M, Theander S, Saltel F, et al. (2008) Synaptotagmin VII splice variants alpha, beta, and delta are expressed in pancreatic beta-cells and regulate insulin exocytosis. FASEB J 22: 194–206. [DOI] [PubMed] [Google Scholar]
- 24. Li Y, Wang P, Xu J, Gorelick F, Yamazaki H, et al. (2007) Regulation of insulin secretion and GLUT4 trafficking by the calcium sensor synaptotagmin VII. Biochem Biophys Res Commun 362: 658–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gustavsson N, Lao Y, Maximov A, Chuang JC, Kostromina E, et al. (2008) Impaired insulin secretion and glucose intolerance in synaptotagmin-7 null mutant mice. Proc Natl Acad Sci USA 105: 3992–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kataoka M, Sekiguchi M, Takahashi M (2009) Identification of a minimal segment of complexin II essential for preferential distribution in axons. J Neurochem 108: 1109–1115. [DOI] [PubMed] [Google Scholar]
- 27. Abderrahmani A, Niederhauser G, Plaisance V, Roehrich ME, Lenain V, et al. (2004) Complexin I regulates glucose-induced secretion in pancreatic beta-cells. J Cell Sci 117: 2239–2247. [DOI] [PubMed] [Google Scholar]
- 28. Maj M, Gartner W, Ilhan A, Neziri D, Attems J, et al. (2010) Expression of TAU in insulin-secreting cells and its interaction with the calcium-binding protein secretagogin. J Endocrinol 205: 25–36. [DOI] [PubMed] [Google Scholar]
- 29. Wagner L, Oliyarnyk O, Gartner W, Nowotny P, Groeger M, et al. (2000) Cloning and expression of secretagogin, a novel neuroendocrine- and pancreatic islet of Langerhans-specific Ca2+-binding protein. J Biol Chem 275: 24740–24751. [DOI] [PubMed] [Google Scholar]
- 30. Gartner W, Vila G, Daneva T, Nabokikh A, Koc-Saral F, et al. (2007) New functional aspects of the neuroendocrine marker secretagogin based on the characterization of its rat homolog. Am J Physiol Endocrinol Metab 293: E347–354. [DOI] [PubMed] [Google Scholar]
- 31. Inada A, Nienaber C, Fonseca S, Bonner-Weir S (2006) Timing and expression pattern of carbonic anhydrase II in pancreas. Dev Dyn 235: 1571–1577. [DOI] [PubMed] [Google Scholar]
- 32. Nishimura W, Kondo T, Salameh T, El Khattabi I, Dodge R, et al. (2006) A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol 293: 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gosmain Y, Cheyssac C, Heddad Masson M, Dibner C, Philippe J (2011) Glucagon gene expression in the endocrine pancreas: the role of the transcription factor Pax6 in alpha-cell differentiation, glucagon biosynthesis and secretion. Diabetes Obes Metab 13 Suppl 131–38. [DOI] [PubMed] [Google Scholar]
- 34. Katz LS, Gosmain Y, Marthinet E, Philippe J (2009) Pax6 regulates the proglucagon processing enzyme PC2 and its chaperone 7B2. Mol Cell Biol 29: 2322–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gosmain Y, Avril I, Mamin A, Philippe J (2007) Pax-6 and c-Maf functionally interact with the alpha-cell-specific DNA element G1 in vivo to promote glucagon gene expression. J Biol Chem 282: 35024–35034. [DOI] [PubMed] [Google Scholar]
- 36. Meur G, Qian Q, da Silva Xavier G, Pullen TJ, Tsuboi T, et al. (2011) Nucleo-cytosolic shuttling of FoxO1 directly regulates mouse Ins2 but not Ins1 gene expression in pancreatic beta cells (MIN6). J Biol Chem 286: 13647–13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bengtsson M, Stahlberg A, Rorsman P, Kubista M (2005) Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res 15: 1388–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ling Z, Heimberg H, Foriers A, Schuit F, Pipeleers D (1998) Differential expression of rat insulin I and II messenger ribonucleic acid after prolonged exposure of islet beta-cells to elevated glucose levels. Endocrinology 139: 491–495. [DOI] [PubMed] [Google Scholar]
- 39. Giddings SJ, Carnaghi LR, Fischer LJ, Miller CP (1991) Differential regulation of rat insulin I and II messenger RNA synthesis: effects of fasting and cyproheptadine. Mol Endocrinol 5: 549–554. [DOI] [PubMed] [Google Scholar]
- 40. Leroux L, Desbois P, Lamotte L, Duvillie B, Cordonnier N, et al. (2001) Compensatory responses in mice carrying a null mutation for Ins1 or Ins2. Diabetes 50 Suppl 1S150–153. [DOI] [PubMed] [Google Scholar]
- 41. Gu C, Stein GH, Pan N, Goebbels S, Hornberg H, et al. (2010) Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metabolism 11: 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cram DS, McIntosh A, Oxbrow L, Johnston AM, DeAizpurua HJ (1999) Differential mRNA display analysis of two related but functionally distinct rat insulinoma (RIN) cell lines: identification of CD24 and its expression in the developing pancreas. Differentiation 64: 237–246. [DOI] [PubMed] [Google Scholar]
