Abstract
The HER2/neu oncogene encodes a receptor-like tyrosine kinase whose overexpression in breast cancer predicts poor prognosis and resistance to conventional therapies. However, the mechanisms underlying aggressiveness of HER2 (human epidermal growth factor receptor 2)-overexpressing tumors remain incompletely understood. Because it assists epidermal growth factor (EGF) and neuregulin receptors, we overexpressed HER2 in MCF10A mammary cells and applied growth factors. HER2-overexpressing cells grown in extracellular matrix formed filled spheroids, which protruded outgrowths upon growth factor stimulation. Our transcriptome analyses imply a two-hit model for invasive growth: HER2-induced proliferation and evasion from anoikis generate filled structures, which are morphologically and transcriptionally analogous to preinvasive patients’ lesions. In the second hit, EGF escalates signaling and transcriptional responses leading to invasive growth. Consistent with clinical relevance, a gene expression signature based on the HER2/EGF-activated transcriptional program can predict poorer prognosis of a subgroup of HER2-overexpressing patients. In conclusion, the integration of a three-dimensional cellular model and clinical data attributes progression of HER2-overexpressing lesions to EGF-like growth factors acting in the context of the tumor's microenvironment.
Keywords: adhesion, breast cancer, EGF, HER2, hypoxia
Introduction
In affluent countries, approximately one in eight women develops breast cancer during her lifetime (Smigal et al., 2006). Breast tumors can be subclassified according to clinical and histological features, including expression of the estrogen receptor (ER) and a kinase called HER2 (human epidermal growth factor receptor 2), but recent genome-wide transcriptome studies have refined this classification (Perou et al., 2000). HER2 is overexpressed in 40–70% of ductal carcinomas in situ (DCIS), premalignant lesions of the breast, whereas overexpression of HER2 (ERBB2/neu) occurs in just 20–25% of invasive ductal carcinomas (IDCs). The latter subgroup exhibits relatively poor clinical prognosis because of increased rates of metastatic spread (Slamon et al., 1987; van de Vijver et al., 1988; Virolle et al., 2003). Whether HER2-overexpressing DCIS develops into relatively virulent IDC is currently debated.
One confounding issue is the inability of HER2 to bind ligands, in contrast to other members of its family, such as the epidermal growth factor receptor (EGFR), which can bind multiple growth factors (GFs). According to one model, the high basal kinase activity of HER2 allows the receptor to signal in the absence of a stimulatory ligand (Lonardo et al., 1990). An alternative model attributes the transforming function of HER2 to its ability to form heterodimers with ligand-activated receptors, thereby prolonging signals (Riese et al., 1995; Pinkas-Kramarski et al., 1996; Klapper et al., 1999). Employing a model system, Muthuswamy et al. (2001) uncovered yet another attribute of HER2. Their system comprised MCF10A mammary cells grown in extracellular matrix (Petersen et al., 1992). Ectopic expression of chimeric HER2 molecules that can be homodimerized by synthetic ligands revealed that HER2 homodimers, unlike EGFR homodimers, can induce the generation of multiacinar structures with filled lumen (Muthuswamy et al., 2001).
Relevant to the possibility that HER2 and GFs collaboratively contribute to cancer progression, analyses integrating clinicopathological and gene expression data indicated that invasion programs associate with prognosis (Desmedt et al., 2008a), and GFs collaborate with HER2 in tumor progression (Muller et al., 1996; Valabrega et al., 2005). Herein, we address such collaborative interactions by using HER2-overexpressing MCF10A cells, as well as data from clinical specimens. When grown in a three-dimensional (3D) matrix, HER2-overexpressing cells formed filled spheroids resembling DCIS, and upon exposure to EGF, they protruded invasive arms. Transcriptomic analyses of this preinvasive to invasive transition identified three upregulated modules, in line with a model that attributes breast cancer progression to GFs acting on oncogene-initiated preinvasive lesions. Importantly, the state of expression of these three modules could stratify patients with HER2-overexpressing IDCs into different prognostic groups, suggesting that ongoing activation of these invasive transcriptional programs is crucially involved in the metastatic spread of HER2-overexpressing mammary tumors.
Results
GFs induce invasion of HER2-overexpressing spheroids
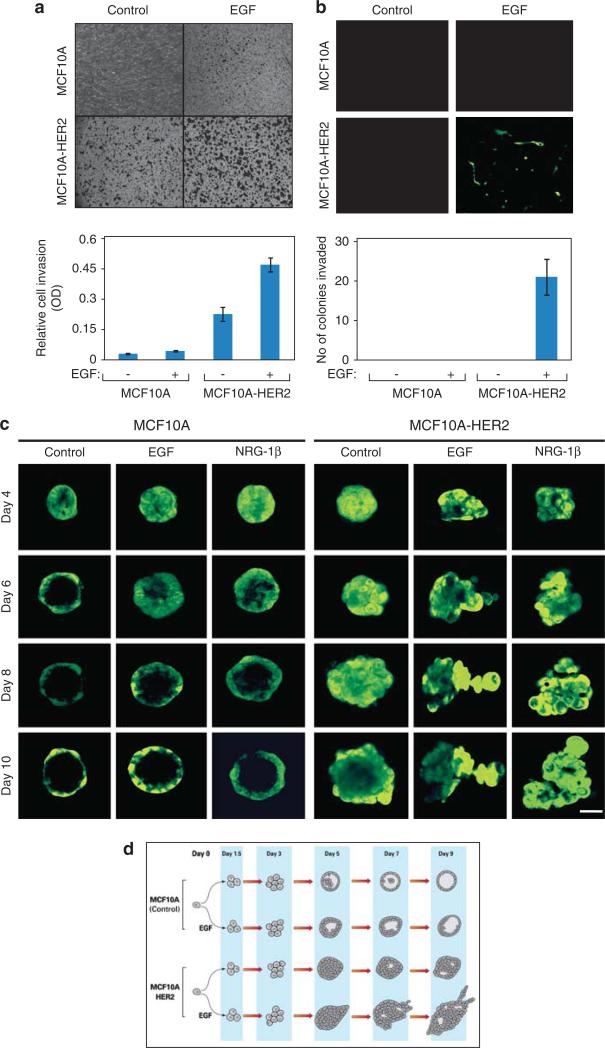
Although monolayers of the nontransformed MCF10A mammary cell line, a well-established cellular model (Muthuswamy et al., 2001), require exogenous EGF for optimal survival, we noted that a significant fraction of cells survived and gave rise to relatively small acini when grown in the absence of EGF, in a natural preparation of extracellular matrix (Matrigel (BD Bioscience, Franklin Lakes, NJ, USA); Supplementary Figure S1A). Because an EGF-blocking antibody, Cetuximab, completely abolished acinus formation, we assume that survival in 3D cultures depends on autocrine or matrix-derived GFs. To examine the functions of HER2 in the absence or presence of GFs, we infected cells with retroviral particles encoding both HER2 and the green fluorescent protein (GFP) (Ueda et al., 2004). A clone that expressed HER2 at levels comparable to those displayed by two commonly used HER2-positive cancer cell lines was selected (Supplementary Figure S1B). Because studies using chimeric versions of EGFR and HER2 concluded that the corresponding heterodimers confer an invasive phenotype (Zhan et al., 2006), we expected EGF to increase invasiveness of HER2-overexpressing cells. To this end, we plated MCF10A cells on Matrigel-coated Transwell filters in the presence or absence of EGF, and stained cells that had migrated through the filter. As expected, the overexpressed HER2 enhanced both basal and EGF-induced cellular invasion in a monolayer (two-dimensional) configuration (Figure 1a).
