Abstract
The INK4/ARF locus regulates senescence and is frequently altered in cancer. In normal cells, the INK4/ARF locus is found silenced by Polycomb repressive complexes (PRCs). Which are the mechanisms responsible for the recruitment of PRCs to INK4/ARF and their other target genes remains unclear. In a genetic screen for transcription factors regulating senescence, we identified the homeodomain-containing protein HLX1 (H2.0-like homeobox 1). Expression of HLX1 extends cellular lifespan and blunts oncogene-induced senescence. Using quantitative proteomics, we identified p16INK4a as the key target mediating the effects of HLX1 in senescence. HLX1 represses p16INK4a transcription by recruiting PRCs and HDAC1. This mechanism has broader implications, as HLX1 also regulates a subset of PRC targets besides p16INK4a. Finally, sampling members of the Homeobox family, we identified multiple genes with ability to repress p16INK4a. Among them, we found HOXA9 (Homeobox A9), a putative oncogene in leukaemia, which also recruits PRCs and HDAC1 to regulate p16INK4a. Our results reveal an unexpected and conserved interplay between homeodomain-containing proteins and PRCs with implications in senescence, development and cancer.
Keywords: HLX1, Homeobox, Polycomb, p16INK4a, senescence
Introduction
The human INK4/ARF locus (CDKN2A and CDKN2B), located on chromosome 9p21, is the most frequently altered region in human cancers (Beroukhim et al, 2010). The INK4/ARF locus encodes two cyclin-dependent kinase inhibitors, p15INK4b and p16INK4a, and an unrelated protein p14ARF. While p15INK4b and p16INK4a regulate Rb phosphorylation, p14ARF behaves as a p53 activator (Gil and Peters, 2006). Besides its credentials in tumour suppression, genome-wide association studies have implicated the INK4/ARF locus in a variety of age-related disorders, including coronary heart disease, type II diabetes and late-onset Alzheimer’s disease (Popov and Gil, 2010). The locus is also important for the regulation of pluripotency and stem-cell renewal in mouse models (Collado et al, 2007) and impedes the reprogramming of human cells into induced human pluripotent stem cells (Banito et al, 2009; Li et al, 2009). The common denominator in these situations is the implementation of senescence by one or more of the INK4/ARF products. Senescence is a highly stable cell-cycle arrest, triggered in response to different stimuli, such as oncogenic signalling. As a consequence, it impacts on numerous pathologies, such as ageing and cancer (Kuilman et al, 2010).
In most primary cells, the INK4/ARF locus is tightly regulated by the Polycomb group (PcG) of transcriptional repressors (Bracken et al, 2007). PcG proteins participate in multi-component complexes termed as Polycomb repressive complex 1 (PRC1) and 2 (PRC2). The PRC2 interacts with histone deacetylases (HDAC) (van der Vlag and Otte, 1999) and crucially establishes trimethylation of histone H3 at lysine 27 (H3K27me3). This epigenetic mark is recognized by the PRC1 maintenance complex (Kerppola, 2009), which then catalyses the monoubiquitination of histone H2A (Morey and Helin, 2010; Beisel and Paro, 2011).
However, it is not clear how the PRCs are recruited to their target genes, and specifically to the INK4/ARF locus. One possibility is an association with long interfering non-coding RNAs (lincRNAs) (Khalil et al, 2009; Yap et al, 2010). The lincRNA ANRIL, which traverses the human INK4/ARF locus, has a role in the recruitment of PRC1 or PRC2 (Yap et al, 2010; Kotake et al, 2011). Another possibility is that specificity is conferred by sequence-specific DNA-binding factors as in Drosophila, where Polycomb responsive elements (PREs) and factors recognizing them have been identified. However, despite the mapping of several thousand PcG-binding sites (Ku et al, 2008), no universal sequence for mammalian PREs has been defined yet. Recently, JARID2, which has an ARID-like DNA-binding domain, has been shown to form a stable complex with PRC2 and to promote its recruitment to a subset of PRC2 targets (Landeira and Fisher, 2011). It was also reported that the transcription factor TWIST1 and the zinc finger protein Zfp277 can repress INK4/ARF in collaboration with PRCs (Negishi et al, 2010; Yang et al, 2010). However, additional transcription factors must still exist with ability to recruit PRC to their targets in different developmental or physiological contexts (Bracken and Helin, 2009; Kerppola, 2009).