- 43. Hald J, Galbo T, Rescan C, Radzikowski L, Sprinkel AE, et al. (2012) Pancreatic islet and progenitor cell surface markers with cell sorting potential. Diabetologia 55: 154–165. [DOI] [PubMed] [Google Scholar]
- 44. Tycko B, Morison IM (2002) Physiological functions of imprinted genes. J Cell Physiol 192: 245–258. [DOI] [PubMed] [Google Scholar]
- 45. Kamiya M, Judson H, Okazaki Y, Kusakabe M, Muramatsu M, et al. (2000) The cell cycle control gene ZAC/PLAGL1 is imprinted--a strong candidate gene for transient neonatal diabetes. Hum Mol Genet 9: 453–460. [DOI] [PubMed] [Google Scholar]
- 46. Ma D, Shield JP, Dean W, Leclerc I, Knauf C, et al. (2004) Impaired glucose homeostasis in transgenic mice expressing the human transient neonatal diabetes mellitus locus, TNDM. J Clin Invest 114: 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hoffmann A, Spengler D (2012) Transient neonatal diabetes mellitus gene Zac1 impairs insulin secretion in mice through Rasgrf1. Mol Cell Biol 32: 2549–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carlsson C, Tornehave D, Lindberg K, Galante P, Billestrup N, et al. (1997) Growth hormone and prolactin stimulate the expression of rat preadipocyte factor-1/delta-like protein in pancreatic islets: molecular cloning and expression pattern during development and growth of the endocrine pancreas. Endocrinology 138: 3940–3948. [DOI] [PubMed] [Google Scholar]
- 49. Friedrichsen BN, Carlsson C, Moldrup A, Michelsen B, Jensen CH, et al. (2003) Expression, biosynthesis and release of preadipocyte factor-1/delta-like protein/fetal antigen-1 in pancreatic beta-cells: possible physiological implications. J Endocrinol 176: 257–266. [DOI] [PubMed] [Google Scholar]
- 50. Chu K, Tsai MJ (2005) Neuronatin, a downstream target of BETA2/NeuroD1 in the pancreas, is involved in glucose-mediated insulin secretion. Diabetes 54: 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Joe MK, Lee HJ, Suh YH, Han KL, Lim JH, et al. (2008) Crucial roles of neuronatin in insulin secretion and high glucose-induced apoptosis in pancreatic beta-cells. Cell Signal 20: 907–915. [DOI] [PubMed] [Google Scholar]
- 52. Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, et al. (2010) The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet 42: 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamagata K, Senokuchi T, Lu M, Takemoto M, Fazlul Karim M, et al. (2011) Voltage-gated K+channel KCNQ1 regulates insulin secretion in MIN6 beta-cell line. Biochem Biophys Res Commun 407: 620–625. [DOI] [PubMed] [Google Scholar]
- 54. Varrault A, Bilanges B, Mackay DJ, Basyuk E, Ahr B, et al. (2001) Characterization of the methylation-sensitive promoter of the imprinted ZAC gene supports its role in transient neonatal diabetes mellitus. J Biol Chem 276: 18653–18656. [DOI] [PubMed] [Google Scholar]
- 55. Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM (2000) The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev 14: 1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 56. Yang BT, Dayeh TA, Volkov PA, Kirkpatrick CL, Malmgren S, et al. (2012) Increased DNA Methylation and Decreased Expression of PDX-1 in Pancreatic Islets from Patients with Type 2 Diabetes. Mol Endocrinol 26: 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Volkmar M, Dedeurwaerder S, Cunha DA, Ndlovu MN, Defrance M, et al. (2012) DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J 31: 1405–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang BT, Dayeh TA, Kirkpatrick CL, Taneera J, Kumar R, et al. (2011) Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia 54: 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuroda A, Rauch TA, Todorov I, Ku HT, Al-Abdullah IH, et al. (2009) Insulin gene expression is regulated by DNA methylation. PLoS One 4: e6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Park JH, Stoffers DA, Nicholls RD, Simmons RA (2008) Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest 118: 2316–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative RT-PCR analysis. Expression of the genes of interest, Chrebp (A), Syt7 (B), Cplx2 (C), Scgn (D), and Tmem59l (E) in Pr-LP, Pr-HP, C4-LP, and C4-HP MIN6 cells was examined by quantitative RT-PCR. These genes were confirmed to be responder genes. n = 4–5. Values are means ± SD. *P<0.05.
(TIF)
PCR primers used in the present study.
(PDF)
Genes differentially expressed between responder and non-responder MIN6 cells.
(PDF)
Genes differentially expressed between Pr-LP and clone 4 MIN6 cells.
(PDF)
Genes differentially expressed between Pr-LP and C4-LP MIN6 cells.
(PDF)
Genes differentially expressed between C4-LP and C4-HP MIN6 cells.
(PDF)