Figure 1.
HER2 delays intraluminal apoptosis and GFs induce invasive arm formation by MCF10A spheroids grown in extracellular matrix. (a) MCF10A and MCF10A/HER2 monolayers (50 000 cells) were plated on Matrigel-coated Transwell chambers and incubated for 12 h in the presence or absence of EGF (20 ng/ml). Thereafter, the opposite face of the Transwell filter was stained using crystal violet. The lower part presents quantification of the number of cells that migrated to the other side of the filter. (b) MCF10A and MCF10A/HER2 cells were suspended in 5% Matrigel and 2000 cells were plated on a Matrigel-coated Transwell filter separating two compartments of a Transwell chamber. After 18 days of incubation in the absence or presence of EGF, spheroids were removed from the upper face of the filter, whereas cells located on the lower face were observed using fluorescence microscopy. The lower part shows quantification of the number of invading colonies per field. Bars represent s.d. of triplicate determinations. (c) Confocal microscopy images of acini of MCF10A and MCF10A/HER2 cells plated in Matrigel for the indicated times in the absence or presence of EGF or neuregulin (NRG)-1β (each at 20 ng/ml). Each series represents time-lapse images from the same acinus (bar, 50 μm). (d) Flow diagrams of RNA sampling for microarray analyses. The schemes present multicellular structures exhibited by MCF10A and MCF10A/HER2 cells grown in extracellular matrix, in the absence or presence of EGF. RNA was isolated at the indicated time points and used for hybridization to DNA arrays.
To test the effect of HER2 and EGF on the ability of spheroids to invade through matrix barriers, cells were suspended in Matrigel and then plated on top of filters. Following 18 days of incubation in the absence or presence of EGF, the spheroids that formed on the filter were removed from the upper face, whereas cell clusters that reached the lower face were photographed. Interestingly, we observed invasiveness only in the case of HER2-overexpressing MCF10A cells that underwent treatment with EGF (Figure 1b). To closely examine this, we cultured HER2-overexpressing MCF10A cells in a reconstituted 3D extracellular matrix and followed individual spheroids in a time-lapse manner. Under these conditions, single MCF10A cells developed clonal aggregates, whose luminal cells gradually underwent apoptosis to form hollow structures surrounded by a single-cell rim and a basement membrane (Figure 1c). When grown in the presence of either EGF or neuregulin β-1, control spheroids displayed thicker walls and a slightly delayed apoptosis, but HER2-overexpressing spheroids exhibited a much longer delay. A similar delay was reported when a chimeric HER2 was stimulated from within the cell using a synthetic ligand (Muthuswamy et al., 2001). Remarkably, when cultured in the presence of GFs, the filled HER2-overexpressing acini progressively protruded arms that invaded into the matrix.
To quantify the effects of EGF and HER2, we developed a morphometric assay (Supplementary Figures S1C and D). Analyses of acini confirmed that HER2 overexpression promoted transformation of circular acini into irregular structures. Yet another alteration, into arm-protruding structures, was induced by GFs. Conceivably, GFs recruit HER2 into heterodimers, to activate the kinase of the ligand-less receptor and promote invasion. In line with this scenario, covalent crosslinking experiments indicated replacement of HER2 homodimers by EGFR/HER2 heterodimers upon EGF treatment (Supplementary Figure S1E). Moreover, acini expressing a catalytically inactive HER2 mutant (D845N; Supplementary Figure S1F) exhibited cleared lumina and failed to protrude invasive arms (Supplementary Figure S1G), suggesting that HER2/EGFR heterodimers, as well as kinase activity, are required for the EGF-induced outgrowths.
Patients’ DCIS and HER2-overexpressing spheroids share proliferation-driving and apoptosis-evading gene programs
We speculated that genetic programs stimulated in MCF10A cells by an overexpressed HER2 bear relevance to DCIS and IDC. Therefore, we extracted RNA at 4 to 5 time points from acini that were grown either in the absence or in the presence of EGF, and hybridized the samples to DNA arrays (see Figure 1d). Comparison of control and HER2-overexpressing MCF10A cells, grown in the absence of EGF, identified a large set of differentially expressed genes (Supplementary Figure S2A, Supplementary File 1; sheet 1), for which enriched Gene Ontology (GO) annotations are shown in Supplementary File 2 (sheets 1 and 2) (Huang da et al., 2009). Consistent with previous reports that attributed to HER2 an ability to inhibit apoptosis while stimulating proliferation of MCF10A cells (Muthuswamy et al., 2001), enriched GO terms in a group of 490 genes, which are higher in MCF10A/HER2 cells, included primarily cell proliferation modules (Supplementary Figure S2B and Supplementary File 2; sheet 2), along with prosurvival genes, whereas a few pro-apoptosis genes were downregulated (Supplementary Figure S2C).
Conceivably, the effect of HER2 on MCF10A proliferation reflects the association of this oncogene with enhanced luminal proliferation in patients with HER2+ DCIS (van de Vijver et al., 1988). In line with this, Gene Set Enrichment Analysis (Subramanian et al., 2005) comparing normal breast epithelia and HER2+ DCIS specimens (Balleine et al., 2008) showed a significant enrichment of genes distinguishing between MCF10A and MCF10A/HER2 cells (Supplementary Figure S3A; false discovery rate (FDR) <0.1%). Moreover, a group of shared genes, exhibiting concordant changes in both MCF10A vs MCF10A/HER2 spheroids and in the comparison of normal breast epithelia vs HER2-positive DCIS, were significantly enriched for functions related to cell proliferation/survival (Supplementary Figures S3B and C, Supplementary File 2; sheet 3). In conclusion, the identification of shared profiles of mRNAs proposes that HER2-overexpressing spheroids not only phenocopy HER2+ DCIS to some extent, but that these structural similarities may also be achieved by similar transcriptional programs.
