Abstract
How regulatory information is encoded in the genome is poorly understood and poses a challenge when studying biological processes. We demonstrate here that genomic redistribution of Oct4 by alternative partnering with Sox2 and Sox17 is a fundamental regulatory event of endodermal specification. We show that Sox17 partners with Oct4 and binds to a unique ‘compressed’ Sox/Oct motif that earmarks endodermal genes. This is in contrast to the pluripotent state where Oct4 selectively partners with Sox2 at ‘canonical’ binding sites. The distinct selection of binding sites by alternative Sox/Oct partnering is underscored by our demonstration that rationally point-mutated Sox17 partners with Oct4 on pluripotency genes earmarked by the canonical Sox/Oct motif. In an endodermal differentiation assay, we demonstrate that the compressed motif is required for proper expression of endodermal genes. Evidently, Oct4 drives alternative developmental programs by switching Sox partners that affects enhancer selection, leading to either an endodermal or pluripotent cell fate. This work provides insights in understanding cell fate transcriptional regulation by highlighting the direct link between the DNA sequence of an enhancer and a developmental outcome.
Keywords: endoderm, enhancer code, lineage specification, pluripotency, Sox/Oct interaction
Introduction
The bulk of mammalian genomes consists of non-coding regions whose function is only poorly annotated despite extensive research (Birney et al, 2007; Gerstein et al, 2010; Ernst et al, 2011). An important portion of the non-coding genome encodes for enhancers that recruit sequence-specific transcription factors (TFs), which in turn trigger changes in the chromatin state, recruit co-activators, and therefore regulate the expression of nearby genes. However, the catalogue of functionally characterized enhancers is slim and the mechanism of enhancer function remains unclear. Specifically, ‘the enhancer code’, the sequence features within enhancers that determine TF recruitment and thereby gene expression and developmental programs, is not well understood. It is believed that TFs partner to combinatorially execute gene expression programs. However, it is debated whether such partnerships require direct and cooperative protein–protein interactions (Wilczynski and Furlong, 2010; Biggin, 2011; Mirny, 2011). If TFs physically cooperate, then the resulting protein complex would target different genomic loci than individual proteins in the absence of interaction partners. Such co-selectivity would have profound consequences for the regulatory output of TFs. Several genomic, biochemical, and structural studies point to direct TF interactions and indicate that such partnerships can alter the genomic binding and even the sequence specificity of the participating factors (Hollenhorst et al, 2009; Slattery et al, 2011). However, a system-wide enquiry on how the genome binding and regulatory role of TFs changes in the presence of alternative partner factors has been missing. To this end, we studied the plasticity of Sox and Oct TF partnerships during lineage commitment in early mammalian development.
The Sox and Oct proteins comprise TF families of 20 and 14 members, respectively, and individual members function in a wide range of biological processes (Phillips and Luisi, 2000; Kondoh and Kamachi, 2010; Bergsland et al, 2011). Several Sox/Oct partnerships have been described in different biological contexts (Kuhlbrodt et al, 1998; Tanaka et al, 2004; Stefanovic et al, 2009); the synergistic action of Sox2 and Oct4 in pluripotent stem cells being the most prominent example of a functionally critical Sox/Oct partnership (Nishimoto et al, 1999; Tomioka et al, 2002; Boyer et al, 2005). Other well-characterized examples are the Sox2/Brn2 partnership during neural development and the Sox11/Brn1 interaction that dictates glial cell identity (Kuhlbrodt et al, 1998; Tanaka et al, 2004). These observations led to the hypothesis that a Sox/Oct partner code underlies many critical cell fate choices (Wilson and Koopman, 2002; Kondoh and Kamachi, 2010).
With the aim of better understanding and identifying Sox/Oct partner codes in pluripotency and endodermal contexts, we investigated the interaction of Oct4 with Sox2 and Sox17 on specific DNA enhancers. Oct4 partnership with Sox2 in embryonic stem cells (ESCs) is well established. However, there are reports suggesting that Oct4 might also be involved in endodermal development, but its mechanisms of action are not yet understood (Niwa et al, 2000). Sox17 is a TF well known to play an important role in both primitive (PrE) and definitive (DE) endoderm identity (Seguin et al, 2008; Morris et al, 2010; Niakan et al, 2010). Interestingly, single-cell gene expression analysis of embryos showed expression of Oct4 in PrE cells at comparable levels as in the pluripotent epiblast (Kurimoto et al, 2006; Guo et al, 2010). Consequently, a state exists in the nascent inner cell mass of the blastocyst, prior to the epiblast and PrE formation, where Sox2, Sox17, and Pou5f1 (aka Oct4) are co-expressed in the same cell (Guo et al, 2010), thereby providing potential competition between Sox2 and Sox17 for Oct4 binding, which would influence subsequent cell lineage decisions. Lineage segregation in the ICM during preimplantation of mouse embryos is dependent on several critical genetic and epigenetic events. The mechanisms that initiate and control these processes are currently central topics of investigation in developmental biology and despite extensive research, very little is known. Indeed, the search for the regulatory mechanisms involved in the control of the earliest stages of mouse development has so far resulted in very few candidates.
In this study, using a genome-wide ChIP (chromatin immunoprecipitation)-sequencing approach we establish that in a pluripotency context, Oct4 binds with Sox2 on a distinctive canonical motif, whereas when Sox17 is introduced into ESCs, Oct4 switches partners and interacts with Sox17 on a compressed motif leading to the induction of a specific endodermal differentiation program. Moreover, we demonstrate that a reengineered Sox17 factor (Jauch et al, 2011) lacks its preference for the compressed motif and thereby redistributes its binding from the compressed to the canonical sites. Apparently, this genomic redistribution turns Sox17EK into a potent inducer of pluripotency. Conversely, the point-mutated Sox2KE has lost its preference for the canonical motif. Moreover, using an in vitro model of PrE development, we show that Oct4 is necessary for PrE induction and that its interaction with Sox17 initiates this specific cell fate choice. Finally, we show that genes with a canonical or compressed motif are expressed in the ICM of late mouse blastocyt where both the EPI and the PrE specifications are occurring. We therefore identified genes known to be important for PrE cell identity but also new candidate genes that will help us to better understand lineage segregation in early mouse preimplantation development.
This work helps in our understanding of cell fate transcriptional regulation and suggests a new partner code for Sox/Oct, whereby the recruitment of Sox17/Oct4 to a compressed Sox/Oct motif specifies the endoderm fate.
Results
Sox2 and Sox17 select disparate genomic loci by selective partnering with Oct4 on canonical and compressed DNA motifs
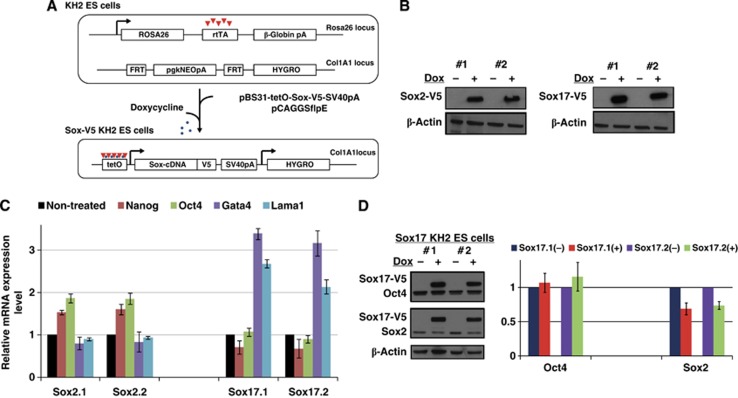
To establish that the genomic binding profile of Oct4 depends on which Sox partner is present in cells, and to show that different Sox/Oct combinations co-select distinctive sets of target genes earmarked by specific composite motifs, we generated ESC lines expressing, epitope-tagged (V5) Sox2 and Sox17 transgenes using a doxycycline-inducible cell line (Beard et al, 2006; Figure 1A). Treatment of ESCs with doxycycline for 48 h induced expression of Sox2-V5 and Sox17-V5 proteins (Figure 1B) and mRNA levels (Supplementary Figure S1). Using quantitative RT–PCR, we observed the upregulation of Nanog and Oct4 in Sox2-V5 expressing cells and the upregulation of Gata4 and Lama1 in Sox17-V5 expressing cells, validating that the transgenic Sox proteins potently stimulate the expression of specific lineage markers (Figure 1C). Genome-wide transcriptional analysis of Sox17-V5 cells showed an increase in the expression of several extra-embryonic endodermal genes including Col4a1, Col4a2, Gata6, and Pdgfra (Supplementary Table S1). To assess how Oct4 levels respond to elevated Sox17 expression, we performed western blots on these cells and found that Oct4 protein levels remained unchanged 48 h after Sox17 induction (Figure 1D). By contrast, Sox2 protein levels decreased slightly (Figure 1D). These data established that 48 h induction was suitable to compare the genomic binding profiles of different Sox/Oct combinations in transitory states occurring early in pluripotent cell differentiation.
Figure 1.
Induced expression of Sox2 and Sox17 in ESC lines. (A) Schematic of the KH2-inducible vector system used to conditionally express epitoge (V5)-tagged Sox proteins in ESCs (FRT (Flipase Recognition Target), pA (Polyadenylation signal), tetO (tetracycline/doxycycline Operator)). (B) Anti-V5 western blot showing the expression of the V5-tagged Sox2 and Sox17 proteins in two independent cell clones treated with (+) or without (−) doxycycline for 48 h. (C) Quantitative RT–PCR of Nanog, Oct4, Gata4 and Lama1 in Sox2- and Sox17-inducible ESCs treated with doxycycline for 48 h in two independent clones. (D) Western blot of Sox17-V5, Oct4 and Sox2 in two Sox17-V5 expressing ESC lines and its quantification for Oct4 and Sox2. Proteins were (i) first incubated with the V5 and the Oct4 antibodies (upper panel), (ii) then the membrane was reprobed with the V5 and Sox2 antibodies (middle panel) and (iii) finally with the β-actin antibody (lower panel).
