Abstract
Aim of the study
Excessive inflammation can lead to tissue damage and dysfunction of vital organs. Hence, regulating inflammatory response is a viable therapeutic approach. In Asian countries, various inflammatory diseases have often effectively been treated with herbal remedies including the root extract of Aralia continentalis Kitagawa (Araliaceae). Here, we investigated the effect of kaurenoic acid (ent-kaur-16-en-19-oic acid: KA), a diterpenoid that is extracted from Aralia continentalis Kitagawa root, on inflammation.
Results
Western blot and RT-PCR analyses show that KA induced the nuclear localization of Nrf2 as low as 1 nM in concentration and that KA treatment induced the expression of Nrf2 dependent genes such as GCLC and HO-1. On the other hand, KA did not affect the degradation of cytoplasmic IκB-α, the nuclear localization of RelA (p65), and NF-κB transcriptional activity in RAW 264.7 cells treated with endotoxin. Consistent with these data, KA treatment failed to suppress gene expression of representative pro-inflammatory mediators including COX-2, nitric oxide, IL-1β, TNF-α, and IL-12, indicating that KA did not have an important impact on NF-κB activation.
Conclusion
Together, these results show that KA was an effective activator of Nrf2, and suggest that the beneficial effects of A. continentalis Kitagawa root extract are, at least in part, mediated by activating Nrf2.
Keywords: Inflammation, Kaurenoic acid, Herbal treatment, anti-inflammation, Nrf2
1. Introduction
Inflammation is a critical component of innate immunity. Proper control of inflammatory reactions has therapeutic benefits because uncontrolled inflammation is closely associated with various infectious and non-infectious inflammatory diseases (Nathan and Ding, 2010). The inflammatory response involves complex interactions among a variety of immune cells including macrophages, neutrophils and lymphocytes, and the interactions among these inflammatory cells differ in the context of the inflammatory milieu ((Soehnlein and Lindbom, 2010)). Therefore, regulation of inflammatory responses has been challenging.
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a key transcription factor that regulate expression of genes that are involved in detoxification of drugs and chemicals (Xu, Li et al. 2005; Thimmulappa, et al. 2002). Normally, Nrf2 resides in the cytoplasm in low abundance. When activated, it increases in abundance and translocates to the nucleus where to bind to its cis -acting antioxidant response element (ARE) sequence, resulting in expression of various phase 2 detoxification genes, such as glutamate-cysteine ligase catalytic subunit (GCLC), NAD(P)H:quinine oxidoreductase-1 (NQO1), and heme oxygenase-1 (HO-1) (Alam, et al. 1999; Bloom and Jaiswal, 2003; Chanas, et al. 2002; Cullinan, et al. 2004)). In addition, Nrf2 seems to have an important role in ameliorating inflammation. Genetic ablation of nrf2 exacerbates inflammation in various inflammatory diseases animal models, including acute lung inflammation, smoke-induced emphysema and asthma (Chan and Kan, 1999; Mochizuki, et al. 2005; Rangasamy, et al. 2004; Rangasamy, et al. 2005).
Inflammatory diseases has long been recognized and treated in Asia by using various herbal extracts as part of traditional Asian medicine. For instance, the roots of Aralia continentalis Kitagawa (Araliaceae) have long been used to treat rheumatism, lumbago, and lameness (Lim, et al. 2009). The methanol extract and dichloromethane fractions of the roots were reported to inhibit IL-8 production in peritoneal macrophages activated by LPS (Lee, et al. 1995). Further analysis of the constituents of the extract showed that the inhibition is attributable to pimaradienoic acid, continentalic acid, and kaurenoic acid (Han, et al. 1983; Tirapelli, et al. 2005). These molecules moderately affected expressions of COX-1 and COX-2 (Dang, et al. 2005). Kaurenoic acid, in particular, inhibits acetic acid-induced colitis in rats (Paiva, et al. 2002; Lim et al., 2009). Thus, it appears that kaurenoic acid (ent-kaur-16-en-19-oic acid), a diterpenoid, has potent anti-inflammatory activity but the mechanism is unknown. In this study, we sought to define how kaurenoic acid exerts its anti-inflammatory function, and our results suggest that kaurenoic acid is a potent Nrf2 activator.
1. Materials and Methods
2.1. Plant material
The plant materials, the roots of A. continentalis, were collected in Imsil, Korea, in November 2004, and identified by Prof. Young-Seung Ju. A voucher specimen (No. JSI 53A) has been deposited in the College of Oriental Medicine, Woosuk Univ,, Korea.
1.2. Isolation of ent-kaur-16-en-19-oic acid (kaurenoic acid: KA)
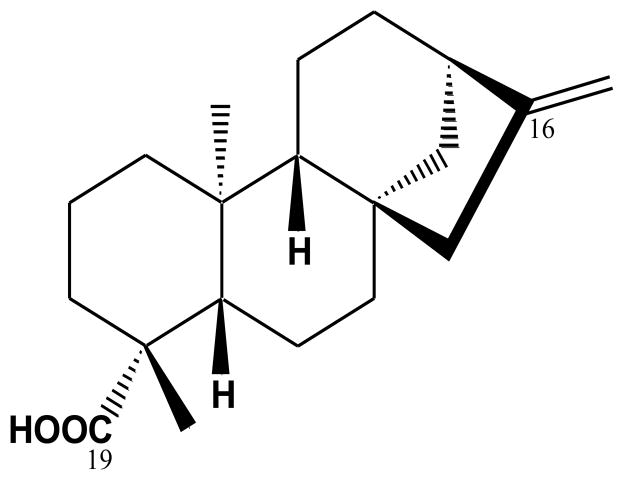
Kaurenoic acid (KA) was isolated as previously described by Dang et al (2005). Briefly, the dried roots of A. continentalis were crushed and extracted three times with MeOH at 60 °C. After a concentration process, the methanol extract was suspended in H2O, and successively partitioned with hexane, CHCl3, EtOAc, and n-BuOH. The CHCl3 layer was subjected to a silica gel column chromatography with increasing polarity (hexane-EtOAc, 100:0 → 1:1) to yield eight fractions (Fr 1-8). The KA was purified by preparative-LC, for which a recycling preparative HPLC (LC-9104) system equipped with a JAI UV detector 3702 and JAIGEL W series column (W252 500 mm plus W251 500 mm, 20.0 mm, i.d., 1000 mm long; Japan Analytical Industry Co., Ltd, Japan) was used with methanol (100% v/v) at a flow rate of 3.5 mL/min and detection at 210 nm. The structure of the KA (Fig. 1) was identified by comparing the NMR spectral data with those in existing literature.
Fig. 1.
Chemical structure of ent-kaur-16-en-19-oic acid (kaurenoic acid: KA).
1.3. ent-kaur-16-en-19-oic acid
1H-NMR (500 MHz, CDCl3) δ 0.92 (3H, s, H-20), 0.98–1.05 (1H, m, H-5), 1.21 (3H, s, H-18), 1.42–1.59 (4H, m, H-7 and 12), 1.56–1.61 (2H, m, H-11), 2.09 (2H, d, J = 3.0 Hz, H-6), 2.61 (1H, br s, H-13), 4,72 (1H, br s, H-17a), 4.77 (br s, H-17b); 13C-NMR (125 MHz, CDCl3): δ 41.38 (C-1), 19.19 (C-2), 37.85 (C-3), 43.92 (C-4), 57.16 (C-5), 21.91 (C-6), 41.38 (C-7), 44.31 (C-8), 55.18 (C-9), 39.78 (C-10), 18.52 (C-11), 33.20 (C-12), 43.85 (C-13), 39.74 (C-14), 49.05 (C-15), 155.91 (C-16), 103.14 (C-17), 29.07 (C-18), 185.10 (C-19), 15.68 (C-20).