EGF alters the morphology of HER2-overexpressing spheroids and promotes breakdown of their basement membrane
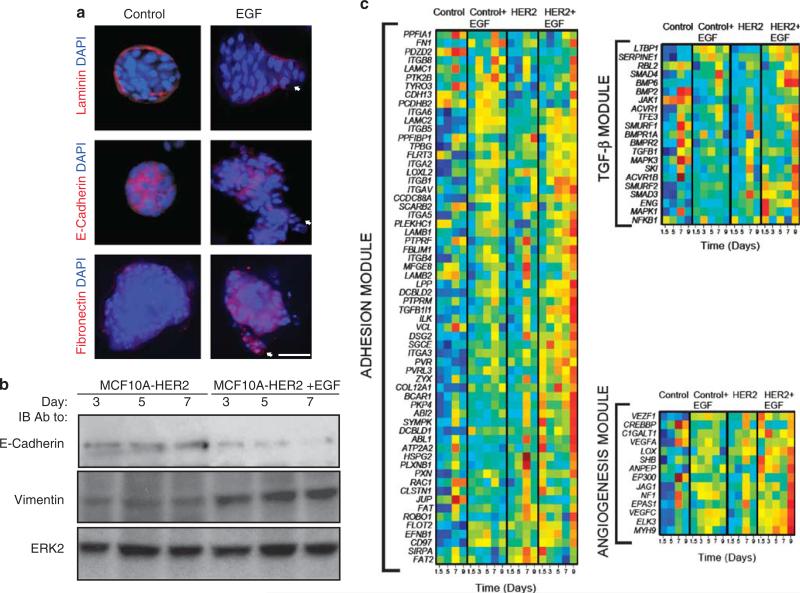
Morphological analyses revealed that unlike untreated HER2 overexpressors, GF-treated spheroids gained vimentin and fibronectin, partly lost E-cadherin, and displayed disrupted Laminin V-containing basement membranes (Figures 2a and b), in line with previous reports (Muthuswamy et al., 2001; Debnath et al., 2002; Zhan et al., 2006). To further study the effects of GFs, we cultured 5-day-old HER2-overexpressing acini with beads that were decorated with immobilized EGF molecules. Images captured 48 h later showed that acini comprising HER2-overexpressing cells extended arms toward nearby located EGF-coated beads, but control MCF10A spheroids displayed no directional outgrowths (Supplementary Figure S4). Taken together, these observations confirm the ability of GFs to regulate adhesion molecules and induce directed invasive growth of HER2-overexpressing acini.
Figure 2.
Morphological and transcriptional effects of HER2 overexpression and EGF treatment of mammary spheroids. (a) Acini of HER2-overexpressing MCF10A cells were grown for 8 days in Matrigel in the absence or presence of EGF, before immunostaining with 4,6-diamidino-2-phenylindole (DAPI) and with the indicated antibodies (scale bar, 50 μm). (b) Acini of HER2-overexpressing MCF10A cells were grown in the absence or presence of EGF for the indicated time intervals, before immunoblotting (IB) with the indicated antibodies. (c) RNA expression levels of genes whose variation of expression during acinar morphogenesis is different in untreated versus EGF-treated MCF10A/HER2 cells and in MCF10A (control) cells. Expression levels were normalized and the genes grouped according to their known functions: adhesion, BMP/TGFβ signaling and angiogenesis/response to hypoxia.
EGF-induced invasive growth of HER2-overexpressing mammary spheroids transcriptionally associates with cellular adhesion, TGFβ signaling and response to hypoxia
To resolve transcriptional modules driving invasive growth, we analyzed EGF-stimulated, HER2-overexpressing cells and identified 336 differentiating genes (Supplementary File 1, sheet 2), along with several enriched pathways and processes (Supplementary File 2; sheets 5 and 6). We focused on a group of 130 up-regulated genes, which exemplify the general ability of HER2 to augment EGF signals. Analysis of GO terms and pathway enrichment indicated that unlike the above described ontological uniformity of HER2-associated transcription, the effect of EGF was characterized by process multiplicity. The most significantly enriched pathway was transforming growth factor-β (TGFβ) signaling (Supplementary File 2; sheet 6). Of the enriched biological processes (FDR <15%; Supplementary File 2; sheet 6), we focused on angiogenesis (also called response to hypoxia) and cell adhesion, because both processes were reflected by treated acini. For each process, we consulted the appropriate database (KEGG for TGFβ signaling; GO for cell adhesion and angiogenesis), and focused on the subset of genes that exhibited notable upregulation on treatment of MCF10A/HER2 cells with EGF. Heatmaps corresponding to the three subsets of genes are presented in Figure 2c.
Transcriptomic similarities between HER2-overexpressing IDCs and EGF-treated spheroids
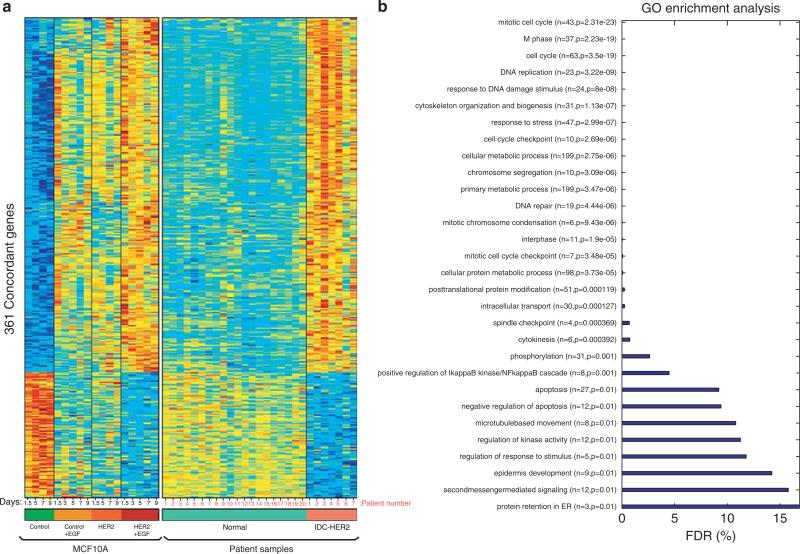
To validate the clinical relevance of the 3D model to IDCs, we analyzed a clinical gene expression data set comprising both HER2+ IDC specimens and histologically normal breast tissues (Chen et al., 2010). As expected, Gene Set Enrichment Analysis showed enrichment (FDR <25%) of genes characteristic to EGF-treated MCF10A/HER2 cells in the set of genes distinguishing HER2+ IDC samples from normal tissues. We identified 361 concordant genes distinguishing both HER2+ IDCs from normal epithelia, as well as EGF-treated MCF10A/HER2 cells from untreated MCF10A cells (Figure 3a; threshold of 5% FDR). Beyond concordant patterns of upregulated and downregulated genes, EGF alone moderately altered expression of the concordant 361 genes in MCF10A acini, but maximal effects were achieved by the combination of EGF treatment and HER2 overexpression. Interestingly, GO analysis of the concordant group revealed predominance of proliferation genes (Figure 3b and Supplementary File 2; sheet 4), but genes contributing to invasive growth were relatively rare. A likely reason may be that the invasive front of breast tumors often represents a minor fraction of clinical specimens.
Figure 3.