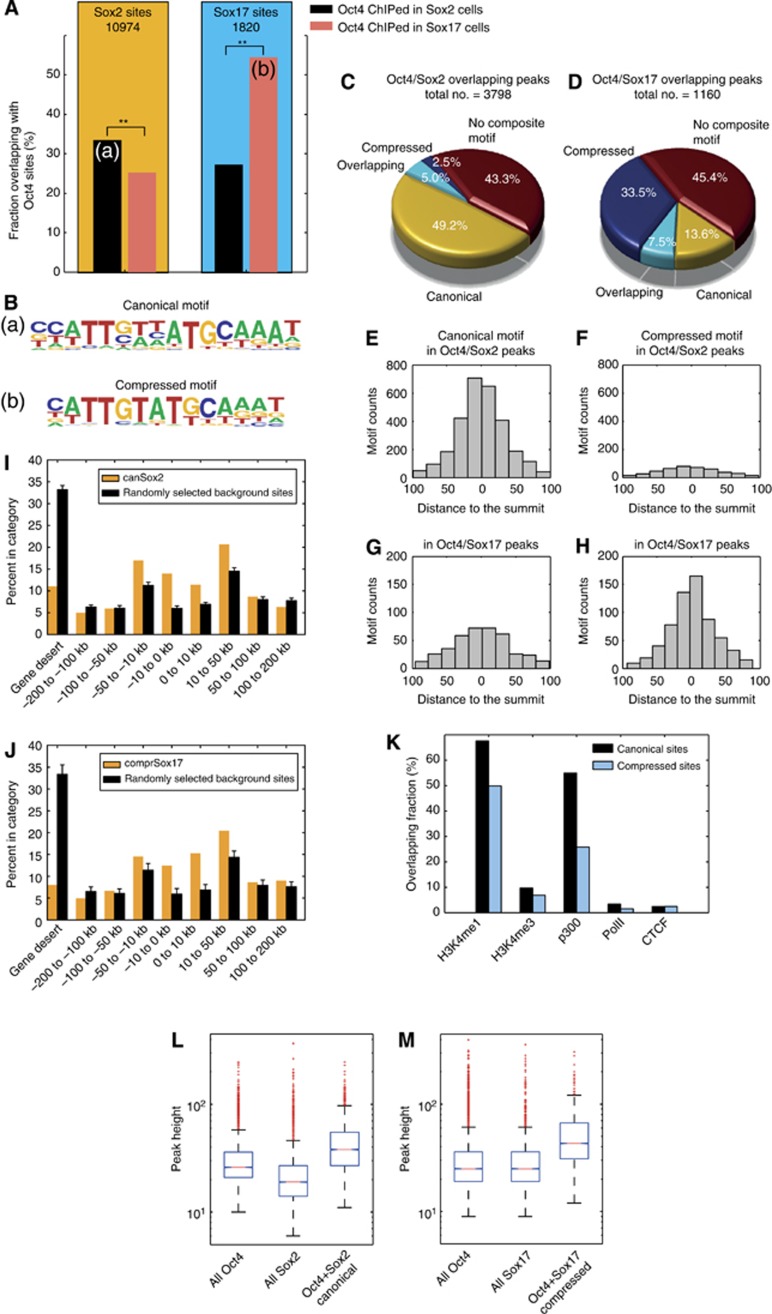
To compare the genomic binding distribution of Sox2/Oct4 and Sox17/Oct4 pairs, we performed chromatin immunoprecipitation assays followed by genome-wide sequencing (ChIP-seq) in two independent cell lines for each Sox factor (Supplementary Table S2). Using antibodies against the exogenous V5-tagged Sox factors and endogenous Oct4, we identified binding sites using a stringent data analysis pipeline (Supplementary Method). We first identified Oct4 sites (i) in the context of exogenous Sox2 expression (Oct4Sox2) and (ii) in the context of exogenous Sox17 expression (Oct4Sox17). Next, we intersected these two Oct4 data sets with Sox2 and Sox17 data sets. We found that Oct4Sox2 (Oct4 ChIP-seq in Sox2 expressing cells) is more likely to co-bind genomic loci with Sox2 when compared with Sox17. Conversely, Oct4Sox17 (Oct4 ChIP-seq in Sox17 expressing cells) is more likely to co-bind with Sox17 when compared to Sox2 (Figure 2A). This indicates that a substantial fraction of Oct4 partnerships is dependent on whether Sox2 or Sox17 is expressed. Hence, changes in the Sox partner lead to a partial genomic redistribution of Sox/Oct4 complexes. That is, Sox2/Oct4 co-select a distinct set of genomic loci which is different from the set co-selected by Sox17/Oct4. Next, we performed a de novo motif analysis on all genomic loci co-bound by either Sox2/Oct4Sox2 or Sox17/Oct4Sox17. For Sox2/Oct4Sox2 sites, we identified the Sox/Oct canonical motif (sequence related to CTTTGTCATGCAAAT) as most highly enriched (Figure 2B(a)). This motif is well characterized and has been functionally validated within the enhancers of several prominent pluripotency genes, including the master regulators Oct4, Sox2, and Nanog, which are jointly targeted by the Sox2/Oct4 pair. However, for Sox17/Oct4Sox17 sites we identified a variation of a Sox/Oct composite motif that is related to a CTTTGTATGCAAAT sequence (Figure 2B(b)). This ‘compressed’ motif shows spacing between the Sox and Oct sites that is reduced by one base pair compared to the canonical motif. We performed motif scans using position weight matrices (PWMs) for canonical and compressed motifs (Figure 2C and D). In agreement with the de novo motif search, we observed a converse distribution of the motifs. More than 50% of the 3798 Sox2/Oct4Sox2 sites contained a canonical motif but only 7.5% contained a compressed motif. In contrast, >40% of the 1160 Sox17/Oct4Sox17 sites contained a compressed motif and 21.1% a canonical motif (Supplementary Table S3). To further assess the significance of the Sox/Oct occupancy at the alternative compressed motif sites, we analysed the location of canonical and compressed motifs with respect to ChIP-seq summits (Figure 2E–H). We found that canonical motifs within Sox2/Oct4Sox2 sites cluster more closely to the summit coordinate, the presumed binding location. The same was observed for the compressed motifs found at Sox17/Oct4Sox17 sites. In contrast, compressed motifs within Sox2/Oct4Sox2 sites and canonical motifs within Sox17/Oct4Sox17 sites show a more widespread pattern, suggesting that a portion of these motifs comprised motifs found in the proximity of the actual binding site but are not directly bound. Given these different motif preferences, and our aim to decipher transcriptional regulation that set functionally different gene sets apart, we focused further analysis on Sox2/Oct4 bound canonical and Sox17/Oct4 bound compressed sites. Earlier ChIP-seq analysis done on multiple pluripotency TFs in mouse ESCs showed that Oct4 and Sox2, together with Nanog and Stat3 tend to cluster quite distally from promoters (Chen et al, 2008). Canonical and compressed sites were likewise found in regions 10–50 kb away from the transcription start sites (Figure 2I and J). Therefore, when assigning motifs to target genes a more extended distance should be considered. Chromatin signatures have been shown to predict regulatory elements in mammalian genomes and allow separation of promoters from enhancers (Birney et al, 2007; Heintzman and Ren, 2007; Heintzman et al, 2007). Likewise, the majority of active mammalian enhancers are associated with the histone acetyl transferase p300 (Visel et al, 2009). To relate the compressed and canonical motifs to annotated regulatory regions, we intersected our motif coordinates with sites defined by the ENCODE consortium for Bruce4 ESC (Figure 2K). We found that a higher proportion of both canonical and compressed binding sites intersected with the enhancer mark H3K4me1 as compared to the promoter mark H3K4me3. Consistent with this, both the canonical and compressed motifs overlapped an additional enhancer mark, p300, but very few with the PolII promoter mark or the CTCF insulator mark. Consistent with its function in pluripotent cells, the canonical sites were proportionally more enriched in ESC enhancers as compared to compressed sites. We next analysed the magnitude of ChIP-seq summits (‘pile-up’). We found that the subset of peaks containing canonical motifs co-bound by Sox2/Oct4Sox2 was significantly higher than the total of Sox2 and Oct4Sox2 peaks (Figure 2L and M). Analogously, the subset of compressed motifs bound by Sox17/Oct4Sox17 was stronger than all Sox17 or Oct4 Sox17 sites. These data suggest a cooperative recruitment of Sox2 and Oct4 to canonical and Sox17 and Oct4 to compressed motifs in vivo.
Figure 2.