1.4. Reagents and antibodies
KA was dissolved in DMSO (Invitrogen, Carisbad, CA, U.S.A.) to 10−3 M. Before experiment, KA was diluted in cell culture media from final 10−4 to 10−9 M. As for vehicle controls, DMSO was similarly diluted in cell culture media from 10 to 0.1% (v/v) so to make control groups treated with the equivalent amount of DMSO used for dissolving the highest concentration of KA in a given experiment. TLR4-specific Escherichia coli LPS was purchased from Alexis Biochemical (San Diego, CA, U.S.A.). Except COX-1 and COX-2 antibodies (Cayman Chemical Co., Ann Arbor, MI, U.S.A.), antibodies against IκB-α, p65 (RelA), Nrf2, YY1, GCLC, β-actin and lamin A/C were from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.).
1.5. Cell culture
A murine macrophage cell line, RAW 264.7 cells, and HEK 293 cells were obtained from the ATCC (American Type Culture Collection, Rockville, MD) and grown in Dulbecco’s Modified Eagle’s Medium (DMEM) containing L-glutamine (200 mg/L) (Hyclone; Logan, UT, USA) supplemented with 10 % (v/v) heat-inactivated fetal bovine serum (FBS) (Invitrogen; Carlsbad, CA, USA) and 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified incubator with 5 % CO 2prior to experiment.
1.6. Assessment of cell viability
The cell viability was assessed with Vybrant MTT assay kit (Invitrogen) per the protocol provided by the company.
1.7. Isolation of total RNA from cells and RT-PCR
Total RNA was isolated with the QIAGEN RNeasy® mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration of RNA was determined by spectrophotometer. Three microgram RNAs were reverse-transcribed by M-MLV reverse transcriptase (Promega, Madison, WI, USA). Single-stranded cDNA was amplified by PCR with specific primers (Table 1).
Table 1.
Oligonucleotide primers used for PCR in this study.
| Target gene | Oligonucleotide sequences (5′ to 3′ direction) | Expected size | Accession number |
|---|---|---|---|
| TNF-α | CTACTCCTCAGAGCCCCCAG AGGCAACCTGACCACTCTCC |
239 bp | NM013693 |
| IL-1β | GTGTCTTTCCCGTGGACCTT TCGTTGCTTGGTTCTCCTTG |
281 bp | NM008361 |
| IL-12 | TGCAACGTTGGAAAGGAAAG CAGCATAAGGCCAAGTGGAA |
227 bp | M86671 |
| COX-1 | GGGTAGACCTTGGCCACATT TCGTGGAGAAGAGCATCAGC |
229 bp | NM008969 |
| COX-2 | CCCAGAGCTCCTTTTCAACC AATTGGCACATTTCTTCCCC |
241 bp | NM011198 |
| GCLC | CACTGCCAGAACACAGACCC ATGGTCTGGCTGAGAAGCCT |
239 bp | NM010295 |
| HO-1 | TGAAGGAGGCCACCAAGGAGG AGAGGTCACCCAGGTAGCGGG |
373 bp | NM010442 |
| GAPDH | GGAGCCAAAAGGGTCATCAT GTGATGGCATGGACTGTGGT |
203 bp | NM008084 |
For PCR amplification, TaqPCRx DNA polymerase, Recombinant (Invitrogen) and the manufacturer’s protocol were used. The reaction conditions were as follows: an initial denaturation at 95 °C for 5 min followed by 30 cycles of denaturation for 40 sec at 95 °C, annealing for 40 sec at 55 °C and extension for 50 sec at 72 °C with a final extension for 7 min at 72 °C. Amplicons were separated in 1.5 % agarose gels in 1× TBE buffer at 100 V for 30 min, stained with ethidium bromide and visualized under UV light. GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) was used as internal controls to evaluate relative expressions of TNF-α, IL-1β, IL-12, GCLC, HO-1 and COX-2.
1.8. Western blot analysis
Total cell extracts of 5×106 cells were prepared as described previously. Nuclear proteins were isolated by NE-PER nuclear extraction kit and the manufacture’s protocol (Thermo Scientific, IL, USA). The amounts of proteins were measured by Bradford (Bio-Rad). Equal amounts of proteins were fractionated by SDS-PAGE and then transferred to PVDF membrane (Bio-Rad). Blots were blocked for at least 1 h with 5 % non-fat dry milk prior to incubation with Nrf2, NF-κB (p65), IκB-α, GCLC, COX-1, COX-2, β-actin, lamin A/C, and YY-1 polyclonal antibodies at 4 °C for overnight. After incubation with secondary antibodies conjugated with HRP for 1 h at room temperature, specific bands of interest were revealed by chemiluminescence (SuperSignal® West Femto, Thermo Scientific).
1.9. Constructs, transfection, reporter cell line, and luciferase assay
The reporter construct harbors four tandem copies of a 36-base enhancer from the 5′ HIV-long terminal repeat (containing two NF-κB binding sites, GGGACTTTCC) placed upstream of the HSV minimal thymidine kinase promoter, which were cloned into pEGFPluc (BD Clontech). The reporter construct (0.5μg) was introduced to RAW 264.7 cells by transfection with Lipofectamin (Invitrogen) and the protocol of the manufacturer, and the transfected cells were selected under G418 (600 μg/ml). Candidate cell lines harboring the NF-κB reporter construct were tested for the induction of luciferase activity. NF-κB mediated luciferase activity was normalized by the amounts of proteins in total cell lysate. For the measurement of Nrf2 activity, 1kb-long promoter region proximal to the initiation site of murine NQO-1 transcription which contains an Nrf2 binding site was cloned into pGL3-basic (Promega). The reporter construct was transiently introduced to HEK293 or RAW264.7 cells by transfection with Lipofectamin (Invitrogen), along with tk-Renilla luciferase construct. Luciferase activity was measured by dual luciferase kit (Promega) and normalized against Renilla luciferase activity. The plasmids encoding Nrf2 and Keap1, an inhibitor of Nrf2, were gifts from Dr. Michael Freeman (Vanderbilt University School of Medicine, Nashville, TN, USA). The recombinant adenoviruses encoding eGFP and IκBDN and infection were described previously (Joo, et al. 2005; Joo, et al. 2007) ).
1.10. Measurement of nitric oxide (NO) production
Produced NO was determined by measuring the stable conversion product of NO, nitrite (NO2−). Briefly, 100 μl of cell culture medium was mixed with 100 μl of Griess reagent (0.1 % N-(1)-naphthyl-ethylenediamine, 1.0 % sulfanilamide, and 2.5 % phosphoric acid) in a 96-well plate and incubated at room temperature for 5 min prior to reading at 540 nm with a microplate reader. Sodium nitrite (NaNO2) was used to generate a standard curve.