Homeobox genes are a superfamily of transcription factors characterized by a conserved 60 amino acid DNA-binding domain, termed as the homeodomain. There are ∼ 260 Homeobox genes in mammalian genomes (Berger et al, 2008) with crucial roles in developmental patterning and in the regulation of processes, such as apoptosis, differentiation, epithelial-to-mesenchymal transition and cell proliferation. Altered expression of Homeobox genes is also a contributory factor in multiple types of cancer (Abate-Shen, 2002). Genome-wide surveys of H3K27me3 and PRC-binding sites have suggested that many Homeobox genes are subjected to PcG-mediated regulation, but there has been less attention to the potential for functional interplay between PcG and Homeobox proteins.
In this study, we identify the Homeobox gene HLX1 (H2.0-like homeobox 1) as a suppressor of senescence and describe how HLX1 can recruit PRCs to repress INK4a. As we identify additional Homeobox proteins with similar properties and the recruitment of PRCs by HLX1 controls a broader subset of PRC-repressed genes, the functional collaboration between PcG and Homeobox proteins could have general relevance in both development and carcinogenesis.
Results
The homeodomain-containing protein HLX1 controls senescence
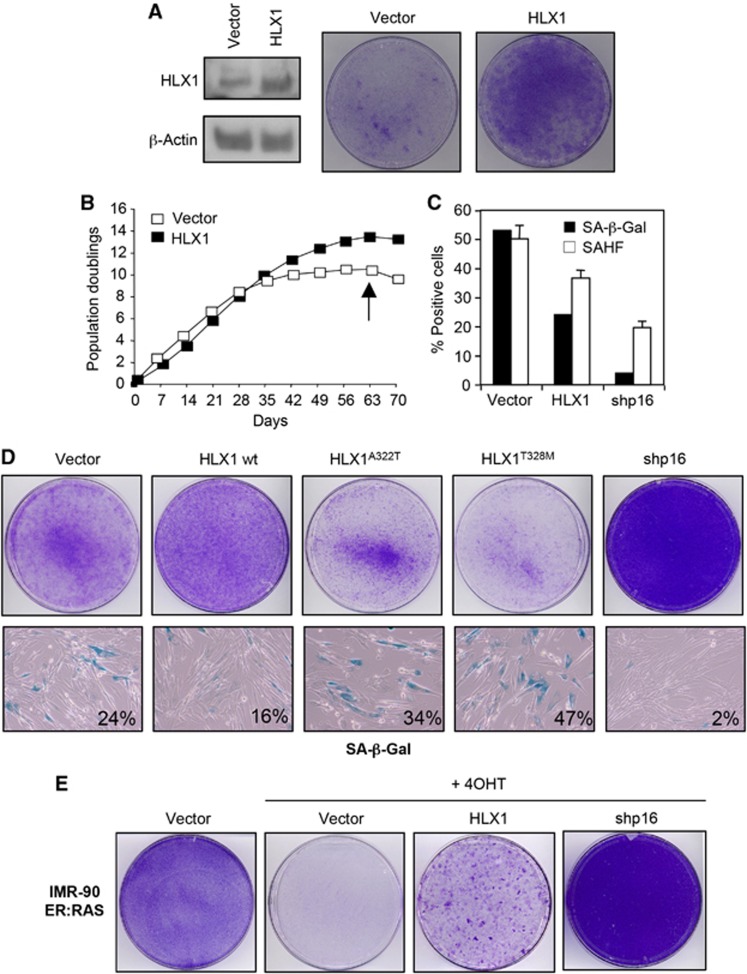
Transcription factors play a crucial role in regulating and implementing the senescence programme (Lanigan et al, 2011). In an effort to identify novel regulators of senescence, we tested a small subset of transcriptional regulators for their ability to extend replicative lifespan of IMR-90 human fibroblasts (Supplementary Figure S1A and B). In this screen, we identified that expression of the Homeobox HLX1 extended the lifespan of IMR-90 human fibroblasts (Figure 1A and B; Supplementary Figure S1). A similar extension of cellular lifespan was observed when HLX1 was expressed in another strain of human fibroblasts, WI-38 (Supplementary Figure S1C). Expression of HLX1 was confirmed by immunoblot (IB; Figure 1A). Consistent with an extension of cellular lifespan IMR-90 cells expressing HLX1 showed increased colony formation (Figure 1A), and delayed senescence, as evaluated by the percentage of cells positive for senescence-associated β-Galactosidase activity (SA-β-Gal) or displaying senescence-associated heterochromatin foci (SAHFs; Figure 1C).