Transcriptomic similarities of HER2-overexpressing IDC specimens and EGF-treated invasive spheroids of MCF10A/HER2 cells. (a) Heatmaps of 361 concordant genes, which are differentially expressed in MCF10A/HER2 spheroids treated with EGF, in comparison with MCF10A spheroids, either untreated or treated with EGF for increasing time intervals. The status of expression of each concordant gene in IDC and in normal human mammary epithelia is shown in the right part. A threshold of 5% FDR was used. Note that each column represents a time point (MCF10A cells) or a patient (clinical samples). (b) Distribution of the 361 concordant genes according to their GO annotation and the respective FDR values (see Supplementary File 2).
Transcriptional programs launched by HER2 and EGF cooperatively enhance BMP/TGFβ signaling
Coordinate upregulation of bone morphogenic proteins (BMPs) and other BMP/TGFβ pathway components, along with downregulation of the antagonist pseudoreceptor BAMBI (BMP and activin membrane-bound inhibitor), was confirmed using quantitative real-time PCR (Figure 4a; refer to red arrows in Figure 4b). Consistently, immunoblotting validated sustained upregulation of the receptor BMPR2 and downregulation of BAMBI (Figure 4c), and stimulation with BMP2 induced phosphorylation of SMAD proteins in HER2 overexpressors (Figure 4d). In line with SMAD-controlled invasive growth, knockdown of SMAD4 (Figure 4e) inhibited invasiveness (Figures 4f and g). Thus, the combination of HER2 overexpression and stimulation with EGF coordinately regulates multiple components of the TGFβ/BMP module. These observations extend previous reports that identified collaborative interactions between HER2 and the TGFβ pathway in breast cancer (Seton-Rogers et al., 2004; Ueda et al., 2004).
Figure 4.
Multicomponent activation of the TGFβ/BMP module of HER2-overexpressing acini upon treatment with EGF. (a) Untreated (blue line) and EGF-treated MCF10A/HER2 cells (red line) were grown in Matrigel. Total RNA was isolated at the indicated time points, subjected to reverse transcription and analyzed by real-time PCR with primers specific to the indicated genes. (b) Schematic presentation of the transcriptionally induced TGFβ/BMP module. Narrow arrows indicate transcriptional upregulation (except for BAMBI, which is downregulated) and P letters symbolize phosphorylation. ACVR, activin receptor; BMPR, BMP receptor. (c) Immunoblot analysis was performed on HER2-overexpressing acini treated without or with EGF for the indicated time intervals, using antibodies to BMPR2 and BAMBI. (d) MCF10A/HER2 cells were grown for 10 days in Matrigel in the presence of EGF. Thereafter, EGF was removed and 24 h later cells were stimulated with BMP2 (10 ng/ml) for the indicated time intervals, before immunoblotting using an antibody specific to the phosphorylated forms of SMAD1, 5 and 8. (e) MCF10A/HER2 cells were transfected with small interfering RNA (siRNA) oligonucleotides specific to SMAD4 (or control siRNA), and grown for 48 h before immunoblotting. (f) MCF10A/HER2 cells were transfected with control siRNA or with SMAD4 siRNA, and 48 h later they were incubated for 12 h in Transwell chambers coated with Matrigel. Cells that invaded into the lower compartment were stained using crystal violet. (g) Quantification of the migration signals of the cells presented in (f).
Lysyl oxidases and other proteins associated with responses to hypoxia are induced by EGF in lumen-filled HER2-ovexpressing acini
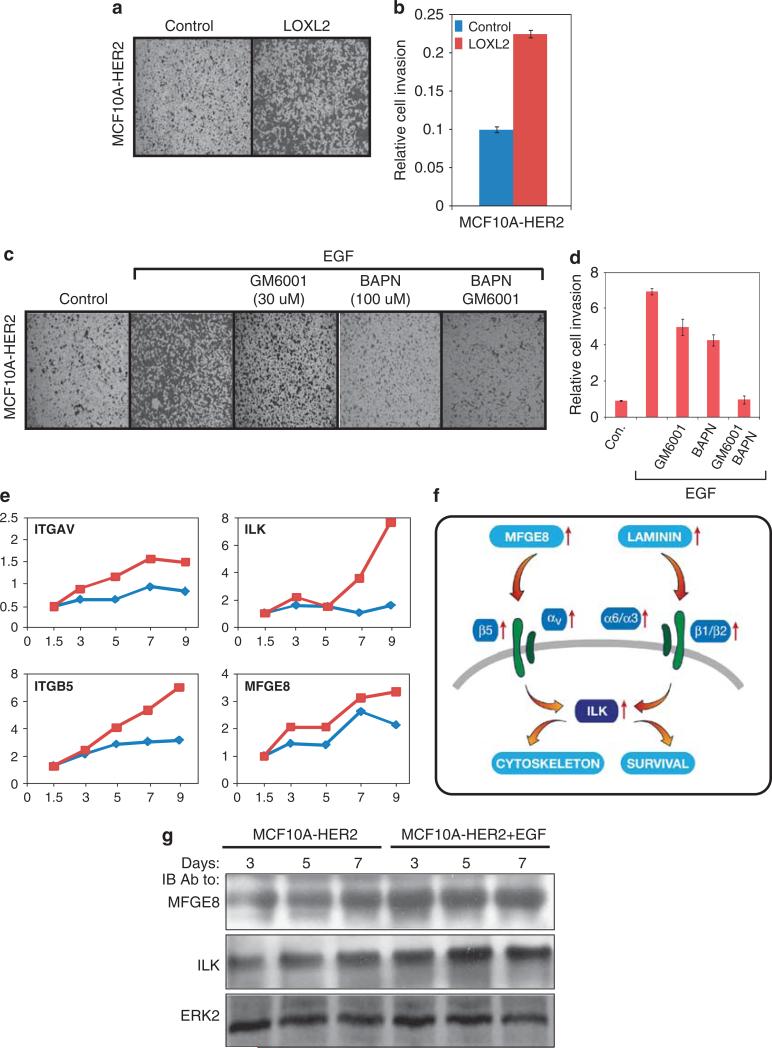
HER2 overexpression upregulated several angiogenic factors in acini of MCF10A cells, and EGF enhanced this induction (Figures 5a and 2c). The inferred angiogenesis module is presented in Figure 5b: coordinate upregulation of several components is likely because of the induction of the endothelial PAS domain protein 1 (EPAS1), a subunit of the hypoxia-inducible transcription factor-2α, the induction of which was confirmed using immunoblotting (Figure 5c). In addition, two lysyl oxidases (LOX and LOXL2), which can oxidize collagen and elastin (Akiri et al., 2003), underwent upregulation at the mRNA level, which we confirmed at the level of the LOXL2 protein (Figure 5c). As several hypoxia-inducible genes are well understood, but the oncogenic roles of the LOXs are currently emerging (Kirschmann et al., 2002; Akiri et al., 2003; Erler et al., 2006), we focused on the latter.