1.11. Statistical analysis
Data is presented as the mean ± SEM (Std. Err.) of at least three separate experiments. For comparison among groups, paired or unpaired T tests and one-way analysis of variance (ANOVA) tests were used (with the assistance of InStat, Graphpad Software, Inc., San Diego, CA). P values less than 0.05 considered be statistically significant. All experiment was performed at least three times independently.
2. Results
3.1. Determination of the effect of KA on cell viability
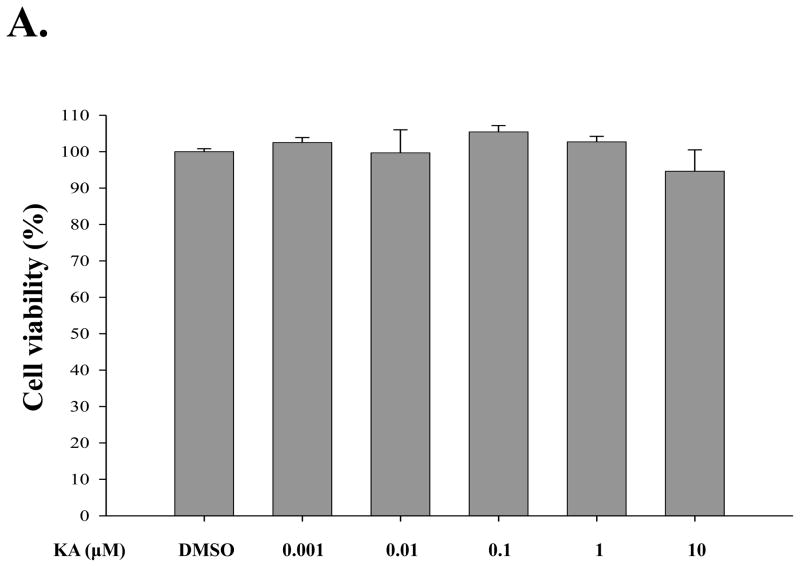
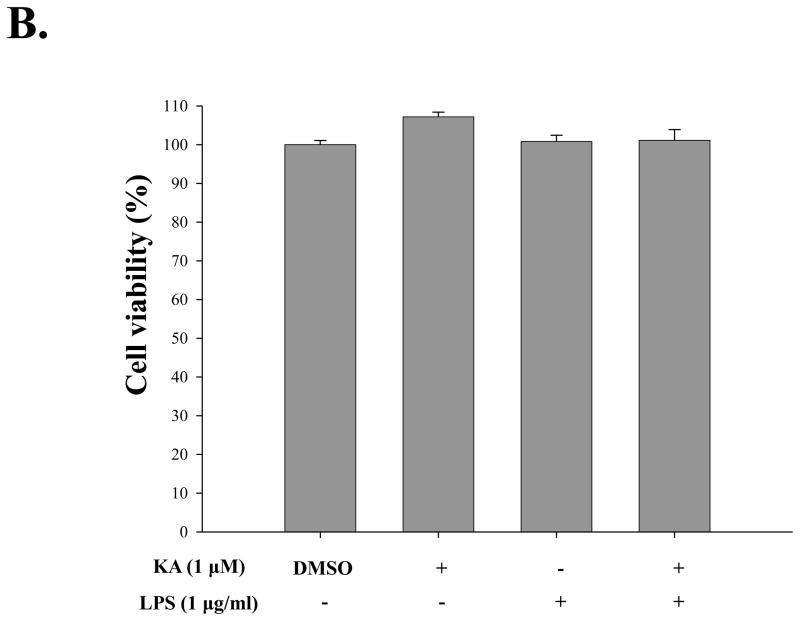
For determination of an optimal dose of KA with the least cellular toxicity, HEK293 cells were treated with different concentrations of KA, from 1 nM to 10 μM, for 16 h, and the viability of the treated cells was measured by MTT assay. As shown in Fig. 2A, KA appeared not to generate cellular toxicity in general, although there was a slight toxicity at 10 μM of KA treatment. Similarly, RAW 264.7 cells, a murine macrophage-like cell line, were treated with 1 μM of KA (Fig. 2B), and there was no significant cellular toxicity by KA. Cellular toxicity was not detectable even when the cells were simultaneously treated with 1 μg/ml of LPS (columns 5 and 6). These results show that over a broad range of concentrations KA had no significant impact on cellular toxicity.
Fig. 2. Effect of kaurenoic acid on cell viability.
Cytotoxicity of kaurenoic acid on HEK 293 cells (A) and RAW 264.7 cells (B) was determined by MTT assay. Results are representative of at least three independent experiments. Data are the mean ± SEM of three independent experiments.
2.2. KA activates Nrf2 and induces the expression of genes that are regulated by Nrf2
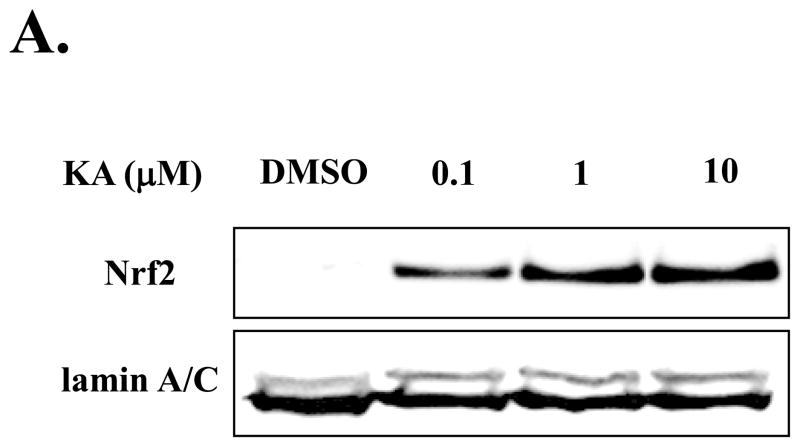
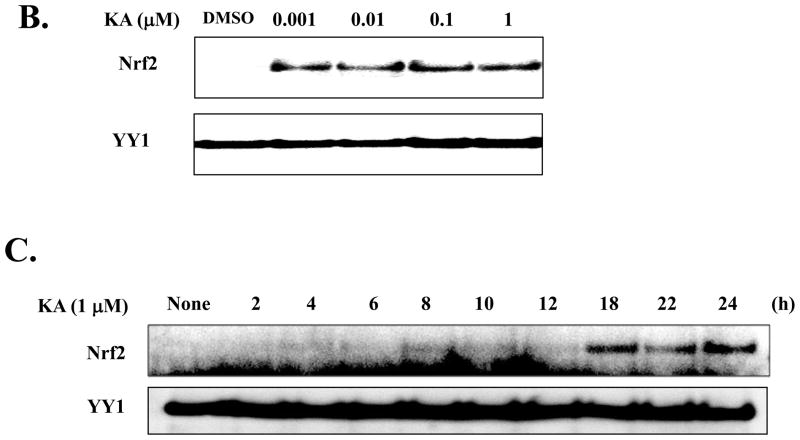
Since Nrf2 has been known as a master transcription factor in down-regulating inflammation, we explored the possibility that the anti-inflammatory function of KA is mediated by activating Nrf2. RAW 264.7 cells were treated with various concentrations of KA for 16 h and the nuclear Nrf2, indicative of Nrf2 activation, was measured by Western blot analysis. As shown in Fig. 3A, Nrf2 was detected in the nucleus of RAW 264.7 cells treated with KA. To determine the lowest dose of KA sufficient to activate Nrf2, we treated HEK 293 cells with various concentrations of KA. As shown in Fig. 3B, KA was capable of activating Nrf2 as low as 1 nM, suggesting that KA has a high potency in Nrf2 activation. To determine the time course for the effect of KA in activating Nrf2, we treated the cells with 1 μM of KA for various periods, and measured the nuclear Nrf2 of the treated cells by Western blot analysis. As shown in Fig. 3C, the nuclear localization of Nrf2 was evident in 18 h after KA treatment. We also performed the similar experiment with RAW 264.7 cells, and obtained similar results (data not shown). Together, our results show that KA is an effective activator for Nrf2.