Sox2/Oct4 and Sox17/Oct4 pairs are recruited at different genomic loci. (A) Oct4 is redistributed to different genomic loci by Sox factors. Co-occurrence of Sox2 or Sox17 with Oct4 in either Sox2-V5 or Sox17-V5 expressing KH2 cells. (a, b) Marks the intersect set of peaks used for de novo motif searching. Sox2 is more likely to co-occur with Oct4 ChiPed in a Sox2 OE background than with Oct4 ChIPed in a Sox17 OE background (P-value 5.1e−91, Z-score test). Inversely, Sox17 predominantly co-occurs on sites also bound by Oct4 ChIPed in Sox17 OE background whereas co-binding with Oct4 ChIPed in a Sox2 OE background much less likely (P-value 5.0e–116, Z-score test). (B) Matrices predicted by de novo motif analysis in (a) Sox2/Oct4Sox2 and (b) Sox17/Oct4Sox17 intersects. (C, D) Fraction of canonical, compressed, both (overlapping) and absence of motifs within (C) Sox2/Oct4Sox2 or (D) Sox17/Oct4Sox17 co-bound loci. (E–H) Motif counts binned by distances of the actual motif coordinate to the summit of the ChIP-seq peak for the canonical and compressed motifs in Sox2/Oct4Sox2 sites and Sox17/Oct4Sox17 sites. (I, J) Genome distribution of canonical motifs found in Sox2/Oct4Sox2 sites and compressed motifs in Sox17/Oct4Sox17 sites with respect to transcription start sites (TSS). (K) Fraction of Sox2/Oct4Sox2 bound canonical motifs in Sox2-V5 KH2 cells and Sox17/Oct4Sox17 bound compressed motifs in Sox17-V5 KH2 cells overlapping with H3K4m1 (enhancer mark), H3K4me3 (promoter mark), p300, PolII and CTCF regions annotated by ChIP-seq for Bruce4 mouse ESC by the ENCODE consortium. (L) The subset of overlapped Oct4Sox2 and Sox2 peaks with a motif is significantly higher than the total of Oct4Sox2 or Sox2 peaks (P-values 5.0e–209 and P<e–200; Wilcoxon rank sum test). (M) Similarly, the subset of peaks co-bound by Sox17/Oct4Sox17 containing a compressed motif is significantly higher than the total of Oct4Sox17 and Sox17 peaks (P-values 2.9e–93 and 1.2e–79; Wilcoxon rank sum test). Solid bars of boxes display the 25th–75th percentile of the peaks with the median indicated as an intersection. The box plots are shown on a logarithmic scale.
Canonical and compressed motifs earmark distinctive sets of genes that are differentially expressed
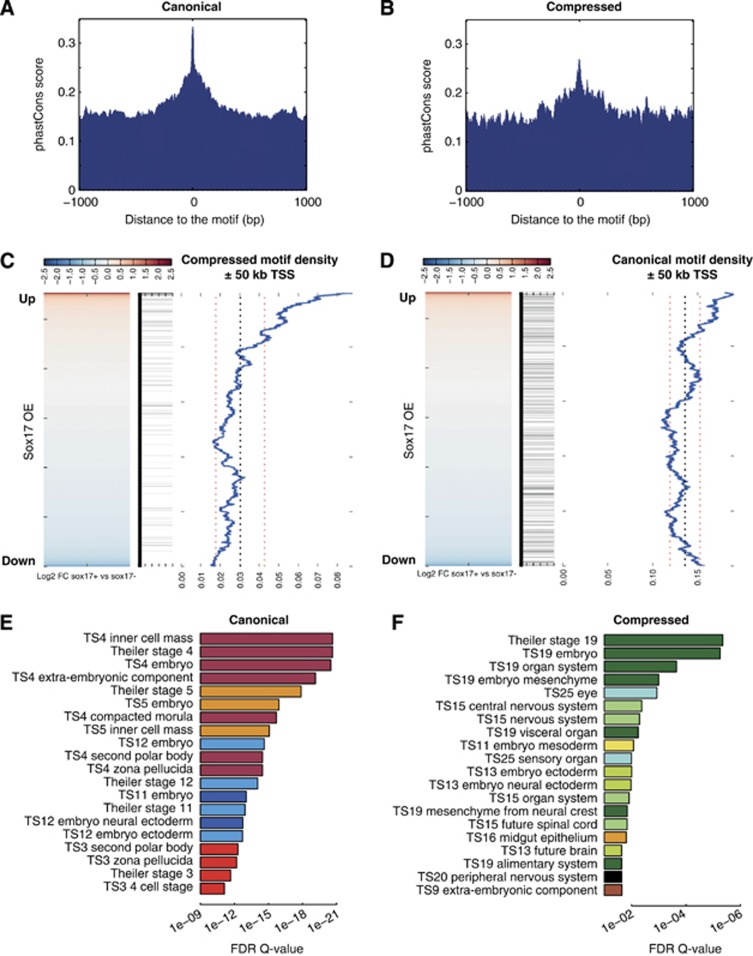
The rewiring of TF binding and gene regulatory networks in general (Schmidt et al, 2010) and of Sox and Oct TFs in particular (Kunarso et al, 2010) was suggested to be a key driver of evolution. Nevertheless, some enhancer elements were found to be under strong negative selection (Visel et al, 2008). We analysed the evolutionary conservation of canonical and compressed motifs using the placental mammalian PhastCons scores (Margulies et al, 2003). Evolutionary conservation was noted for the canonical, and to a lesser extent the compressed motif, indicating that a subset of the observed binding events are conserved within mammals (Figure 3A and B). To study gene expression changes during endodermal differentiation, gene expression profiling analysis was performed in ESCs before and after the induction of Sox17 expression (Supplementary Table S1). The genes were ranked by log2 transformed fold expression changes and scanned for the occurrence of compressed and canonical motifs in a distance ±50 kb from the TSS (Figure 3C and D). A significant enrichment was observed for the compressed motif near the TSS of genes upregulated during endodermal differentiation; this was not observed for downregulated or unaltered genes (Figure 3C). Among the upregulated genes containing a Sox17/Oct4 bound compressed motif are genes annotated to play a role during extra-embryonic endoderm development such as Col4a1, Col4a2, Lama1, Sall4, Pdgfra, Emb, and Hesx1 but also novel candidate regulators such as Tyro3 and Nr2f6. Canonical motif densities were less correlated with expression changes (Figure 3D). This observation strongly suggests that the cooperative binding of Sox17 and Oct4 drives the expression of genes during endodermal differentiation. To assess the functionality of gene sets earmarked by canonical and compressed motifs, gene ontology analysis was performed using GREAT. As expected, the canonical motif predominates within regulatory domains of genes expressed in the compacted morula, inner cell mass and very early stages of mouse development (Figure 3E). In contrast, compressed motifs were found near a different set of genes, many of which act at slightly later stages of development and contribute to germ layer and organ system differentiation (Figure 3F). Because so few genes are annotated with respect to the development of extra-embryonic endoderm and early definitive endoderm in gene ontology analysis, it is unlikely to find such ontology terms. However, many of the terms associated with the compressed motif are related to internal organs derived from endoderm. Indeed, genes annotated as expressed in TS19 are also known to function during extra-embryonic and endoderm development. Consistently, the ontology ‘extra-embryonic component; endoderm’ was found to be significantly enriched albeit not among the top 20 (Q-value 0.026). Namely genes such as Cyp26a1, Dab2, Hesx1, Hnf1b, Fst, Lama1, Lama2, Otx2, Pdgfa, Pdgra, Sall4, Smad2, and Srgn are jointly annotated by endodermal and TS19 ontology terms and have a Sox/Oct compressed motif nearby.
Figure 3.
Canonical and compressed motifs regulate different sets of genes. (A, B) The 30-Way PhastCons scores of 20 placental mammals tracks downloaded from the UCSC Table Browser were ascertained, using a window extended ±1000, bp from the centre of each motif, for 2269 canonical motifs in Sox2-V5 KH2 cells (A) and 523 compressed motifs in Sox17-V5 KH2 cells (B). (C, D) Microaray probes from KH2 cells expressing Sox17-V5 were ranked from most upregulated (red) to downregulated (blue) and the presence of a canonical or compressed motif was scored if found within 50 kb of the TSS of the gene (black horizontal bars indicate the presence of the motif). The density of compressed motif and canonical motif was measured using a sliding window comprising 10% of the total number of microarray probes. Presence of a motif gives the gene a value of 1 and absence of the motif the value 0. A sliding window is then used to calculate a density plot, black dotted line is the mean, and the two red dotted lines are one standard deviation away from the mean. (E, F) GREAT (great.stanford.edu) gene ontology analysis for 2269 canonical motifs bound by Sox2 and Oct4Sox2 (E) or 523 compressed motifs bound by Sox17 and Oct4Sox17 (F). Top 20 Mouse Genome Information (MGI) expression ontology terms are shown ranked by binomial multiple testing corrected Q-values and colour coded according to the theiler stage.
These analyses highlight the functional divergence of the Sox2/Oct4 canonical and Sox17/Oct4 compressed motifs to drive cell fate choices. It appears that each motif governs the expression of a distinct set of genes required for the specification of either the endoderm lineage for the compressed motif, or pluripotency for the canonical motif.