Figure 5.
HER2 overexpression enhances transcriptional induction of an angiogenesis/hypoxia module upon treatment with EGF. (a) Total RNA was subjected to analysis of the indicated genes by PCR, as in Figure 4a. (b) Schematic presentation of the angiogenesis module. Narrow arrows indicate transcriptional upregulation. AREG, amphiregulin. (c) Immunoblot analysis was performed on HER2-overexpressing acini treated without or with EGF for the indicated time intervals, using hypoxia-inducible transcription factor-2α (HIF2α) and LOXL2 antibodies. (d, e) MCF10A/HER2 cells were plated in Matrigel and grown for 4 days in the presence of EGF. Thereafter, the cultures were grown for 8 additional days in the absence or presence of the indicated concentrations of a lysyl oxidase inhibitor, β-aminopropionitrile (BAPN). Capturing of photos and morphometric analyses of 50 acini were performed 8 days later. (f) MCF10A/HER2 cells were plated in Matrigel in the absence or presence of a recombinant LOXL2 enzyme (1 μm), and then incubated for 18 days, before phase microscopy (top) and staining with 4,6-diamidino-2-phenylindole (DAPI) and an anti-Laminin antibody (bottom; bars, 100 μm). White arrows mark outgrowths across the Laminin shell.
Inhibition of LOX activity using β-aminopropionitrile abrogated formation of invasive acini of MCF10A/HER2 cells (Figures 5d and e) without affecting cell proliferation, implying that secretion of LOXs is required for invasiveness, probably because these enzymes control matrix stiffness (Levental et al., 2009). Accordingly, treatment of MCF10A/HER2 cells with a recombinant LOXL2 induced their invasive outgrowths (Figure 5f), even in the absence of EGF, as well as enhanced invasion through a filter coated with collagen type I (Figures 6a and b). To address potential collaboration between LOXs and matrix metalloproteinases, we combined β-aminopropionitrile and a general inhibitor of matrix metalloproteinases (GM6001), and observed complete inhibition of invasion of MCF10A/HER2 cells (Figures 6c and d). In aggregate, these observations propose that EGF-induced activation of an angiogenesis-like switch contribute to the invasive potential of HER2-overexpressing acini.
Figure 6.
LOXL2 and secreted integrin ligands are involved in the enhanced migratory response of HER2-overexpressing cells to EGF. (a, b) MCF10A/HER2 cells (50 000 cells) were plated on type I collagen-coated Transwell chambers and then incubated for 24 h in the absence or presence of recombinant LOXL2 enzyme (1 μm). Cells that invaded into the lower chamber of the Transwell membrane were stained using crystal violet, photographed and relative cell invasion quantified in triplicates. (c, d) MCF10A and MCF10A/HER2 cells (50 000 cells) were plated on type-I collagen-coated Transwell chambers and incubated for 24 h with EGF in the absence or presence of a general matrix metalloproteinase (MMP) inhibitor (GM6001) or a LOX inhibitor (β-aminopropionitrile (BAPN)). Cells that invaded into the lower part of the Transwell chamber were stained using crystal violet, and relative cell invasion was quantified in triplicates. (e) Total RNA was subjected to analysis of the indicated genes by PCR, as in Figure 4a. (f) A scheme showing functional relationships within the upregulated matrix-adhesion module. Narrow red arrows indicate transcriptional upregulation. (g) Immunoblot analysis was performed on HER2-overexpressing acini treated without or with EGF for the indicated time intervals, using MFGE8 or ILK antibodies.
The combination of HER2 overexpression and EGF treatment enhances expression of a large module of cell adhesion mediators
EGF signaling has previously been implicated in evasion from anoikis, a form of programmed cell death associated with disturbed integrin signaling (Collins et al., 2005). In accordance, we found that the combination of EGF treatment and HER2 overexpression strongly induced more than 60 adhesion-related genes, including the integrin-linked kinase (ILK), a wide spectrum of integrins (for example, αV, α2, α3, α5, α6, β1, β4, β5 and β8) and their respective ligands, such as Laminin and Milk Fat Globule EGF factor 8 (MFGE8/lactadherin; Figure 2c). Notably, it is known that β4 signaling is necessary for HER2-initiated mammary tumorigenesis (Guo et al., 2006), and ILK plays a critical role in progression of such tumors (Pontier et al., 2010). Transcriptional induction of ILK, MFGE8, αV and β5 was confirmed by using quantitative real-time PCR (Figure 6e; see scheme in Figure 6f), and increased levels of ILK and MFGE8 were also verified by immunoblotting (Figure 6g).
Secretion of MFGE8, which binds heterodimers of integrin αV and integrin β5 or β3 and activates ERK, is required for branching morphogenesis of the mammary gland (Ensslin and Shur, 2007). In line with this report, we found that exogenous MFGE8 activates ERK phosphorylation (Figure 7a) and enhances invasiveness of HER2-overexpressing cells (Figures 7b and c). Moreover, exogenously added MFGE8 specifically enhanced formation of invasive outgrowths by acini of HER2-overexpressing spheroids (Figure 7d). As expected, inhibition of MFGE8 action by using an integrin αV blocking antibody attenuated invasive growth (Figure 7d), and stable knockdown of MFGE8 (Figure 7e) inhibited the EGF-induced formation of invasive structures (Figures 7f and g). These effects of MFGE8 were extended to HER2-overexpressing human breast cancer cells grown in Matrigel. EGF treatment of SKBR3 and BT474 cells increased the size, irregularity and branching of their acini, but MFGE8-blocking antibodies (to integrin αV) or a LOX inhibitor reversed this phenotype, similar to the effect of the HER2-specific antibody trastuzumab and the EGFR/HER2 dual kinase inhibitor GW2974 (Supplementary Figure S5). In conclusion, transcriptional induction of specific ligands and their cognate integrins is essential for EGF-induced invasion of HER2-overexpressing mammary cells.
Figure 7.
Effects of MFGE8 on invasion of HER2-overexpressing mammary cells. (a) MCF10A and MCF10A/HER2 cells were serum starved for 12 h, stimulated for 10 min with MFGE8 (100 ng/ml) and cell extracts immunoblotted with the indicated antibodies, including an antibody the phosphorylated form of ERK. (b, c) MCF10A/HER2 cells (50 000 cells) were plated on Matrigel-coated Transwell chambers and incubated for 12 h in the absence or presence of EGF (20 ng/ml) and/or MFGE8 (100 ng/ml). Cells that invaded into the lower side of the Transwell chambers were stained using crystal violet, photographed and counted. (c) Quantification of the cell migration data shown in (b). Bars represent s.d. of triplicates. (d) Phase microscopy analysis of MCF10A/HER2 acini plated in Matrigel and grown for 10 days in the absence or presence of EGF (20 ng/ml), ectopic MFGE8 (10 ng/ml) or a blocking antibody to integrin αV (bar, 50 μm). (e) Monolayers of MCF10A/HER2 cells were stably infected with short hairpin RNA (shRNA) particles, either control (C) or shRNA targeting MFGE8 (clones 74, 77 and 78). Cell clones were tested for MFGE8 expression using immunoblotting. (f, g) MCF10A/HER2 cells expressing the indicated shRNAs were cultured for 10 days in Matrigel. Phase contrast micrographs are shown (bar, 50 μm), along with morphometric analysis of acini. Shown are averages±s.d. values of triplicates.