Fig. 3. KA activates Nrf2.
(A) Nuclear Nrf2, indicative of activated Nrf2, was measured by Western blot analysis. A minimal concentration of KA sufficient for Nrf2 activation (B) and the efficacy of KA (C) were determined. Lamin A/C or YY1, a transcription factor resided in nucleus, was similarly measured as internal controls. Results are representative of at least three independent experiments.
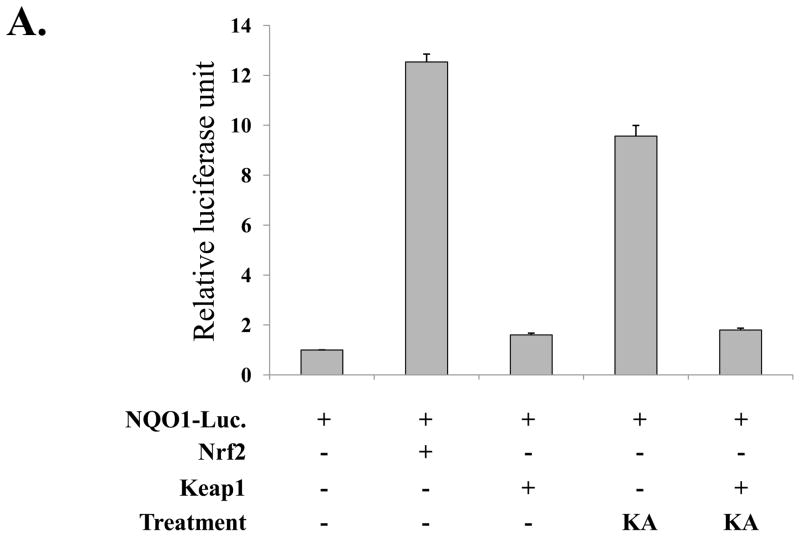
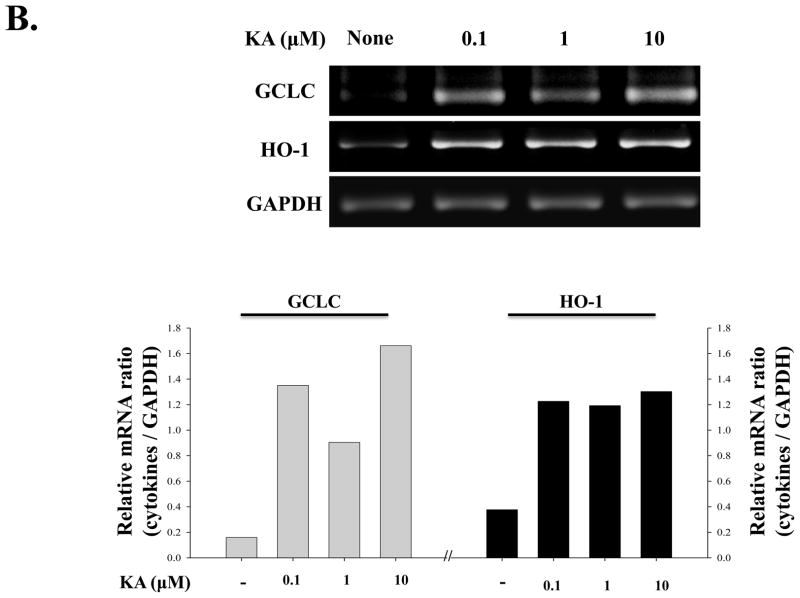
Since KA induced the nuclear localization of Nrf2, we further tested whether KA induces Nrf2-dependent gene expression. First, we tested whether KA increases Nrf2 transcriptional activity. HEK293 cells were transfected with an Nrf2-reporter construct along with plasmids encoding Nrf2 and Keap1, an inhibitor for Nrf2, and subsequently treated with KA (0.1μM) for 16h. As shown in Fig. 4A, over-expression of Nrf2 increased luciferase activity, which was blunted by Keap1 expression (2nd and 3rd columns from the left), suggesting that NQO-1 promoter is responsive to Nrf2. When the transfected cells were treated with KA, Nrf2 transcriptional activity was increased but was similarly blunted by Keap1 (4th and 5th columns). Next, for the test of Nrf2-dependent gene expression by KA, RAW 264.7 cells were treated with various amounts of KA for 16 h and total RNA was extracted from the treated cells for RT-PCR analysis of GCLC and HO-1, Nrf2 dependent genes. As shown in Fig. 4B, KA treatment of RAW 264.7 cells induced the expression of Nrf2 dependent genes. Taken together, these results show that KA activated Nrf2, resulting in Nrf2-dependent gene expression.
Fig. 4.
KA treatment induces the expression of Nrf2-dependent genes. (A) HEK293 cells were transfected with an Nrf2-luciferase reporter (NQO-1-luciferase) along with Nrf2 and Keap1 expressing vectors. The transfected cells were treated with KA, and the effect of KA on Nrf2 transcriptional activity was measured by luciferase assay. (B) RAW 264.7 cells were treated with three different concentrations of KA, and the expression of GCLC and HO-1 was measured by semi-quantitative RT-PCR. Intensity of each PCR bands was analyzed by densitometer and shown as relative increases of each Nrf2-dependent genes against GAPDH. Results are representative of at least three independent experiments.