The genomic distribution of Sox factors can be altered by mutagenesis of the Oct4 interface
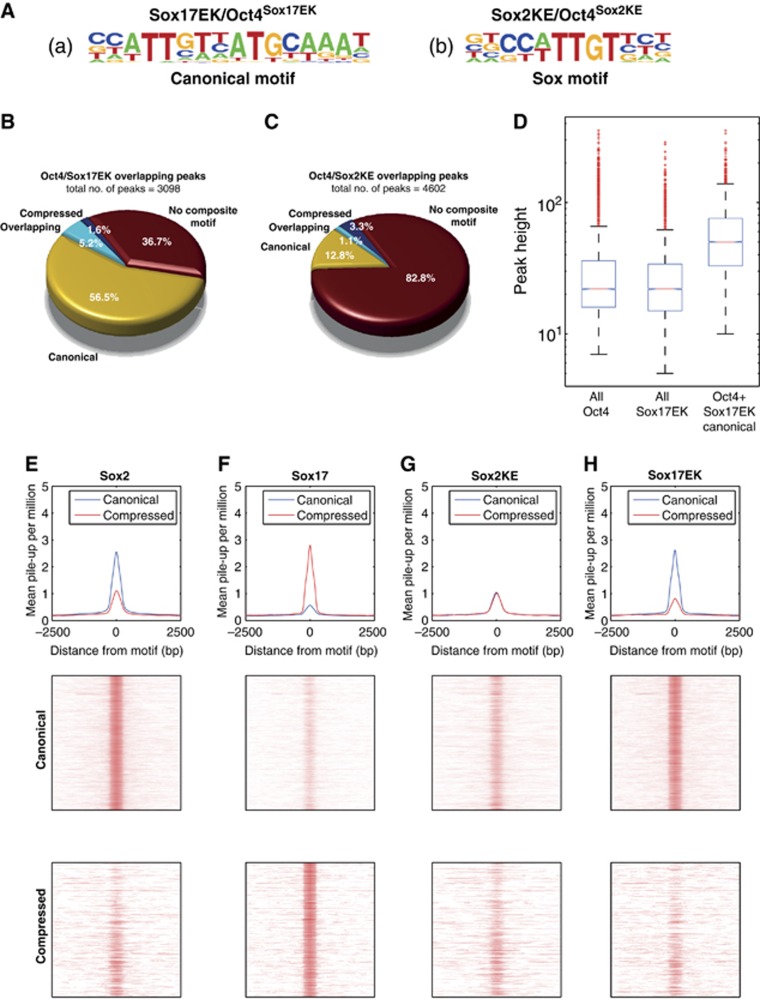
We have previously demonstrated that strategic point mutations introduced within Sox2 and Sox17 at the Oct4 interaction domains altered their biochemical binding activities in vitro (Jauch et al, 2011). To assess the genomic binding profile of the reengineered Sox factors in vivo, we expressed Sox2KE-V5 or Sox17EK-V5 in ESCs (Supplementary Figure S2) and performed ChIP-seq analysis of the V5-tagged Sox proteins, and of endogenous Oct4 in the context of the expression of either Sox proteins (Oct4Sox2KE or Oct4Sox17EK) (Supplementary Table S2). First, we conducted de novo motif analysis on loci co-bound by either Sox2KE/Oct4Sox2KE or Sox17EK/Oct4Sox17EK. In stark contrast to wild-type Sox17/Oct4Sox17 (Figure 2B(b)), we found the canonical motif to be most enriched in Sox17EK/Oct4Sox17EK co-bound sites (Figure 4A(a)). Moreover, the Sox2KE/Oct4Sox2KE pair no longer partners on canonical motif, but a single Sox element was found to be the top scoring motif (Figure 4A(b)). This is consistent with our previous quantitative electromobility shift assay (EMSA) experiments indicating that Sox2KE does not bind on the compressed motif as effectively as Sox17 (Ng et al, 2012). Apparently, Sox2 requires further mutations to engineer Sox17-like DNA recognition in vitro and in ES cells. While simple homology models do not reveal candidate amino acids that need to be modified to install Sox17-like binding activities into Sox2, future structural or molecular dynamics studies of Sox/Oct4 complexes on the compressed motif can potentially resolve this question.
Figure 4.
Reengineered Sox2KE and Sox17EK factors target different loci than their wild-type counterparts. (A) Position weight matrices (PWMs) of motifs found de novo in peaks marked co-bound by Sox17EK/Oct4Sox17EK or Sox2KE/Oct4Sox2KE revealing a canonical Sox/Oct motif and a single Sox site. (B, C) Fraction of Sox17EK/Oct4Sox17EK or Sox2KE/Oct4Sox2KE peaks containing compressed, canonical, or both motifs (‘overlapping’). The total numbers of overlapped Oct4 and Sox2KE/Sox17EK peaks are shown. (D) Comparison of peak heights for canonical motifs co-bound by Sox17EK/Oct4Sox17EK, Oct4Sox17EK or Sox17EK. Solid bars of boxes display the 25th–75th percentile of the peaks with the median indicated as an intersection. The subset peaks where Sox17EK/Oct4Sox17EK binds canonical motifs are significantly higher than the total of Oct4 peaks (P-value <1e–200, Wilcoxon rank sum test). (E–H) Mean pile-up per million reads for both canonical and compressed data in Sox2, Sox17, Sox2KE and Sox17EK expressing KH2 cells. Middle and bottom rows show density maps. Rows were ranked by motif quality scores with the highest scoring motifs at the top. The top panels show the mean pile-up per million reads for both canonical and compressed data.
Motif scans showed an occurrence of over 60% of the canonical motif within Sox17EK/Oct4Sox17EK sites compared to only 6.8% of the compressed motif (Figure 4B). Notably, Sox17EK/Oct4Sox17EK sites are more likely to contain canonical motifs than Sox2/Oct4Sox2 sites (Figure 2C). This is consistent with our previous finding that Sox17EK cooperates even more strongly with Oct4 than wild-type Sox2 on the canonical motif (Ng et al, 2012). Likewise, Sox17EK has consistently produced a larger number of pluripotent colonies in iPS experiments than Sox2 (Jauch et al, 2011). This suggests that Sox17EK is a more potent dimerization partner for Oct4 on ESC enhancers, which leads to an enhanced ability to trigger reprogramming to pluripotency. More than 80% of the Sox2KE/Oct4Sox2KE sites did not contain either of the two composite motifs, though the canonical motif was still more abundant than the compressed motif (13.9% versus 4.4% of the co-bound sites) (Figure 4C). Collectively, these results clearly indicate that a modification of the dimerization preferences of these Sox TFs in vitro induces a dramatic change in the genomic binding profiles of the Sox2KE/Oct4Sox2KE and Sox17EK/Oct4Sox17EK pairs. The subset of peaks containing a Sox17EK/Oct4Sox17EK bound canonical motif was on average substantially higher than the total of Sox17EK or Oct4Sox17EK peaks (Figure 4D). This observation underscores that a single point mutation swaps the preference of Sox17 from the compressed to the canonical motif. Finally, we analysed the pile-up of reads from the four duplicate Sox ChIP-seq libraries over motif coordinates obtained from Sox2/Oct4Sox2 and Sox17/Oct4Sox17 intersects (Figure 2C, D and 4E–H). Expectedly, Sox2 and Sox17 exhibited an inverse read distribution; Sox2 reads piling on the canonical motif and Sox17 reads piling on the compressed motif (Figure 4E and F). Sox2KE, however, showed an equally weak pile-up on both composite motifs (Figure 4G). In contrast, Sox17EK reads piled up strongly on the canonical motif, but were only weakly enriched on the compressed motif (Figure 4H). These data comprise the first ChIP-seq analysis of reengineered TFs and show that by modifying the Oct4 interaction surface of Sox factors it is possible to completely change the motif specificity, and thus the genomic distribution, of both Sox2 and Sox17 in vivo.