Figure 1.
Expression of the homeodomain-containing protein HLX1 controls senescence. (A) Expression of HLX1 extends cellular lifespan. IMR-90 cells (passage 17) were infected with pBABE or pBABE-HLX1 vectors and after selection, cells were seeded for colony formation assays (right). Immunoblot (IB) showing HLX1 expression is shown as a control (left). (B) Growth curves of IMR-90 cells infected with pBABE or pBABE-HLX1 vectors. (C) Effect of HLX1 expression on SA-β-Gal activity and SAHF formation in IMR-90 cells. Cells were analysed at passage 27 (marked by an arrow in B). shp16, shRNA targeting p16INK4a. (D) Effect of point mutations in the HLX1 homeodomain on cell growth and senescence in IMR-90 cells was assessed by crystal violet (top) and SA-β-Gal staining (bottom). Cells are shown at passage 24, as explained in Supplementary Figure S2. (E) Expression of HLX1 diminishes the arrest observed during OIS. Crystal violet staining of IMR-90 ER:RAS cells infected with the indicated vectors and treated with 4OHT where indicated (+4OHT).
HLX1 is a transcription factor containing a homeodomain responsible for its binding to DNA. We generated two HLX1 point mutants (A322T and T328M) by mutating residues predicted to be critical for preserving the homeodomain structure. These mutants were expressed at similar levels to HLX1 wild type (wt; Supplementary Figure S2A), but rather than extending the lifespan of IMR-90 cells, homeodomain mutants caused decreased cell proliferation and a premature entry into senescence (Figure 1D; Supplementary Figure S2B).
We next used IMR-90 ER:RAS cells, in which oncogene-induced senescence (OIS) can be triggered by 4-hydroxy-tamoxifen (4OHT) activation of an ER:RAS chimera (Barradas et al, 2009). Ectopic expression of HLX1 partially prevented the OIS arrest, as noted by enhanced colony formation (Figure 1E) and a higher percentage of cells incorporating BrdU (data not shown), although the effect was not as striking as that achieved upon p16INK4a depletion.
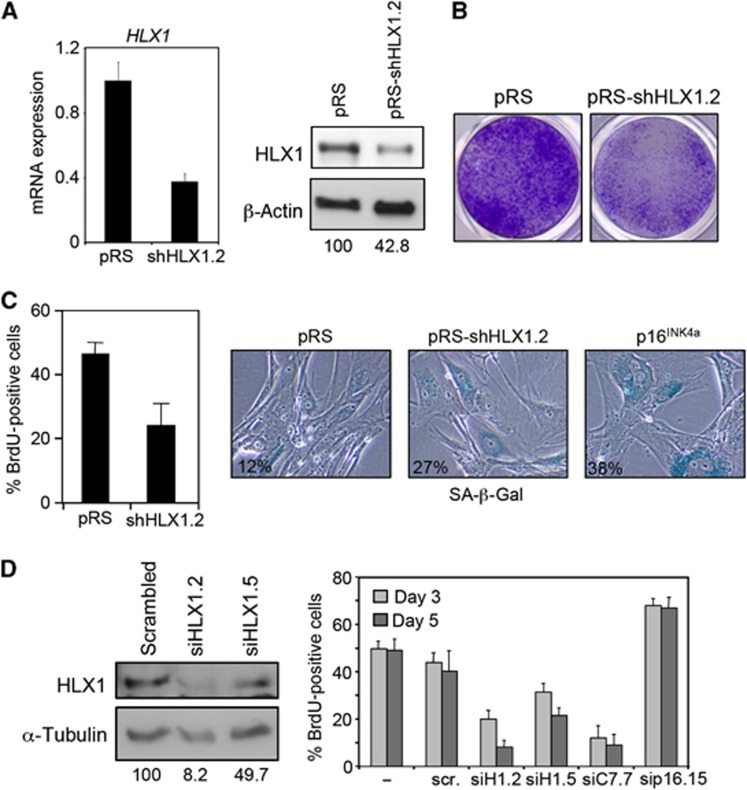
To further understand the relevance of HLX1 on senescence, we investigated the effect of knocking down HLX1 levels with four independent siRNAs or shRNAs (summarized in Supplementary Figure S3A). HLX1 depletion using a retroviral shRNA (pRS-shHLX1.2) was confirmed by IB and quantitative RT–PCR (qRT–PCR; Figure 2A) and caused decreased colony formation in IMR-90 cells (Figure 2B). The impaired colony formation was due to a senescence-like response, as evidenced by decreased BrdU incorporation and an increase in the percentage of SA-β-Gal-positive cells (Figure 2C). Similar results were obtained using a lentiviral shRNA vector targeting HLX1 (Supplementary Figure S3B). Transfection of two independent siRNAs targeting HLX1 further confirmed the results, causing also a decrease in the growth of IMR-90 human fibroblasts, primary human keratinocytes (NHK) and prostate epithelial cells (HPrEC, Figure 2D; Supplementary Figure S4A). Overall, the above results show that HLX1 regulates senescence.