2.3. KA does not affect NF-κB activity
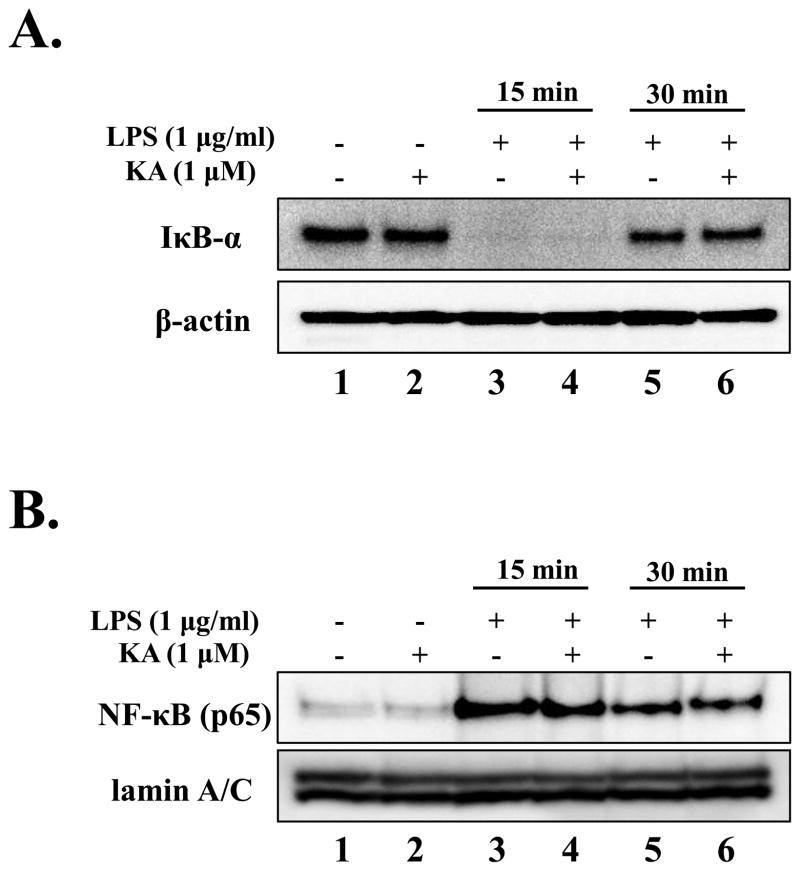
Since expression of many pro-inflammatory cytokine genes is regulated by NF-κB, we examined if KA affects the activation of NF-κB. RAW 264.7 cells were pre-treated with KA (1 μM) for 16 h and subsequently treated with LPS (1 μg/ml) for 15 and 30 min. Cytoplasmic and nuclear proteins were fractionated, and the absence of cytoplasmic inhibitory κB-α(IκB-α) and the presence of nuclear p65, the canonical markers of NF-κB activation, were determined by Western blot analysis. As shown in Fig. 5A, the cytoplasmic IκB-α disappeared rapidly in response to LPS by 15 min (lane 3) and was restored within 30 min after LPS treatment (lane 5), which was not affected by KA treatment (lanes 4 and 6). Consistent with these data, as shown in Fig. 5B, nuclear p65 was detected at 15 min after LPS treatment (lane 3), which was not affected by KA treatment (lanes 4 and 6).
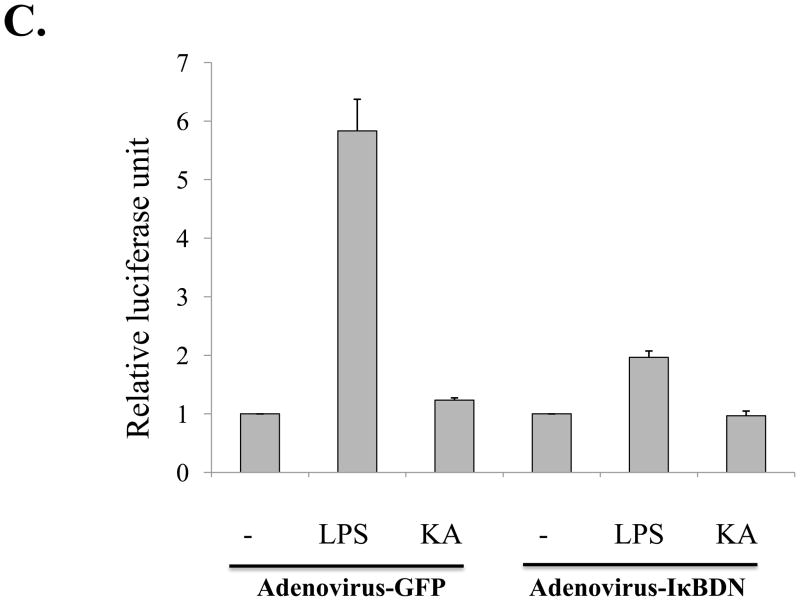
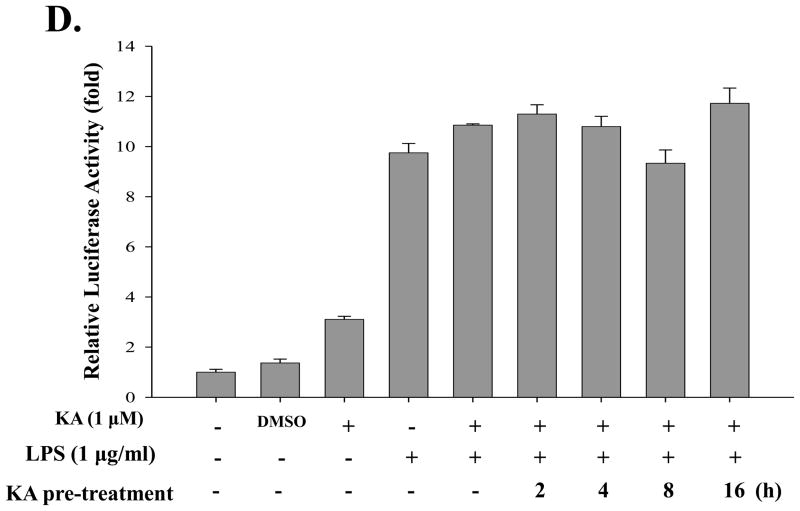
Fig. 5. Kaurenoic acid does not affect NF-κB activity.
The effect of KA on NF-κB was determined by measuring the degradation of the cytoplasmic IκB-α (A) and the presence of the nuclear p65 (B) by Western blot analysis. β-actin and lamin A/C were measured as internal controls for cytoplasmic and nucleoplasmic proteins, respectively. The effect of KA on NF-κB transcriptional activity was measured by using an NF-κB reporter cell line derived from RAW 264.7 cell. (C) The cells, either infected with recombinant adenoviruses encoding either eGFP or IκBDN with multiplicity of infection of 1 for 24h, were treated with KA. (D) The cells were treated with KA simultaneously with, or prior to, LPS treatment. Data are the mean ± SEM of three independent experiments. All results are representative of three independent experiments.
Although KA seemed not to affect the changes of the cytoplasmic IκB-α and the nuclear p65, it is possible that KA affects NF-κB mediated transcriptional activity. To address this, we generated a RAW 264.7 cell line that stably harbors an NF-κB-luciferase reporter construct (see Materials and Methods). First, we tested whether KA increases NF-κB transcriptional activity (Fig. 5C). When treated with LPS (1 μg/ml) for 16 h, the cell line yielded a strong luciferase activity, which was however blunted by NF-κB inhibitor IκBDN expressed by a recombinant adenovirus encoding the gene, indicating that LPS treatment induced NF-κB mediated transcription in the cell line (2nd and 6th columns). Consistent with the results of Fig. 5A and 5B, treatment with 1 μM of KA did not affect NF-κB activity (the 3rd column). Next, to examine a possible suppressive effect of KA, the cell line was treated with LPS along with KA. As shown in Fig. 5D, simultaneous treatment with KA did not affect LPS-induced NF-κB activity (4th and 5th columns). To test the effect of pre-treatment of KA on NF-κB mediated transcription, we treated the cell line with KA for various periods, from 2 h up to 16 h, prior to LPS treatment for 16 h, showing that pretreatment with KA did not significantly affect NF-κB mediated transcription (columns 6 to 9). We performed similar experiment in which LPS-treated cells were subsequently treated with KA for various periods, and results show no effect of KA on NF-κB transcriptional activity either (data not shown). In addition, when similar experiment was performed with an independently isolated, different cell line, we obtained similar results (data not shown). Taken together, these results indicate that KA did not affect NF-κB transcriptional activity.