An in vitro-based cumulative gene signature predicts survival of HER2-positive breast cancer patients
Our results demonstrated that coexpression of genes belonging to three transcriptional modules, including several genes known to precondition and nourish the metastatic niche (for example, LOX and vascular endothelial growth factor), accompanies the acquisition and maintenance of an invasive phenotype in vitro. Assuming that persistent expression of this molecular switch critically determines the clinical outcome of patients with early-stage invasive breast cancer, we evaluated the prognostic value of these genes by using nine publicly available microarray data sets as a training set (Supplementary File 3; sheet 1). After excluding patients who received systemic adjuvant treatment, genes from all three modules were individually assessed through a Cox model with relapse-free survival (RFS) in the HER2 subtype (Supplementary File 3; sheet 2), and 25 genes of high prognostic value (P<0.1) were selected (Supplementary File 3; sheet 3) as a combined signature (hereafter HER2/EGF signature). This signature provided a strong predictor of RFS in patients with HER2+ tumors only (Figure 8a; upper row). Performance of the signature was evaluated in an independent test set comprising 344 patients who received no systemic adjuvant therapy (Wang et al., 2005; Minn et al., 2007). This analysis corroborated the ability of the HER2/EGF signature of 25 genes to discriminate patients with a favorable outcome from those with high relapse rates, specifically in the HER2+ subtype (hazard ratio = 4.1, 95% confidence interval: 1.1–14.5, P = 0.03; see Figure 8a; lower row).
Figure 8.
An invasive signature predicts survival of HER2-overexpressing patients. (a) Kaplan–Meier analyses of RFS of breast cancer patients grouped into three major subtypes, according to ER and HER2 status. Nine breast cancer microarray data sets were used as a training set (upper row), along with an independent test set (lower row; VDX data set; Supplementary File 3, sheet 1). Tumors were stratified according to high (red), medium (green) or low (blue) expression of the HER2-associated invasive signature comprising 25 genes (Supplementary File 3, sheet 3). Hazard ratios (HRs; average and 95% confidence interval), patient numbers and P-values are indicated. Note that the prognostic value of the HER2/EGF signature is confined to the HER2 subtype. (b) Forest plots summarizing HRs of relapse in the test set of breast cancer patients (VDX data set). The area of each square and its arms respectively represent the number of patients and the 95% confidence intervals corresponding to the listed clinical parameters (top part) and published gene signatures (lower part), including the HER2/EGF signature (highlighted bar) we describe herein. Note that the red boxes indicate statistical significance and that the number of relapse events in low-grade ER–/HER– tumors was too small for analysis.
Using the test set, we compared the prognostic performance of the HER2/EGF signature relative to both clinical parameters and previously published gene signatures (Figure 8b) for each breast tumor subtype. As expected, the resulting Forest plots indicated that several first-generation signatures, which were developed to derive prognostic guidance in patients with ER-positive breast cancers, retained prognostic significance in the ER+/HER2– group, but none performed well in the ER–/HER2– subtype (Figure 8b). In contrast, HER2/EGF along with three recent gene signatures, reliably predicted RFS in the HER2+ group, outperforming not only histopathological characteristics, but also the first-generation signatures. In conjunction with the invasive phenotype that we observed in vitro, the prognostic power of the HER2/EGF signature supports a two-hit progression model for HER2-overexpressing lesions, as we discuss below.
Discussion
HER2 is amplified in a large fraction of DCIS cases, but only 20–25% of IDCs exhibit overexpression (Slamon et al., 1987; van de Vijver et al., 1988). These observations established the notion that HER2 acts as a first hit, which is followed by a second, invasion-promoting hit affecting only a fraction of DCIS lesions. Previous reports proposed that TGFβ (Seton-Rogers et al., 2004) and 14-3-3-ζ (Lu et al., 2009) serve as second hits. Similarly, a transgenic mouse model indicated that HER2 and TGFα cooperate in mammary tumorigenesis (Muller et al., 1996). On the basis of an in vitro 3D cellular system and clinical data, the present study identifies EGF-like factors as a potential second hit.
The following sequence of events may provide biochemical grounds for the proposed two-hit model:
(i) HER2 induces proliferation and evasion from apoptosis
Normally, mammary ducts acquire cavities through cell divisions with the metaphase plates perpendicular to the apical surface (Jechlinger et al., 2009), and by disengagement of inner cell layers from the basement membrane (Simpson et al., 2008). We found that overexpression of HER2 endows cells with transcriptional attributes that evade luminal apoptosis, in line with a previous report (Debnath et al., 2002). Concomitantly, HER2 signaling disrupts apical–basal polarity (Figure 2a), likely by associating with components of the PAR complex (Aranda et al., 2006).
(ii) GFs enhance an intraluminal response to hypoxia
It is conceivable that decreased intraluminal oxygen levels within filled structures initiate a rudimentary hypoxic reaction, which is exaggerated by GFs (Figure 5b). Moreover, hypoxia is known to enhance metastatic potential through the induction of matrix-modifying enzymes, which increase matrix stiffness (Erler and Weaver, 2009; Levental et al., 2009).
(iii) Collaborative induction of invasive growth
Neither HER2 nor EGF can launch invasion across basement membranes, but their collaboration is very effective. The underlying mechanisms likely require amplification of EGF-induced intracellular signals and coordinate activation of the BMP/TGFβ, angiogenesis and integrin modules. Congruently, ectopic expression of TGFβ in MCF10A cells expressing activated HER2 strongly induced migration (Seton-Rogers et al., 2004; Ueda et al., 2004).
Based on results obtained with the 3D model system and their reflection in clinical outcome (Figure 8a), we propose that neither HER2 amplification nor the presence of GFs is sufficient for development of IDCs, but their co-occurrence can instigate metastasis. According to the proposed model, expansion of foci of ductal hyperplasia is limited by intraluminal apoptosis, unless they overexpress HER2, which drives proliferation and forms DCIS. The more virulent scenario combines HER2 amplification with GFs, thereby switching a robust, autostimulatory program. This model predicts that exposure to GFs can identify a relatively aggressive class of HER2-overexpressing tumors. Such GFs may derive from autocrine secretion, or from paracrine sources including stromal cells, surgical wounds (Tagliabue et al., 2003) and tumor-associated myeloid cells (Rilke et al., 1991).