Figure 2.
Knockdown of HLX1 induces premature senescence. (A) IMR-90 cells (passage 12) were infected with a control retroviral vector (pRS) or a vector targeting HLX1 (pRS-shHLX1.2). The efficiency of HLX1 knockdown was monitored by qRT–PCR (left) and IB (right). The numbers under the blots correspond to quantification of HLX1 signal normalized to β-actin. (B) Knockdown of HLX1 using shHLX1.2 results in reduced colony formation. (C) HLX1 knockdown causes premature senescence. The effect of knocking down HLX1 in IMR-90 cells (passage 15) was monitored by BrdU incorporation (left) and SA-β-Gal staining (right). (D) Knockdown of HLX1 by siRNA transfection causes growth arrest. IMR-90 cells were transfected with the indicated siRNAs, and siRNA efficiency was evaluated by immunoblot (left). The numbers under the blots correspond to quantification of HLX1 signal normalized to α-tubulin. The percentage of BrdU-positive cells was evaluated by IF (right). scr., scrambled; siH1, siRNA targeting HLX1; siC7, siRNA targeting CBX7; sip16, siRNA targeting INK4a.
HLX1 represses p16INK4a expression
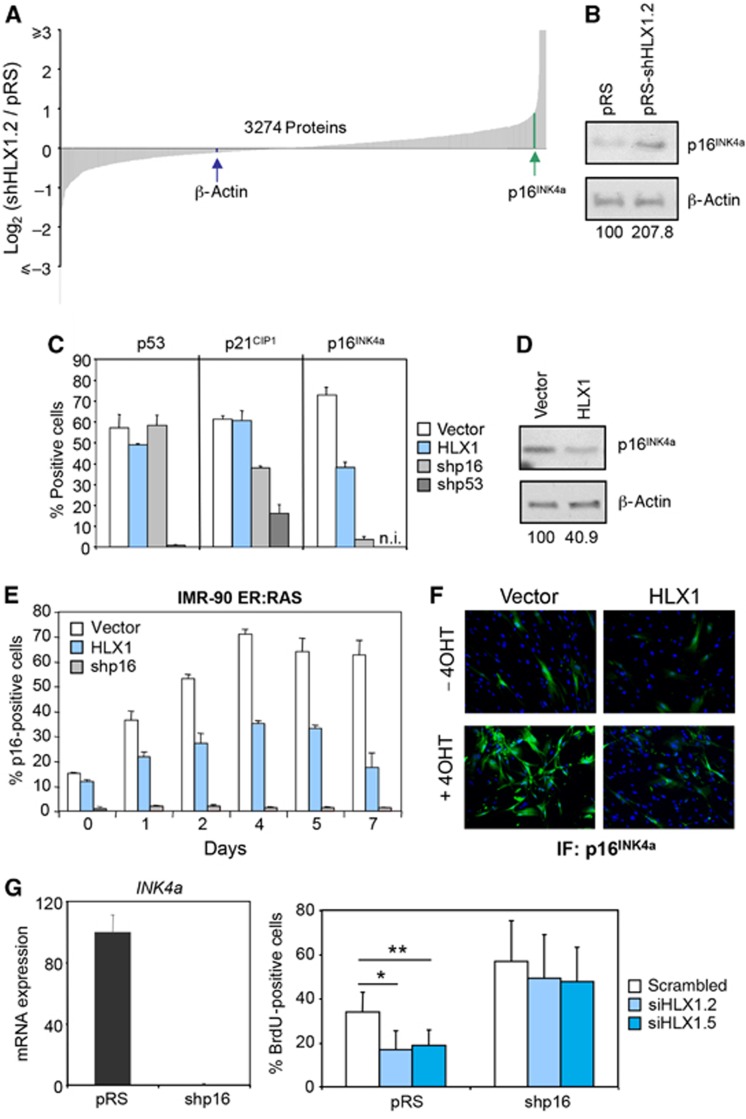
We next used stable isotope labelling with amino acids in cell culture (SILAC) to measure changes in protein levels upon HLX1 knockdown. Using stringent criteria for protein identification and quantification, we detected a total of 3274 proteins. Although the expression of several senescence-related proteins changed (e.g., PAI1, MMP1, GLB1 and CCN1 were upregulated and CSIG was downregulated), the most interesting change among them was that of p16INK4a. The expression of p16INK4a was upregulated ∼2-fold upon HLX1 knockdown (Figure 3A; a complete list of proteins detected is presented in Supplementary Table S1). In addition, we did not detect a change in previously described p16INK4a regulators that could explain the increase in p16INK4a expression. The upregulation of p16INK4a upon HLX1 depletion was confirmed by IB and immunofluorescence (IF), reproduced using different shRNAs or siRNAs and observed not only in IMR-90 cells, but also in HPrEC and NHK (Figure 3B; Supplementary Figures S3C, S4B and C). While we did not observe significant changes in p53 or p21CIP1 expression, we did note that p16INK4a levels were lower in cells expressing HLX1, as assessed by IF and IB (Figure 3C and D; Supplementary Figure S5). Expression of HLX1 also prevented partially the p16INK4a induction caused by RAS (Figure 3E and F). Moreover, the HLX1 homeodomain mutants not only failed to repress, but in fact induced p16INK4a expression (Supplementary Figure S2C and D).