2.4. KA does not affect the expression of pro-inflammatory genes
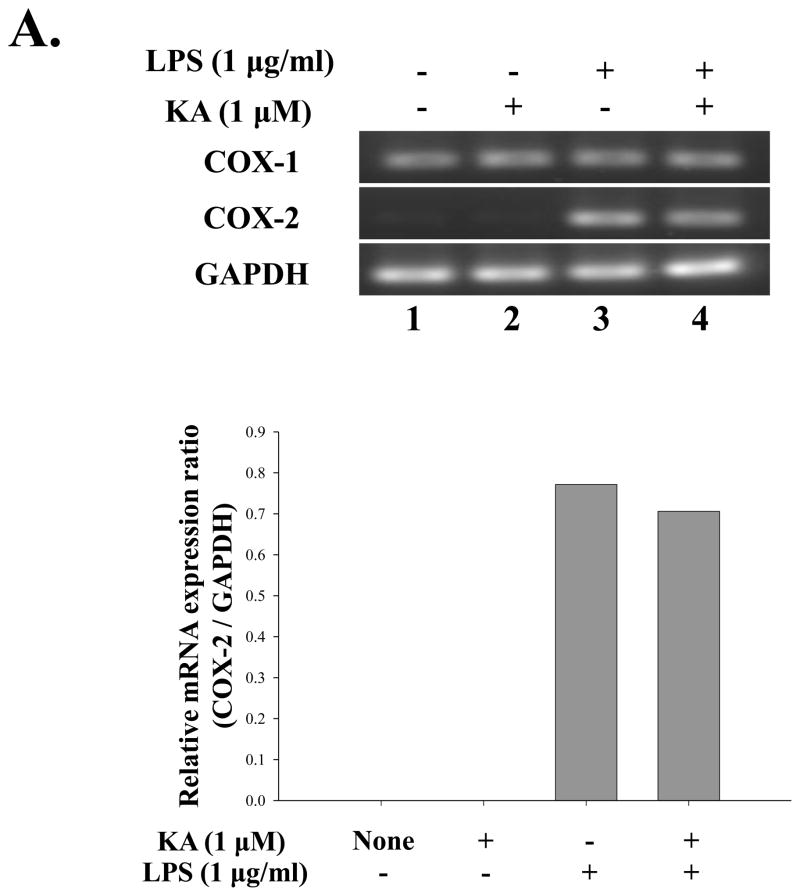
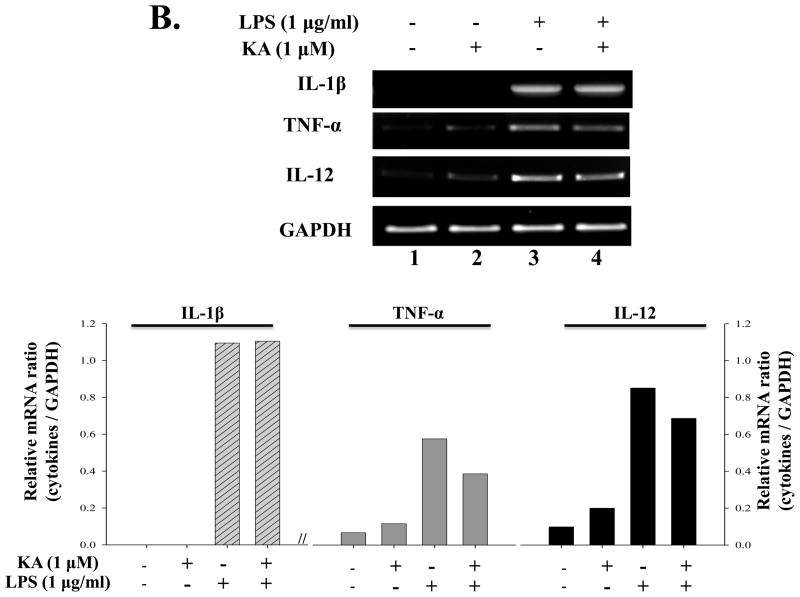
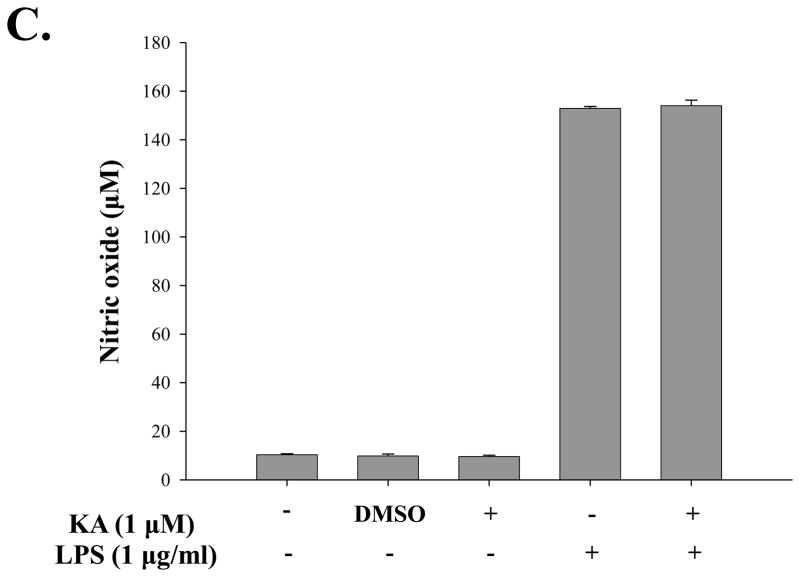
Although expression of many pro-inflammatory genes is regulated by NF-κB, other transcription factors also play an important role. Therefore, it is possible that KA suppresses expression of pro-inflammatory genes through affecting other factor. To test this possibility, we treated RAW 264.7 cells with KA for 16h and then with LPS (1 μg/ml) for various periods, and measured expressions of pro-inflammatory genes including COX-2, IL-1β, TNF-α, and IL-12. To examine the effect of KA on COX-2 expression, we pretreated RAW 264.7 cells with KA and subsequently with LPS. Total RNA was prepared for RT-PCR analysis. As shown in Fig. 6A, LPS treatment induced COX-2 expression, which was, however, not significantly reduced by KA pretreatment (lanes 7 and 8). To examine the effect of KA on the expression of pro-inflammatory genes, we similarly measured mRNA expression of IL-1β, TNF-α, and IL-12. As shown in Fig. 6B, the expression of these genes was not significantly affected by KA treatment either. Finally, since NO is known to be a pro-inflammatory factor in diseases (Payne, 2003; Tao and Chen, 2009), we examine the effect of KA on NO produced by RAW 264.7 cells. As shown in Fig. 6C, LPS treatment increased NO production (the 4th column from the left), which was, however, not changed by KA treatment (the 5th column). Taken together, these results show that KA did not affect the expression of the major pro-inflammatory genes.
Fig. 6. Effect of kaurenoic acid on the expression of pro-inflammatory genes.