Although it is based on an in vitro cell-stroma system, the predictive power of the HER2/EGF signature exceeds the prognostic value of clinical parameters such as tumor size and grade, but it remains restricted to patients belonging to the HER2+ group (Figure 8b). In addition, the newly defined signature outshines all previously described gene signatures, except two good prognosis immune signatures (STAT1 and IRMODULE) (Teschendorff et al., 2007; Desmedt et al., 2008a) and the Decorin (DCN) signature (Farmer et al., 2009). Future studies will examine the possibility that GFs can serve as the long-awaited prognostic markers of high-risk DCIS lesions. Already, the gene signature we identified provides a novel tool to predict prognosis in patients with HER2-overexpressing IDCs, based upon on-going expression of an invasive program. Whether this signature can also identify therapeutic targets in patients with HER2-positive breast cancers is an intriguing possibility that requires further investigation.
Materials and methods
Reagents, cells and buffers
MCF-10A cells were obtained from the ATCC (Manassas, VA, USA) and maintained in medium containing 10 μg/ml insulin, 0.1 μg/ml cholera toxin, 0.5 μg/ml hydrocortisone, 5% horse serum and 10 ng/ml EGF. Recombinant MFGE8 was from R&D Systems (Minneapolis, MN, USA).
Retroviral infection
pBMN-HER2-IRES-EGFP (from Carlos Arteaga, Nashville, TN, USA) was co-transfected with a retroviral packaging plasmid pSV-ψ-env-MLV into 293T retrovirus-packaging cells using FuGENE (Roche Applied Science, Indianapolis, IN, USA). Cells were transduced with virus-containing medium 72 h later and after 5 passages cells stably expressing GFP were sorted by flow cytometry.
Cell proliferation assay
The MTT (3-(4,5-dimethylthiazol-z-yl)-2,5-diphenyl tetrazolium bromide) was used as previously described (Pinkas-Kramarski et al., 1996).
Morphogenesis assay
Trypsinized cells were re-suspended in Dulbecco's modified eagle (DME)/F12 medium supplemented with 2% horse serum, insulin, cholera toxin and hydrocortisone. Eight-chambered plates (BD Biosciences) were coated with 35 μl growth factor reduced Matrigel (BD Bioscience) per well. The cells were mixed 1:1 with assay medium containing Matrigel (4%) and EGF (20 ng/ml), and 400 μl added to each chamber.
Indirect immunofluorescence
Acini were fixed in methanol–acetone and slides blocked in goat serum (10%) containing buffer. Secondary blocking was performed in buffer containing goat anti-mouse F(ab′)2 fragments (20 μg/ml). Primary antibodies were incubated 15–18 h at 4 °C and secondary antibodies for 1 h. Confocal microscopy was performed using Bio-Rad Radiance 2000 microscope (Bio-Rad, Oberkochen, Germany).
Quantitative PCR and oligonucleotide microarrays
RNA was isolated using a Versagene kit (Gentra Systems, Minneapolis, MN, USA). Complementary DNA was generated using Invitrogen SuperScriptII (Invitrogen, Carlsbad, CA, USA). Real-time PCR was performed using SYBR Green I (Carlsbad, CA, USA). All experiments were carried out in triplicates, and results were normalized to β2 microglobulin. For microarrays, 1.0 μg RNA was labeled, fragmented and hybridized to Affymetrix HuGENE 1.0 ST oligonucleotide arrays (Applied Biosystems, Foster City, CA, USA). Primer sequences are listed under Supplementary File 3 (sheet 4).
Microarray data analysis
CEL files were normalized according to the ‘sketch quantile RMA’ algorithm (Affymetrix Expression Console). The value 5 (log2 scale) served as threshold. Because of technical reasons we excluded samples from day 3 of the EGF-untreated group. Pairwise comparisons consisted of two tests. (1) Find (t-test; FDR 5%) genes whose average expression in the experiments involving the two cell types was different. (2) Identify genes whose expression exhibited significant variation over time. (3) Differentiating genes are the union of the two lists obtained by (1) and (2).
Analysis of public gene expression data sets
Ten breast cancer microarray data sets were used. Hybridization probes were mapped to Entrez GeneID as described (Shi et al., 2006). RFS was used as an end point. If RFS was not available, we used distant metastasis-free survival. We censored the survival data at 10 years to have comparable follow-up (Haibe-Kains et al., 2008; Desmedt et al., 2008a). Molecular subtypes were defined by model-based clustering in a two-dimensional space defined by ESR1 (ER) and ERBB2 (HER2) scores (Desmedt et al., 2008b).
Development of the HER2/EGF signature
The HER2/EGF metastasis index was computed for each sample:
where S is the set of genes used to compute the index, and xi is the expression of a gene included in S. Genes in S were identified if they were significantly associated with prognosis in the HER2+ subgroup (training set). Each index was rescaled such that quantiles 2.5 and 97.5% are equaled to –1 and +1 respectively. We estimsted hazard ratios using Cox regression (Cox, 1972). For ternary classification of individual gene expressions, signature scores or the HER2 metastasis index, the cutoffs were selected to be the tertiles in each data set and in each subtype separately. The resulting ternary classification was encoded as 0, 0.5 and 1 for the low, intermediate and high groups, respectively. The Kaplan–Meier product-limit estimator was used to display time to event curves based on this classification. Log-rank tests were used to test differences between curves (Therneau and Grambsch, 2000). Sensitivity and specificity for binary classifications were estimated through the nearest neighbor estimation method used in time-dependent receiver operating characteristic curves (Heagerty et al., 2000).