Oct4 is important for the differentiation of F9 cells into primitive endoderm
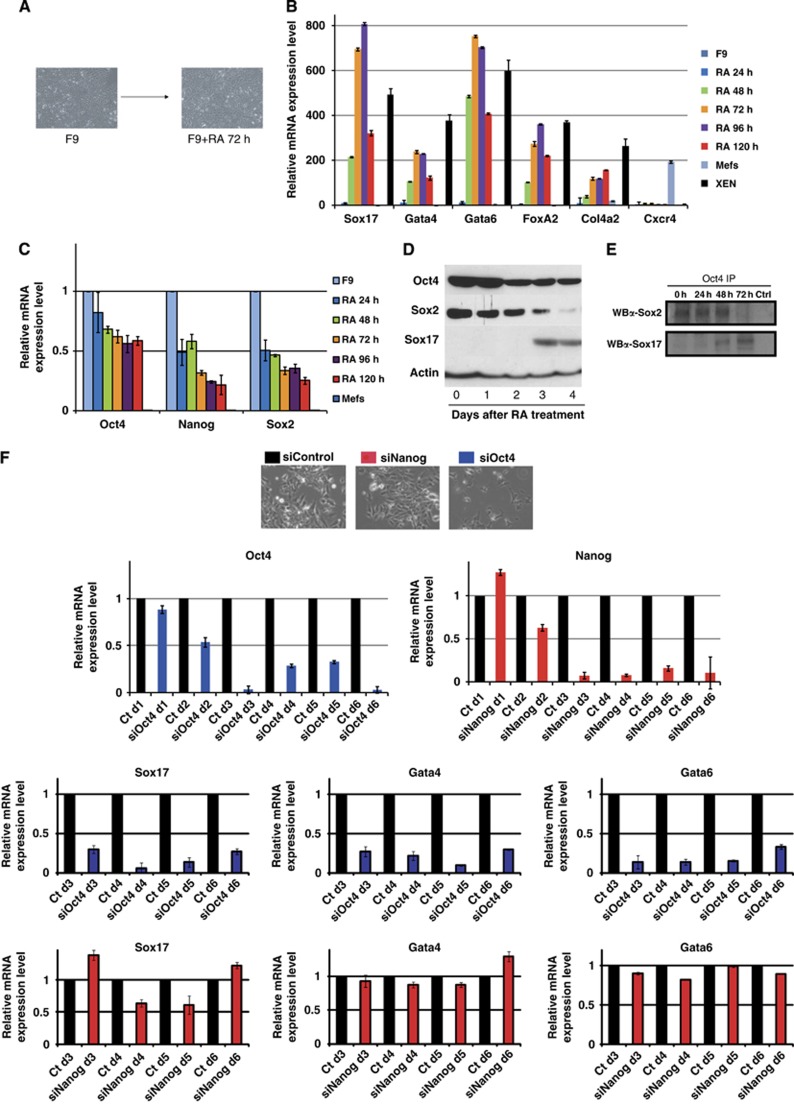
The intriguing findings of disparate motif preferences for Sox17/Oct4Sox17 versus Sox2/Oct4Sox2 pairs prompted the study of Sox17/Oct4 partnership in the context of endoderm formation. To this end, we investigated the Sox17/Oct4 partnership in F9 embryonic carcinoma (EC) cells, which are widely used as an in vitro model of extra-embryonic endoderm formation (Strickland and Mahdavi, 1978). F9 is a mouse EC cell line that expresses Oct4, Sox2, and Nanog and shares with ESC the core molecular machinery that governs self-renewal. However, these cells can be readily differentiated into extra-embryonic endoderm cells by treatment with retinoic acid (RA). After 2 days of RA treatment, the cell morphology changes (Figure 5A) and the expression of extra-embryonic endodermal markers Sox17, Gata4, Gata6, FoxA2 and Col4a2 is elevated, whereas the expression of the definitive endoderm marker Cxcr4 (Yasunaga and Nisikawa, 2007) remains low (Figure 5B). Ectodermal (Nestin, Sox1, GFAP) and mesodermal (T, Mixl1, Flk1) markers remain constant in their expression and are comparable to XEN cells (Figure S3), which confirm the specific differentiation of F9 cells into PrE after treatment with RA. In contrast, the expression of pluripotency markers Oct4, Sox2 and Nanog decreases (Figure 5C). However, while Sox2 and Nanog expression decreases continuously with RA treatment, Oct4 expression reaches a plateau after 2 days of differentiation and remains at roughly 50% of its original level (Figure 5C). Similarly, Oct4 protein levels initially decrease after RA treatment, but then do not further decline after day 2, whereas Sox2 protein levels continue to decrease, becoming barely detectable by day 4 (Figure 5D). However, the Sox17 protein appeared at a stage when Sox2 was already in low abundance. Co-immunoprecipitation experiments done on F9 cells induced to differentiate into PrE for 72 h showed that Oct4 co-precipitated Sox2 in undifferentiated F9 cells, but this interaction was lost by day 3 (Figure 5E), closely matching the Sox2 protein level (Figure 5D). Conversely, Oct4/Sox17 complexes were detected as early as 48 h after RA treatment and robustly at 72 h (Figure 5E), which was related to the protein levels of Sox17 observed in Figure 5D. Next, we sought to determine if Oct4 is important for primitive endoderm induction of F9 cells. We performed a knock-down of Oct4 or Nanog in undifferentiated F9 cells and observed >80% decrease in their expression level at day 3 (Figure 5F). We induced them to differentiate after day 1 into PrE to correlate the increase of Sox17 expression with the loss of Oct4 expression. We analysed the cells by qRT–PCR and observed a robust decrease in the expression of PrE markers including Sox17, Gata4 and Gata6 in Oct4 knocked down cells (Figure 5F), whereas cells knocked down for Nanog expressed these markers at comparable levels as control cells transfected with a non-targeting siRNA (Figure 5F). These results collectively indicate that knock-down of Oct4 impairs the differentiation of F9 cells towards an extra-embryonic endoderm cell fate. Further indirect evidence supports this function of Oct4: (i) in zebrafish, the Oct4 homologue spg (pou2) is essential for endoderm (Reim et al, 2004), (ii) Oct4 is robustly expressed in the developing mouse PrE (Palmieri et al, 1994; Kurimoto et al, 2006; Guo et al, 2010), (iii) in ESCs, overexpression of Oct4 results in differentiation towards PrE (Niwa et al, 2000) and finally (iv) XEN-P cells (extra-embryonic precursor cells) expressing high levels of Oct4 have been derived from rat blastocysts (Debeb et al, 2009). Altogether, these data with the Oct4 knock-down experiment in F9 cells suggest that Oct4 is indeed important for the induction of PrE differentiation.
Figure 5.
Oct4 expression is important for the differentiation of F9 cells into primitive endoderm (PrE). (A) Brightfield pictures of undifferentiated F9 cells and differentiated F9 cells after retinoic acid (RA) treatment for 72 h. Relative mRNA expression as determined by Q-RT–PCR for PrE markers (B) Sox17, Gata4, Gata6, FoxA2, Col4a2 and Cxcr4; and pluripotency markers (C) Oct4, Nanog and Sox2 in F9 cells treated with RA for 24–120 h and XEN cells. (D) Western blot of Oct4, Sox2 and Sox17 in F9 cells treated with RA for 1–4 days. (E) Oct4/Sox2 and Oct4/Sox17 co-immunoprecipations performed in untreated and RA-treated F9 cells. An Oct4 antibody was used for immunoprecipitation and western blot was done using Sox2 and Sox17 antibodies. (F) Brightfield photos of F9 cells transfected with Oct4, Nanog and Non-targeting control siRNAs and induced to differentiate at d1 after knock-down. Analysis of the expression levels of Oct4 Nanog, Sox17, Gata4 and Gata6 by Q-RT–PCR.
Sox17/Oct4 co-binding to compressed motifs triggers an endodermal development program
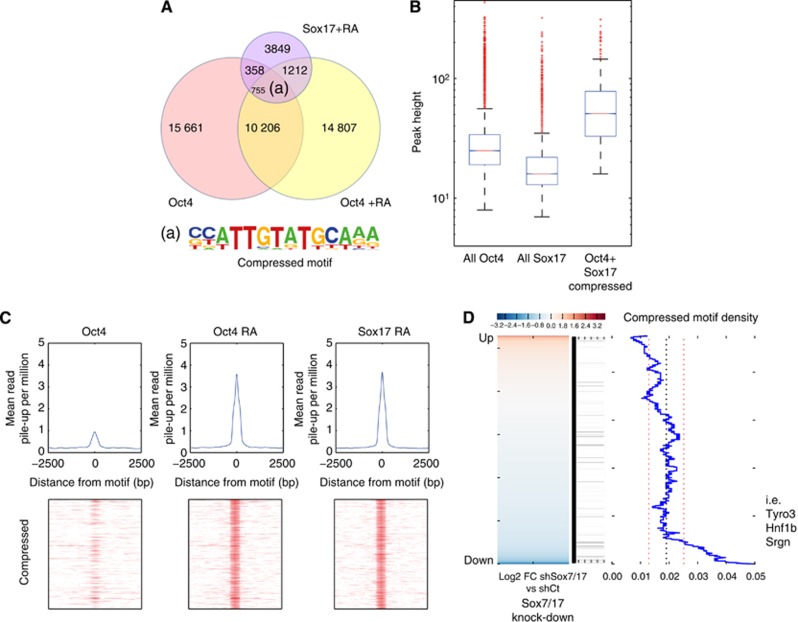
Our biological system provided the opportunity to identify the genes selectively targeted by the Oct4/Sox17 pair in PrE-like cells and to assess the dynamics of Oct4 expression during differentiation. We performed an Oct4 ChIP-seq experiment on undifferentiated F9 cells (Oct4F9) as well as on differentiated F9 cells (Oct4F9+RA), and a Sox17 ChIP-seq on differentiated F9 cells (Sox17F9+RA) (Supplementary Table S2). A higher proportion of Sox17 peaks overlapped with Oct4F9+RA sites, as compared to Oct4F9 sites, suggesting a partial redistribution of Oct4 during PrE differentiation in a Sox factor-dependent manner (Figure 6A). Importantly, de novo motif analysis of Sox17F9+RA/Oct4F9+RA sites revealed the compressed motif as the top scoring sequence (Figure 6A; Supplementary Table S3). The subset of peaks where Sox17F9+RA/Oct4F9+RA co-bind on the compressed motif is on average substantially higher than the total of Sox17F9+RA or Oct4F9+RA peaks, which indicates strong cooperative interactions of both TFs on the compressed motif (Figure 6B). These results are consistent with what was observed in our experiments with ESC. Read pile-up analysis revealed that Oct4 is only marginally enriched on compressed motifs before RA induction, suggesting that endoderm differentiation induces Sox17 and Oct4 co-recruitment to the compressed motif (Figure 6C). By analysing the Oct4/Sox17 genomic loci, we found that both TFs bind via the compressed motif to genes involved in primitive endoderm fate, including Hnf1b, Pdgfra, Sall4 and Col4a1 (Barbacci et al, 1999; Lim et al, 2008; Artus et al, 2010). In addition we identified a new set of potential endoderm-inducing genes, some of which have previously been shown to be expressed during endoderm specification, including Smad2, Elf3 or Hhip (Kawahira et al, 2003; Liu et al, 2004; Kwon et al, 2009). In order to assess the possible link between gene expression and TF binding sites, we used gene expression data obtained from F9 cells knocked down for both Sox7 and Sox17 (Supplementary Table S4). This double knock-down strategy avoids any possible redundancy between these two Sox factors as they have been shown to regulate common pan-endodermal genes (Seguin et al, 2008). By comparing the compressed motif density to the genes that were upregulated or downregulated in double knock-down F9 cells, a significant increase was observed in the compressed motif density for genes that were downregulated after Sox7 and Sox17 double knock-down (Figure 6D). This indicates that Sox17/Oct4 co-binding to compressed motifs is necessary for the transcriptional activation of endodermal genes. We compared Sox17 ChIP-seq data obtained from F9+RA cells with Sox17 ChIP-chip data performed in XEN cells which are embryo-derived PrE cells. In all, 1079 genes were found to intersect between the two studies (Supplementary Figure S4), but only 37 out of these 1079 genes harbour a compressed motif (Supplementary Table S6), suggesting that these sites are only bound by Sox17 and not by Oct4. This is not surprising as Oct4 is expressed at low levels in XEN cells compared to F9 cells induced to differentiate. These data indicate that Oct4 and Sox17 co-recruitment on the compressed motif is important for the induction of PrE cell fate, but is likely unnecessary for continued maintenance of PrE.