Total RNA was extracted from the differentially treated RAW 264.7 cells and analyzed by semi-quantitative RT-PCR for the expression of COX-1 and -2 (A) and of pro-inflammatory cytokines including IL-1β, TNF-α and IL-12 (B). Intensity of each PCR bands was analyzed by densitometer and shown as relative increases of each pro-inflammatory cytokines against GAPDH. (C) The effect of KA on the production of pro-inflammatory NO was determined. Data are the mean ± SEM of three independent experiments. All results are representative of three independent experiments.
3. Discussion
Inflammation is an essential part of innate immunity that protects from invading pathogens. In self-limiting inflammation, inflammatory response elicited by pathogens quickly subsides and returns to a homeostatic level. In pathologic conditions, however, the inflammatory response persists, resulting in tissue damage (Serhan, et al. 2008). Using herbal extracts to treat various inflammatory diseases has been a major part of traditional medicine in many Asian countries (Lim et al., 2009). In U.S.A., the National Center for Complementary and Alternative Medicine has supported research that probes these traditional Asian remedies as an alternative and complementary medicine. Despite the long history of usage of these remedies, the mechanisms by which the remedies exert their effects on inflammation remain largely unknown, which has hampered proper usage of them in conjunction with mainstream medical treatments for various inflammatory diseases. Thus, uncovering the mechanisms provides potentially significant health benefits to patients who suffer from acute or chronic inflammatory diseases.
Aralia continentalis has been used as a major ingredient of Asian traditional herbal remedies prescribed for the treatment of inflammatory diseases because of its suppressive effect on inflammation (Moon, et al. 2007; Cheon, et al. 2009). As effector molecules in the plant, continentalic acid and kaurenoic acid (KA) were suggested (Han, et al. 1983; Lim, et al. 2009). While precise mechanisms by which these molecules control inflammation remain unknown, it was suggested that the anti-inflammatory activity of KA is attributable to suppression of NF-κB, a key transcription factor that regulates inflammation, and of the expression of NF-κB dependent genes (Choi, et al. 2011). In this study, we explored other possible mechanism that KA controls inflammation via Nrf2, a transcription factor that down regulates inflammation. We found that KA potently activated Nrf2, suggesting that KA is one of the active constituents in the root extract of A. continentalis Kitagawa (Araliaceae) that suppresses inflammation.
Unlike previous reports, our results show that KA affects neither the NF-κB transcriptional activity nor the expression of the major pro-inflammatory molecules including COX-2, nitric oxide, IL-1β, TNF-α, and IL-12, expressions of which are largely dependent on NF-κB. Although it is highly likely that the apparent disparity to other studies is due to different experimental settings, it is of note that we used a highly purified, TLR-4 specific LPS for the stimulation of macrophages. Therefore, it is possible that KA suppresses NF-κB activated via other TLRs but not via TLR4. In addition, since we used low concentrations of KA, it is possible that the amounts used for the current study were insufficient to suppress NF-κB activity. Supportive to this notion, we observed the suppression of NF-κB activity when treated cells with high concentrations of KA (data not shown).
Nrf2 is a transcription factor that regulates diverse pathophysiology. Nrf2 is responsible for the expression of various detoxifying phase II enzymes, relieving oxidative stress. Nrf2 also plays a key role in suppressing inflammation because genetic ablation of Nrf2 that exacerbates inflammation in various inflammatory diseases animal models (Chan and Kan, 1999; Mochizuki, et al. 2005; Rangasamy, et al. 2004; Rangasamy, et al. 2005). In addition, given that oxidative stress and inflammation are associated with certain types of cancer, Nrf2 has been considered as a promising target for the treatment of cancer (Ohta, et al. 2008). Recently, 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl] imidazole (CDDO-IM), a triterpenoids derivative, was developed as a potent agonist of Nrf2 and has been tested as an anti-cancer drug (Liby, et al. 2005). Our results show that KA in a nanomolar concentration could activate Nrf2 and induced the expression of Nrf2 dependent genes, which suggests that KA is an effective Nrf2 activator. Given the diverse roles of Nrf2 in pathophysiology, it is possible that KA could be an excellent natural compound for the treatment of inflammatory diseases and certain cancer, although the role of Nrf2 in cancer is controversial and its effect may hinge on the types of cancer.
Although a growing body of evidence supports the anti-inflammatory function of Nrf2, it is unclear how Nrf2 controls inflammatory responses. One of possible mechanisms is that Nrf2 directly suppresses the expression of pro-inflammatory genes by interfering with key transcription factors such as NF-κB and AP-1 (Li, et al. 2008; Liu, et al. 2008; Segal, et al. 2010). We tested this by over-expressing Nrf2 in macrophages that were subsequently treated with LPS. The results suggest that Nrf2 activation may not directly override the expression of representative pro-inflammatory genes including IL-1β and TNF-α (data not shown). Similarly, when macrophages were pretreated with KA to activate Nrf2 and subsequently treated with LPS, the expression of IL-1β and TNF-α was not altered (data not shown). It is noteworthy that although sulforaphane, a potent activator of Nrf2, has been known to suppress the expression of pro-inflammatory cytokines, it suppresses the expression of the genes by preventing oligomerization of TLR4 and thereby blocking TLR4 signaling (Youn, et al. 2010). Nevertheless, our results suggest that Nrf2 is not a factor that directly suppresses the expression of pro-inflammatory genes.
Given our results, another possible mechanism is conceivable, in which Nrf2 induces the expression of genes that are capable of compromising or nullifying the pro-inflammatory responses in the context of the inflammatory milieu. For instance, inflammation is often associated with oxidative stress caused by produced reactive oxygen species (ROS) (Reuter, et al. 2010). It is now well documented that ROS is a potent activator of Nrf2, and activated Nrf2 induces the production of proteins such as NQO-1 and GCLC that scavenge ROS (Reuter, et al. 2010). Thus, it is likely that these Nrf2 dependent gene products lessen the degree of inflammatory response by relieving oxidative stress, contributing to resolving inflammation. Since our previous study showed that LPS can activate Nrf2 without involvement of ROS (Kim, et al. 2011), it is also possible that pro-inflammatory cytokines act on parenchymal cells, in autocrine and paracrine ways, to activate Nrf2 as a countermeasure to prevent prolonged inflammation.
4. Conclusion
In this study, we show that KA, isolated from the root extract of A.continentalis Kitagawa (Araliaceae) that has long been prescribed to control various inflammatory diseases as a traditional Asian medicine, activated Nrf2 and induced Nrf2-regulated gene expression without affecting NF-κB activity. Since Nrf2 is involved in regulation of inflammation, our results suggest that Nrf2 activated by KA in the root extract is a mechanism that contributes to the regulation of inflammation. Our results could help elucidate a mechanism of traditional Asian remedies, contributing to implementing them as a complementary and alternative medicine.