Supplementary Material
Acknowledgements
We thank Drs Carlos Arteaga and Ittai Ben-Porath for reagents and helpful insights. Our laboratories are supported by research grants from the National Cancer Institute (Grant CA72981), the M.D. Moross Institute for Cancer Research, Kekst Family Institute for Medical Genetics, Women's Health Research Center funded by Bennett-Pritzker Endowment Fund, Marvelle Koffler Program for Breast Cancer Research, Harry and Jeanette Weinberg Women's Health Research Endowment, Oprah Winfrey Biomedical Research Fund, Arresto Biosciences, the European Commission, and the German Research Foundation. YY is the incumbent of the Harold and Zelda Goldenberg Professorial Chair. ED is the incumbent of the Henry J Leir Professorial Chair.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Akiri G, Sabo E, Dafni H, Vadasz Z, Kartvelishvily Y, Gan N, et al. Lysyl oxidase-related protein-1 promotes tumor fibrosis and tumor progression in vivo. Cancer Res. 2003;63:1657–1666. [PubMed] [Google Scholar]
- Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, et al. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat Cell Biol. 2006;8:1235–1245. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]
- Balleine RL, Webster LR, Davis S, Salisbury EL, Palazzo JP, Schwartz GF, et al. Molecular grading of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14:8244–8252. doi: 10.1158/1078-0432.CCR-08-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DT, Nasir A, Culhane A, Venkataramu C, Fulp W, Rubio R, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat. 2010;119:335–346. doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NL, Reginato MJ, Paulus JK, Sgroi DC, Labaer J, Brugge JS. G1/S cell cycle arrest provides anoikis resistance through Erk-mediated Bim suppression. Mol Cell Biol. 2005;25:5282–5291. doi: 10.1128/MCB.25.12.5282-5291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR. Regression models and life tables. J R Statist Soc B. 1972;4:102–118. [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008a;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- Desmedt C, Ruiz-Garcia E, Andre F. Gene expression predictors in breast cancer: current status, limitations and perspectives. Eur J Cancer. 2008b;44:2714–2720. doi: 10.1016/j.ejca.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Ensslin MA, Shur BD. The EGF repeat and discoidin domain protein, SED1/MFG-E8, is required for mammary gland branching morphogenesis. Proc Natl Acad Sci USA. 2007;104:2715–2720. doi: 10.1073/pnas.0610296104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- Erler JT, Weaver VM. Three-dimensional context regulation of metastasis. Clin Exp Metastasis. 2009;26:35–49. doi: 10.1007/s10585-008-9209-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- Guo W, Pylayeva Y, Pepe A, Yoshioka T, Muller WJ, Inghirami G, et al. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell. 2006;126:489–502. doi: 10.1016/j.cell.2006.05.047. [DOI] [PubMed] [Google Scholar]
- Haibe-Kains B, Desmedt C, Piette F, Buyse M, Cardoso F, Van't Veer L, et al. Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics. 2008;9:394. doi: 10.1186/1471-2164-9-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;65:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jechlinger M, Podsypanina K, Varmus H. Regulation of transgenes in three-dimensional cultures of primary mouse mam-mary cells demonstrates oncogene dependence and identifies cells that survive deinduction. Genes Dev. 2009;23:1677–1688. doi: 10.1101/gad.1801809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschmann DA, Seftor EA, Fong SF, Nieva DR, Sullivan CM, Edwards EM, et al. A molecular role for lysyl oxidase in breast cancer invasion. Cancer Res. 2002;62:4478–4483. [PubMed] [Google Scholar]
- Klapper LN, Glathe S, Vaisman N, Hynes NE, Andrews GC, Sela M, et al. The ErbB-2/HER2 oncoprotein of human carcinomas may function solely as a shared coreceptor for multiple stroma-derived growth factors. Proc Natl Acad Sci USA. 1999;96:4995–5000. doi: 10.1073/pnas.96.9.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardo F, Di Marco E, King CR, Pierce JH, Segatto O, Aaronson SA, et al. The normal erbB-2 product is an atypical receptor-like tyrosine kinase with constitutive activity in the absence of ligand. New Biol. 1990;2:992–1003. [PubMed] [Google Scholar]
- Lu J, Guo H, Treekitkarnmongkol W, Li P, Zhang J, Shi B, et al. 14-3-3zeta Cooperates with ErbB2 to promote ductal carcinoma in situ progression to invasive breast cancer by inducing epithelial-mesenchymal transition. Cancer Cell. 2009;16:195–207. doi: 10.1016/j.ccr.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Padua D, Bos P, Nguyen DX, Nuyten D, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci USA. 2007;104:6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller WJ, Arteaga CL, Muthuswamy SK, Siegel PM, Webster MA, Cardiff RD, et al. Synergistic interaction of the Neu proto-oncogene product and transforming growth factor alpha in the mammary epithelium of transgenic mice. Mol Cell Biol. 1996;16:5726–5736. doi: 10.1128/mcb.16.10.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- Pontier SM, Huck L, White DE, Rayment J, Sanguin-Gendreau V, Hennessy B, et al. Integrin-linked kinase has a critical role in ErbB2 mammary tumor progression: implications for human breast cancer. Oncogene. 2010;29:3374–3385. doi: 10.1038/onc.2010.86. [DOI] [PubMed] [Google Scholar]
- Riese DJ, II, van Raaij TM, Plowman GD, Andrews GC, Stern DF. The cellular response to neuregulins is governed by complex interactions of the erbB receptor family. Mol Cell Biol. 1995;15:5770–5776. doi: 10.1128/mcb.15.10.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilke F, Colnaghi MI, Cascinelli N, Andreola S, Baldini MT, Bufalino R, et al. Prognostic significance of HER-2/neu expression in breast cancer and its relationship to other prognostic factors. Int J Cancer. 1991;49:44–49. doi: 10.1002/ijc.2910490109. [DOI] [PubMed] [Google Scholar]
- Seton-Rogers SE, Lu Y, Hines LM, Koundinya M, LaBaer J, Muthuswamy SK, et al. Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc Natl Acad Sci USA. 2004;101:1257–1262. doi: 10.1073/pnas.0308090100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett. 2008;272:177–185. doi: 10.1016/j.canlet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Smigal C, Jemal A, Ward E, Cokkinides V, Smith R, Howe HL, et al. Trends in breast cancer by race and ethnicity: update 2006. CA Cancer J Clin. 2006;56:168–183. doi: 10.3322/canjclin.56.3.168. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabue E, Agresti R, Carcangiu ML, Ghirelli C, Morelli D, Campiglio M, et al. Role of HER2 in wound-induced breast carcinoma proliferation. Lancet. 2003;362:527–533. doi: 10.1016/S0140-6736(03)14112-8. [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer Verlag; New York: 2000. [Google Scholar]
- Ueda Y, Wang S, Dumont N, Yi JY, Koh Y, Arteaga CL. Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor beta-induced cell motility. J Biol Chem. 2004;279:24505–24513. doi: 10.1074/jbc.M400081200. [DOI] [PubMed] [Google Scholar]
- Valabrega G, Montemurro F, Sarotto I, Petrelli A, Rubini P, Tacchetti C, et al. TGFalpha expression impairs Trastuzumab-induced HER2 downregulation. Oncogene. 2005;24:3002–3010. doi: 10.1038/sj.onc.1208478. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, Peterse JL, Mooi WJ, Wisman P, Lomans J, Dalesio O, et al. Neu-protein overexpression in breast cancer. Association with comedo-type ductal carcinoma in situ and limited prognostic value in stage II breast cancer. N Engl J Med. 1988;319:1239–1245. doi: 10.1056/NEJM198811103191902. [DOI] [PubMed] [Google Scholar]
- Virolle T, Krones-Herzig A, Baron V, De Gregorio G, Adamson ED, Mercola D. Egr1 promotes growth and survival of prostate cancer cells. Identification of novel Egr1 target genes. J Biol Chem. 2003;278:11802–11810. doi: 10.1074/jbc.M210279200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Zhan L, Xiang B, Muthuswamy SK. Controlled activation of ErbB1/ErbB2 heterodimers promote invasion of three-dimensional organized epithelia in an ErbB1-dependent manner: implications for progression of ErbB2-overexpressing tumors. Cancer Res. 2006;66:5201–5208. doi: 10.1158/0008-5472.CAN-05-4081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.