Figure 6.
Oct4 and Sox17 co-bound the compressed motif in F9 cells induced to differentiate into primitive endoderm. (A) Fractional overlap of endogenous Sox17 binding sites in RA-treated F9 cells co-occupied with Oct4 either before (red circle) or after adding RA (yellow circle). Upon differentiation, Oct4 co-occupies significantly more sites with Sox17 than prior the RA addition (1987 versus 1133; P-value=1.9e−143; Z-score test) indicating that Sox17 and Oct4 are co-recruited to different genes. Importantly, many of these new genes are earmarked by compressed motifs as found by de novo motif searching. (B) Distribution of peak heights in F9 cells treated with RA for all Oct4 sites, all Sox17 sites, and overlapped Oct4 and Sox17 sites. Solid bars of boxes display the 25th–75th percentile of the peaks with the median indicated as an intersection. The box plots are shown in a logarithmic scale. The subset of peaks when Oct4 co-binds with Sox17 to compressed motifs comprises significantly higher cohort than the total of Oct4 peaks (P=2.3e−95, Wilcoxon rank sum test). (C) Mean pile-up per million for compressed motifs in Oct4, Oct4 RA and Sox17 RA. Bottom row shows density maps of ChIP-seq data. (D) Compressed motif correlation with microarray gene expression of Sox7/Sox17 double knocked-down F9 cells. Motifs were assigned to genes if they are found within 50 kb of a Refgene TSS and the motif density plot is generated as in Figure 3.
Endodermal genes are regulated by cooperative binding of Sox17 and Oct4 to compressed motifs
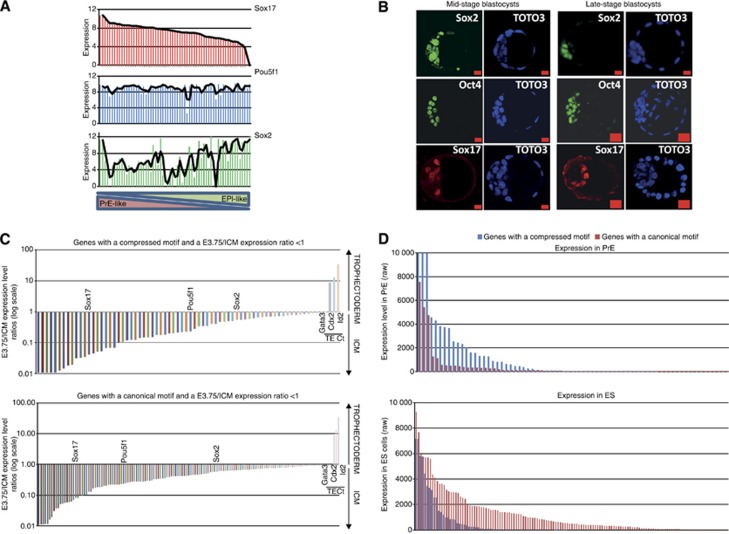
There is evidence suggesting that Oct4 may play a role in the establishment of the PrE within the mouse blastocyst as it is expressed there and its forced expression in ESC leads to a PrE differentiation program (Palmieri et al, 1994; Niwa et al, 2000). Based on our results, we asked whether Oct4 may differentially target EPI and PrE genes within the ICM in the particular context of its binding partners Sox2 and Sox17 which are also both expressed in the ICM. We reexamined our previously generated single-cell gene expression data (Guo et al, 2010) and focused this analysis on Oct4, Sox2 and Sox17 expression within the ICM of the ∼64-cell stage blastocyst. We ranked these ICM cells according to their expression level of Sox17, the cells expressing high levels of Sox17 are presumably presumptive of PrE, whereas cells expressing low levels of Sox17 are presumptive of EPI (Figure 7A). We observed that the expression pattern of Sox2 is inversely correlated to Sox17 looking across all cells. However, the expression level of Oct4 remained the same in all cells regardless whether they are PrE-like or EPI-like. This mRNA expression pattern is consistent with Oct4, Sox2 and Sox17 protein levels, with Oct4 uniformly expressed in all cells of the ICM, while Sox2 is initially expressed in all cells of the ICM but then becomes restricted only to the epiblast and Sox17 being initially expressed in a mixed sub-population of ICM cells that later becomes restricted to the blastocoel surface of the ICM (Figure 7B). These data suggest that Oct4 may preferentially bind the canonical Sox/Oct element in partnership with Sox2 in presumptive EPI while in PrE, it may bind the compressed Sox/Oct element in partnership with Sox17. We therefore generated RNA-seq data from whole mouse embryos at E3.75 (stage where the three cell types: TE, PrE and EPI are well resolved) and from dissected ICMs in order to identify genes expressed specifically in the ICM (Supplementary Table S5). For each gene identified, we measured the ratio of their expression level, determined by the RPKM (reads per kilobase per million mapped reads) value, in E3.75 blastocyst versus ICM. Therefore, genes with a ratio lower than 1 were assigned as ICM specific, whereas genes with a ratio higher than 1 were assigned as TE specific. We found that among the ICM-specific genes 67 are earmarked by a compressed motif and 133 by a canonical motif (Supplementary Table S5). These gene sets are candidates for the differential transcriptional regulation by Sox2/Oct4 versus Sox17/Oct4. The ratios are shown for each motif in Figure 7C, in which were plotted as controls Sox2, Sox17 and Oct4 for ICM-specific genes, and Cdx2, Gata3 and Id2 for TE-specific genes. We then looked at the expression level of these genes in ESC and PrE-like cells by using the raw expression values obtained from genome-wide gene expression analysis of mouse ESC and F9 cells differentiated into PrE-like cells to better resolve EPI and PrE genes within the ICM (Figure 7D; Supplementary Table S5). The genes with a compressed or a canonical motif represented in the histograms are listed in Supplementary Table S5. We found the compressed motif to be more strongly associated with the presumed PrE subset of genes while the canonical motif was more strongly associated with presumed EPI genes (Figure 7C). This argues that the earmark with a canonical motifs leads to Sox2/Oct4-mediated EPI expression program whereas an earmark with a compressed motif enables a Sox17/Oct4-mediated PrE expression program.
Figure 7.
Oct4/Sox2 and Oct4/Sox17 complexes regulate specific genes in mouse embryos at the blastocyst stage. (A) Sox17, Sox2, Oct4, Nanog and Gata4 expression levels distribution across each cell of a 64-cell stage mouse embryo obtained by single-cell gene expression analysis (Guo et al, 2010). A background of Ct=28 was used to obtain an absolute expression level. (B) Immunostainings of Oct4 and Sox17 in mouse embryos at mid and late blastocyst stage. Scale bar is 20 μm. (C) Histogram plots representing E3.75 blastocyst versus ICM expression level ratios for genes harbouring a compressed or a canonical motif in their enhancer region. Expression levels were determined by RNA-seq in RPKM (reads per kilobase per million mapped reads). (D) Histogram plot of the raw expression levels of each genes in ES and PrE cells with a compressed or a canonical motif and an E3.75 blastocyst/ICM lower than 1.