Acknowledgments
This work was financially supported by grants from KRIBB Research Initiative Program, Korea Research Council of Fundamental Sciences & Technology (NTM1000913), and Ministry of Education Science and Technology (KGM2250911). We also thank Korea Institute of Oriental Medicine for the support of the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. The Journal of Biological Chemistry. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. The Journal of Biological Chemistry. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proceedings of the National Academy of Sciences of the United State of America. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, Elcombe CR, Henderson CJ, Wolf CR, Moffat GJ, Itoh K, Yamamoto M, Hayes JD. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochemical Journal. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon MS, Yoon T, Yasukawa K, Yu SY, Kim SJ, Choi G, Moon BC, Lee A, Choo BK, Kim HK. Effect of Aralia continentalisand Angelica biserrata on inflammatory response in lipopolysaccharide-induced RAW 264.7 macrophages and phorbol ester-induced ear edema. Journal of the Korean Society for Applied Biological Chemistry. 2009;52:157–162. [Google Scholar]
- Choi RJ, Shin EM, Jung HA, Choi JS, Kim YS. Inhibitory effects of kaurenoic acid from Aralia continentalis on LPS-induced inflammatory response in RAW264.7 macrophages. Phytomedicine. 2011;18:677–682. doi: 10.1016/j.phymed.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Molecular and Cellular Biololgy. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang NH, Zhang X, Zheng M, Son KH, Chang HW, Kim HP, Bae K, Kang SS. Inhibitory constituents against cyclooxygenases from Aralia cordata Thunb. Archives of Pharmacological Research. 2005;28:28–33. doi: 10.1007/BF02975131. [DOI] [PubMed] [Google Scholar]
- Han BH, Han YN, Han KA, Park MH, Lee EO. Studies on the anti-inflammatory activity of Aralia continentalis (I) – Characterization of continentalic acid and its anti-inflammatory activity. Archives of Pharmacological Research. 1983;6:17–23. [Google Scholar]
- Joo M, Hahn YS, Kwon M, Sadikot RT, Blackwell TS, Christman JW. Hepatitis C virus core protein suppresses NF-kappaB activation and cyclooxygenase-2 expression by direct interaction with IkappaB kinase beta. The Journal of Virology. 2005;79:7648–7657. doi: 10.1128/JVI.79.12.7648-7657.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M, Kwon M, Sadikot RT, Kingsley PJ, Marnett LJ, Blackwell TS, Peebles RS, Jr, Urade Y, Christman JW. Induction and function of lipocalin prostaglandin D synthase in host immunity. The Journal of Immunology. 2007;179:2565–2575. doi: 10.4049/jimmunol.179.4.2565. [DOI] [PubMed] [Google Scholar]
- Kim KH, Lyu JH, Koo ST, Oh SR, Lee HK, Ahn KS, Sadikot RT, Joo M. MyD88 is a mediator for the activation of Nrf2. Biochemical and Biophysical Research Communications. 2011;404:46–51. doi: 10.1016/j.bbrc.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Lee GI, Ha JY, Min KR, Nakagawa H, Tsurufuji S, Chang IM, Kim Y. Inhibitory effects of Oriental herbal medicines on IL-8 induction in lipopolysaccharide-activated rat macrophages. Planta Medica. 1995;61:26–30. doi: 10.1055/s-2006-957992. [DOI] [PubMed] [Google Scholar]
- Li W, Khor TO, Xu C, Shen G, Jeong WS, Yu S, Kong AN. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochemical Pharmacology. 2008;76:1485–1489. doi: 10.1016/j.bcp.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liby K, Hock T, Yore MM, Suh N, Place AE, Risingsong R, Williams CR, Royce DB, Honda T, Honda Y, Gribble GW, Hill-Kapturczak N, Agarwal A, Sporn MB. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Research. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- Lim H, Jung HA, Choi JS, Kim YS, Kang SS, Kim HP. Anti-inflammatory activity of the constituents of the roots of Aralia continentalis. Archives of Pharmacological Research. 2009;32:1237–1243. doi: 10.1007/s12272-009-1909-3. [DOI] [PubMed] [Google Scholar]
- Liu GH, Qu J, Shen X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochimica et Biophysica Acta. 2008;1783:713–727. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Mochizuki M, Ishii Y, Itoh K, Iizuka T, Morishima Y, Kimura T, Kiwamoto T, Matsuno Y, Hegab AE, Nomura A, Sakamoto T, Uchida K, Yamamoto M, Sekizawa K. Role of 15-deoxy delta(12,14) prostaglandin J2 and Nrf2 pathways in protection against acute lung injury. American Journal of Respiratory and Critical Care Medicine. 2005;171:1260–1266. doi: 10.1164/rccm.200406-755OC. [DOI] [PubMed] [Google Scholar]
- Moon PD, Jeong HJ, Um JY, Kim HM, Hong SH. LPS-induced inflammatory cytokine production was inhibited by HyungbangJihwangTang through blockade of NF-kappaB in peripheral blood mononuclear cells. International Journal of Neuroscience. 2007;117:1315–1329. doi: 10.1080/00207450600936692. [DOI] [PubMed] [Google Scholar]
- Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, Shibata T, Yamamoto M, Hirohashi S. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Research. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- Paiva LA, Gurgel LA, Silva RM, Tome AR, Gramosa NV, Silveira ER, Santos FA, Rao VS. Anti-inflammatory effect of kaurenoic acid, a diterpene from Copaifera langsdorffi on acetic acid-induced colitis in rats. Vascular Pharmacology. 2002;39:303–307. doi: 10.1016/s1537-1891(03)00028-4. [DOI] [PubMed] [Google Scholar]
- Payne DN. Nitric oxide in allergic airway inflammation. Current Opinion in Allergy and Clinical Immunology. 2003;3:133–137. doi: 10.1097/00130832-200304000-00007. [DOI] [PubMed] [Google Scholar]
- Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. The Journal of Clinical Investigation. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. The Journal of Experimental Medicine. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radical Biology and Medicine. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal BH, Han W, Bushey JJ, Joo M, Bhatti Z, Feminella J, Dennis CG, Vethanayagam RR, Yull FE, Capitano M, Wallace PK, Minderman H, Christman JW, Sporn MB, Chan J, Vinh DC, Holland SM, Romani LR, Gaffen SL, Freeman ML, Blackwell TS. NADPH oxidase limits innate immune responses in the lungs in mice. PLoS One. 2010;5:e9631. doi: 10.1371/journal.pone.0009631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nature Reviews Immunology. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soehnlein O, Lindbom L. Phagocyte partnership during the onset and resolution of inflammation. Nature Reviews Immunology. 2010;10:427–439. doi: 10.1038/nri2779. [DOI] [PubMed] [Google Scholar]
- Tao HM, Chen GZ. Endothelial NO synthase gene polymorphisms and risk of ischemic stroke: a meta-analysis. Neuroscience Research. 2009;64:311–316. doi: 10.1016/j.neures.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Research. 2002;62:5196–5203. [PubMed] [Google Scholar]
- Tirapelli CR, Ambrosio SR, Coutinho ST, de O, da Costa FB, de Oliveira AM. Pharmacological comparison of the vasorelaxant action displayed by kaurenoic acid and pimaradienoic acid. Journal of Pharmacy and Pharmacology. 2005;57:997–1004. doi: 10.1211/0022357056578. [DOI] [PubMed] [Google Scholar]
- Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Archives of Pharmacological Research. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- Youn HS, Kim YS, Park ZY, Kim SY, Choi NY, Joung SM, Seo JA, Lim KM, Kwak MK, Hwang DH, Lee JY. Sulforaphane suppresses oligomerization of TLR4 in a thiol-dependent manner. The Journal of Immunology. 2010;184:411–419. doi: 10.4049/jimmunol.0803988. [DOI] [PubMed] [Google Scholar]