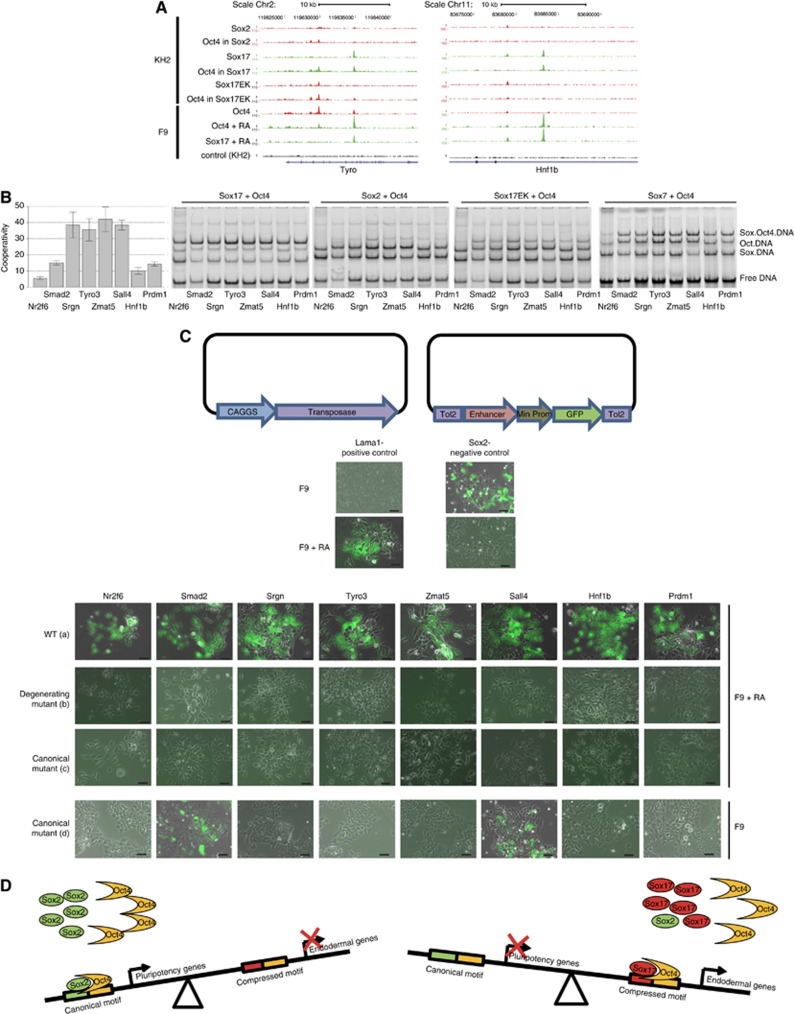
To further study the function of the compressed motif during endodermal differentiation, we selected eight genes with prominent and unique Sox17 and Oct4 peaks in KH2 and/or F9 cells after RA induction (Figure 8A; Supplementary Figure S5). Criteria for selecting genes for validation was response during exogenous Sox17 expression in KH2 cells (Nr2f6, Prdm1, Zmat5) or downregulation after Sox7/17 knock-down in F9 cells (Tyro3, Hnf1b, Srgn), the presence of a compressed motif and, in some cases, literature support for their function during extra-embryonic endoderm development (Sall4, Smad2) (Liu et al, 2004; Lim et al, 2008). First, we conducted EMSA experiments and showed that Oct4 and Sox17 bound cooperatively on these selected enhancer elements whereas the cooperation of Sox2 or Sox17EK with Oct4 was abolished or markedly diminished (Figure 8B). We also showed that Sox7 cooperatively binds with Oct4 on the compressed motif (Figure 8B), like Sox17, which can explain the redundancy between Sox7 and Sox17 in regulating endodermal genes. Next, we conducted an in vivo reporter assay (Kawakami et al, 2007). Selected enhancer regions were cloned into the Tol2 vector, comprised of a minimal promoter linked to GFP, and co-transfected with a vector expressing a transposase for stable integration in F9 cells that were induced, or not, to differentiate into PrE-like cells (Figure 8C). An increase in GFP expression was observed after 4 days of transfection only in F9 cells differentiated into PrE-like cells (Figure 8C(a)) but not in undifferentiated F9 cells (data not shown), indicating a specific expression pattern of these genes during endodermal differentiation. Moreover, when we introduced rational mutations that degenerated the compressed motif of the described enhancers by the replacement of multiple critical nucleotides within the Sox and Oct half-sites (degenerating mutants) reporter activity upon differentiation was completely abolished (Figure 8C(b)). Next, to more rigorously assess the relevance of the compressed motif for reporter expression we generated mutations by inserting a single nucleotide between Sox and Oct half-sites to transform the compressed motif into a ‘canonical motif’ (canonical mutants, Figure 8C(c); Supplementary Figure S6). Notably, even this subtle disturbance of the enhancer architecture led to a complete loss of the expression of GFP in differentiating F9 cells. This indicates that cooperative binding of Sox17/Oct4 to the compressed motif is indeed necessary to drive the expression of these endodermal genes. In a reverse experiment, we also tested the ‘canonical mutants’ in undifferentiated F9 cells and observed a gain in GFP expression for Sall4- and Smad2-driven reporters that would otherwise have been silent (Figure 8C(d); Supplementary Figure S6). These results indicate that a single nucleotide insertion can dramatically alter enhancer activities by recruiting alternative Sox/Oct4 complexes.
Figure 8.
Sox17 and Oct4 cooperatively bind on the compressed motif and regulate endodermal genes transcription. (A) Binding profiles of Sox factors and Oct4 in KH2 and F9 cells at Hnf1b and Tyro3 genomic loci are shown. (B) EMSAs were performed using recombinant, DNA binding domains of Sox17, Sox2, Sox17EK and Sox7 with Oct4 on eight different DNA elements containing the compressed motif. Co-operativity factors for Sox17/Oct4 were represented as bar plots with ±standard deviations error bars. (C) GFP reporter assay done on F9 cells treated (a–c) or not (d) with RA with Wild-type (a), Degenerating mutants (b) and Canonical mutants (c, d) enhancers cloned in the Tol2 vector (Kawakami, 2007) (scale is 50 μm). (D) Model describing the Sox2/Oct4 and Sox17/Oct4 partnerships in ESC. In undifferentiated ESC, where Oct4 and Sox2 expression levels are high, both factors cooperate and target specifically the canonical motif to regulate the expression of specific pluripotency genes. When the balance of Sox factors shifts in ES cells Oct4 switches from an interaction with Sox2 on canonical mofits towards an interaction with Sox17, and targets specific genes containing a compressed motif to trigger endodermal specification.
Discussion
In this study, the cooperation of Oct4 with Sox2 or Sox17 during the maintenance of the undifferentiated state of ESC and PrE induction was explored. We found that Sox17/Oct4 partner to co-select specific target genes during commitment to primitive endoderm. We employed a model of endodermal specification through forced expression of Sox17 in ESC. Sox17 has been shown to play a major role in endodermal differentiation of both human (Seguin et al, 2008) and mouse (Niakan et al, 2010) ESC. We focused in this study more specifically on Oct4. Oct4 is well known to play a major role in maintaining the pluripotent state of ESC but Oct4 has also been shown to play a role in PrE differentiation of these cells; however, the mechanisms involved in this process are still poorly understood. By analysing Sox17/Oct4 co-binding sites in this system, we first showed that these two TFs co-operate and bind to a specific compressed Sox/Oct motif. We identified a few hundred specific enhancer sites bound by both Oct4 and Sox17. We found that one of the earliest events occurring during endodermal induction is the redistribution of Oct4 binding sites from the canonical motif in undifferentiated ESC that express high levels of Sox2 to the newly discovered compressed motif when the cells are induced to differentiate into endoderm due to increasing levels of Sox17. We subjected the compressed motif to careful analysis to ascertain whether it contributes to an endodermal enhancer code. We now provide evidence that the genomic binding profile of Oct4 critically depends on which Sox partner factor is present and that the enhancer selection often relies on the nature of the composite motif.
To study the molecular mechanism of enhancer selection, we performed ChIP-seq analysis using rationally engineered Sox factors. These mutants harboured a single amino-acid mutation at the Oct4 interaction site that inverts their biological functions (Jauch et al, 2011). ChIP-seq experiments done on these mutant factors inducibly expressed in ESC using the same systems as their wild-type counterparts showed a clear redistribution of Oct4 binding sites from the compressed to the canonical motif for Sox17EK. Sox2KE lost its ability to bind to the canonical motif but did not acquire the ability to bind to the compressed motif. Potentially, Sox2KE may only be kick starting differentiation towards an endoderm fate by stimulating an as yet-unidentified endoderm specifier (possibly one or all of Smad2, Nr2f6, Srgn, Tyro3, Zmat4, Sall4, Hnf1 or Prdm1). Alternatively, Sox2KE may simply be interfering with the normal function of Sox2 while Oct4 remains broadly available for other Sox factors to bind, predisposing the ESC to differentiate towards endoderm once Sox17 is upregulated.
Overall, these results show that Sox/Oct partnerships target specific loci in order to regulate cell fate determination and that a strong cooperation between these TFs is important to bind to their specific enhancer sites. As both Sox17 and Oct4 are expressed in the primitive layers of embryos at the blastocyst stage, we hypothesized that they might play a role in the segregation of ICM cells into primitive endoderm cells. We therefore utilized a well-characterized PrE in vitro model, which consisted of F9 cells treated with RA. Oct4 was again recruited to enhancers containing compressed motifs in the presence of Sox17. Moreover, GFP-reporter assays further established that co-binding of Sox17/Oct4 to the compressed motif is necessary to drive the expression of genes during endoderm induction. Among the genes earmarked by compressed motifs are several genes with reported roles during endoderm specification, including Hnf1b, Pdgfra, Smad2 and Sall4 but also a few hundreds of new candidate genes, including Prdm1, Srgn, Tyro3, Nr2f6 and Zmat5. Finally, we correlated the results we obtained using ESC and F9 cells with data we generated from mouse embryos in order to gain more insight into lineage segregation during embryonic development. The inner cell mass of the mouse blastocyst contains cells with two distinct lineage potential: the epiblast and the primitive endoderm. The epiblast gives rise to the embryo proper, whereas the PrE specifies extra-embryonic tissues that support the development of the embryo and act as a signalling source to pattern the embryonic tissues prior to gastrulation. Till now, the mechanisms that govern segregation between the epiblast and the PrE remained unclear. ESC and F9 cells represent tractable developmental systems to uncover these mechanisms as they can be converted to PrE-like cells. At embryonic day 3.5, two populations of cells can be distinguished in the ICM of mouse blastocysts, one that is Gata6+ and supposedly at the origin of PrE cells and one Nanog+ that will give rise to epiblast cells. In a single-cell study, Kurimoto et al (2006) identified several genes specifically expressed in each of these sub-populations. When we compared these lists with our list of genes bound by Sox17/Oct4 on a compressed motif we found 11 common genes, 10 of which were expressed specifically in the Gata6+ cell population (Pdgfra, Emb, Dusp4, Lhfpl2, Lama1, Pfkl, Pgam2, Tfpi, Rap2c and an unknown gene) whereas only one (Bcl7a) was expressed in the Nanog+ cell population. Additionally, from our ICM-specific RNA-seq data the compressed motif is enriched in PrE genes and the canonical motif in pluripotent genes. These data emphasize the importance of the genes we identified as Sox17/Oct4 targets and further functional characterization of these genes will help for a better understanding of PrE specification and segregation in vivo. Mechanistically, we show here that one of the earliest events occurring during endodermal induction is the redistribution of Oct4 binding sites from the canonical motif in undifferentiated ESC that expresses high levels of Sox2 to the newly discovered compressed motif when the cells are induced to differentiate into endoderm due to increasing levels of Sox17.
Overall, these results show that Sox/Oct partnerships target specific loci in order to regulate cell fate determination and that a strong cooperation between these TFs is important to bind to their specific enhancer sites. We propose here a model by which, in pluripotent cells, Oct4 and Sox2 expression levels being high, both factors cooperate and target specifically the canonical motif to regulate the expression of genes involved in self-renewal and pluripotency (e.g., Nanog). When these cells are subjected to an endodermal differentiation signal such as FGF4 within the ICM, Sox17 levels increase leading to a switch of Oct4 from an interaction with Sox2 to an interaction with Sox17, and thereby targets specific genes containing a compressed motif to trigger the endodermal expression program (Figure 8D). Further functional characterization of these newly identified genes will help for a better understanding of PrE specification and segregation in vivo. Together, we provide a conceptual framework for the enhancer code that governs early cell fate decision and demonstrate the molecular mechanism of selective Sox-Oct partnerships during this process.
Materials and methods
Generation of inducible KH2 ES cells expressing V5-tagged Sox factors
KH2 ES cells were obtained from Open Biosystems and cultured in DMEM supplemented with 15% fetal bovine serum (Invitrogen), 0.055 mM β-mercaptoethanol, 2 mM L-glutamine, 0.1 mM non-essential amino acid and 1000 U/ml of LIF. Cells were maintained at 37°C with 5% CO2. These cells were utilized to insert a single copy of Sox2, Sox17, Sox2KE and Sox17EK cDNAs tagged with a V5 epitope using the method described in Beard et al (2006). Doxycycline was used at a final concentration of 1 μg/ml.
F9 cell culture
F9 cells were grown in media with DMEM (Invitrogen), 10% FBS, 1% L-glutamine (Invitrogen) and 1% penicillin/streptomycin. To induce their differentiation into primitive endoderm cells, cells were plated at a density of 300 000 cells/10 cm dish and grown in medium containing 1 μM of RA.
Microarray hybridization and data analysis
Mouse Ref-8 v2.0 Expression BeadChip microarrays (Illumina) were used for genome-wide expression analysis. For hybridization, cRNAs, from duplicate or triplicate samples, were synthesized and labelled using TotalPrep RNA Amplification Kit (Ambion), following manufacturer’s instructions. Scanned data from the BeadChip raw files for all samples were retrieved and background corrected using BeadStudio, and subsequent analyses were completed in GeneSpring GX. Data were normalized both within and between arrays, and corrected for multiple testing according to Benjamini–Hochberg. We defined genes as significantly differentially expressed when the FDR is <0.05. The data may be obtained via Gene Expression Omnibus (GSE43234).
RNA sequencing
RNA of E3.75 mouse embryo blastocysts and of immunosurgically dissected ICMs (Solter and Knowles, 1975) were extracted using the PicoPure RNA Isolation Kit. Reverse transcription and amplification steps were done using the Nugen Ovation RNA-Seq system. The SOLiD fragment library construction kit was used for the library construction from 1 μg of amplified cDNA. The resulting libraries were sequenced using SOLiD technology. The respective manufacturer’s protocol was followed at all stages. Sequenced reads were aligned to the mm9 mouse genome using the ABI MapReads program. Gene expression in the form of RPKM values was measured by normalizing the total reads mapping to a gene with its length and sequencing depth. All the transcripts were used for this calculation. The data may be obtained via Gene Expression Omnibus (GSE44553).
Reporter assays
Approximately 105 cells untreated or treated with RA were transfected in 6-well plates with 2.5 μg Tol2 GFP reporter plasmid, in which selected enhancers have been cloned using the primers listed in Supplementary Table S5, and 2.5 μg of Transposase expressing plasmid using Lipofectamine 2000 reagent. After overnight incubation, the transfection mix was replaced with F9 media with or without RA. The experiments were repeated twice. Mutant vectors were generated by using the Quick XL mutagenesis kit (Stratagene) according to manufacturer’s instructions.
Electromobility shift assay
EMSAs were carried out using purified recombinant proteins according to the method as described previously (Ng et al, 2012).
Supplementary Material
Acknowledgments
We thank S Prabhakar and TL Huber for valuable comments on the manuscript. We are grateful to Dr K Kawakami for providing the Tol2 system. This work is supported by the Agency for Science, Technology and Research (A*STAR, www.a-star.edu.sg) Singapore.
Author contributions: IA and RJ contributed to conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; CJ, MD, UD, GKB, RT, CKLN and WH contributed to collection of data; APH contributed to data analysis and interpretation; PR, PK and LWS contributed to financial support, administrative support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Artus J, Panthier JJ, Hadjantonakis AK (2010) A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development 137: 3361–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci E, Reber M, Ott MO, Breillat C, Huetz F, Cereghini S (1999) Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development 126: 4795–4805 [DOI] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R (2006) Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28 [DOI] [PubMed] [Google Scholar]
- Bergsland M, Ramskold D, Zaouter C, Klum S, Sandberg R, Muhr J (2011) Sequentially acting Sox transcription factors in neural lineage development. Genes Dev 25: 2453–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD (2011) Animal transcription networks as highly connected, quantitative continua. Dev Cell 21: 611–626 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Debeb BG, Galat V, Epple-Farmer J, Iannaccone S, Woodward WA, Bader M, Iannaccone P, Binas B (2009) Isolation of Oct4-expressing extraembryonic endoderm precursor cell lines. PLoS ONE 4: e7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, Alves P, Chateigner A, Perry M, Morris M, Auerbach RK, Feng X, Leng J, Vielle A, Niu W, Rhrissorrakrai K et al. (2010) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P (2010) Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18: 675–685 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Ren B (2007) The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell Mol Life Sci 64: 386–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ (2009) DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet 5: e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch R, Aksoy I, Hutchins AP, Ng CK, Tian XF, Chen J, Palasingam P, Robson P, Stanton LW, Kolatkar PR (2011) Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells 29: 940–951 [DOI] [PubMed] [Google Scholar]
- Kawahira H, Ma NH, Tzanakakis ES, McMahon AP, Chuang PT, Hebrok M (2003) Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development 130: 4871–4879 [DOI] [PubMed] [Google Scholar]
- Kawakami K (2007) Tol2: a versatile gene transfer vector in vertebrates. Genome Biol 8(Suppl 1): S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H, Kamachi Y (2010) SOX-partner code for cell specification: Regulatory target selection and underlying molecular mechanisms. Int J Biochem Cell Biol 42: 391–399 [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M (1998) Cooperative function of POU proteins and SOX proteins in glial cells. J Biol Chem 273: 16050–16057 [DOI] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G (2010) Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634 [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M (2006) An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res 34: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon MC, Koo BK, Kim YY, Lee SH, Kim NS, Kim JH, Kong YY (2009) Essential role of CR6-interacting factor 1 (Crif1) in E74-like factor 3 (ELF3)-mediated intestinal development. J Biol Chem 284: 33634–33641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Tam WL, Zhang J, Ang HS, Jia H, Lipovich L, Ng HH, Wei CL, Sung WK, Robson P, Yang H, Lim B (2008) Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell 3: 543–554 [DOI] [PubMed] [Google Scholar]
- Liu Y, Festing M, Thompson JC, Hester M, Rankin S, El-Hodiri HM, Zorn AM, Weinstein M (2004) Smad2 and Smad3 coordinately regulate craniofacial and endodermal development. Dev Biol 270: 411–426 [DOI] [PubMed] [Google Scholar]
- Margulies EH, Blanchette M, Haussler D, Green ED (2003) Identification and characterization of multi-species conserved sequences. Genome Res 13: 2507–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA (2011) Nucleosome-mediated cooperativity between transcription factors. Proc Natl Acad Sci USA 107: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Teo RT, Li H, Robson P, Glover DM, Zernicka-Goetz M (2010) Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci USA 107: 6364–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CK, Li NX, Chee S, Prabhakar S, Kolatkar PR, Jauch R (2012) Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res 40: 4933–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Ji H, Maehr R, Vokes SA, Rodolfa KT, Sherwood RI, Yamaki M, Dimos JT, Chen AE, Melton DA, McMahon AP, Eggan K (2010) Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev 24: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto M, Fukushima A, Okuda A, Muramatsu M (1999) The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol 19: 5453–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR (1994) Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol 166: 259–267 [DOI] [PubMed] [Google Scholar]
- Phillips K, Luisi B (2000) The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol 302: 1023–1039 [DOI] [PubMed] [Google Scholar]
- Reim G, Mizoguchi T, Stainier DY, Kikuchi Y, Brand M (2004) The POU domain protein spg (pou2/Oct4) is essential for endoderm formation in cooperation with the HMG domain protein casanova. Dev Cell 6: 91–101 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT (2010) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin CA, Draper JS, Nagy A, Rossant J (2008) Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell 3: 182–195 [DOI] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, Mann RS (2011) Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell 147: 1270–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solter D, Knowles BB (1975) Immunosurgery of mouse blastocyst. Proc Natl Acad Sci USA 72: 5099–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Abboud N, Desilets S, Nury D, Cowan C, Puceat M (2009) Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. J Cell Biol 186: 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland S, Mahdavi V (1978) The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell 15: 393–403 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H (2004) Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol 24: 8834–8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A (2002) Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res 30: 3202–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA (2008) Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40: 158–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA (2009) Genomic views of distant-acting enhancers. Nature 461: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynski B, Furlong EE (2010) Dynamic CRM occupancy reflects a temporal map of developmental progression. Mol Syst Biol 6: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M, Koopman P (2002) Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev 12: 441–446 [DOI] [PubMed] [Google Scholar]
- Yasunaga M, Nisikawa S (2007) Production of endoderm-derived visceral organ cells from ES cells. Tanpakushitsu Kakusan Koso 52: 57–66 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.