Figure 3.
HLX1 regulates p16INK4a expression. (A) SILAC identifies p16INK4a as a HLX1 target. IMR-90 cells were infected with a control pRS vector or a vector knocking down HLX1 expression (pRS-shHLX1.2). Cells were cultured in ‘Heavy’ (H) or ‘Light’ (L) SILAC medium, respectively. Cell extracts were prepared and global changes in protein levels analysed by mass spectrometry. (B) Immunoblot confirming the p16INK4a upregulation upon HLX1 knockdown. The numbers under the blots correspond to quantification of HLX1 signal normalized to β-actin. (C) Decreased p16INK4a expression in IMR-90 cells expressing HLX1. Analysis of the expression of p16INK4a, p21CIP1 and p53 by IF in IMR-90 cells infected with the indicated vectors. The percentages of positive cells are plotted and representative images of this experiment are shown in Supplementary Figure S5. n.i., not included in the experiment. (D) The repression of p16INK4a by HLX1 expression was confirmed by IB. The numbers under the blots correspond to quantification of HLX1 signal normalized to β-actin. (E, F) HLX1 prevents the induction of p16INK4a during OIS. Analysis of p16INK4a levels by IF in IMR-90 ER:RAS cells. Representative images (F) and quantification (E) are shown. (G) p16INK4a depletion rescues the growth arrest caused by HLX1 knockdown. IMR-90 cells were infected with a shRNA vector targeting p16 (shp16) and p16INK4a knockdown was confirmed by qRT–PCR (left). Control (pRS) or p16INK4a-depleted cells (shp16) were transfected with the indicated siRNAs and BrdU incorporation analysed (right). *P<0.05, **P<0.01.
To test whether p16INK4a is an important mediator of the effect of HLX1 on senescence, we generated IMR-90 cells with abolished p16INK4a expression (Figure 3G, left). While knocking down HLX1 levels resulted in a significant arrest of control cells, it only caused a small decline in BrdU incorporation in IMR-90 cells infected with shp16 (Figure 3G, right). Similar results were observed when the oncoviral protein E7, which targets RB and other pocket proteins, was used (Supplementary Figure S6). Although these results do not exclude that HLX1 regulates senescence by additional mechanisms, they clearly show that p16INK4a is key to mediate the senescence response regulated by HLX1.
HLX1 binds to the INK4a promoter
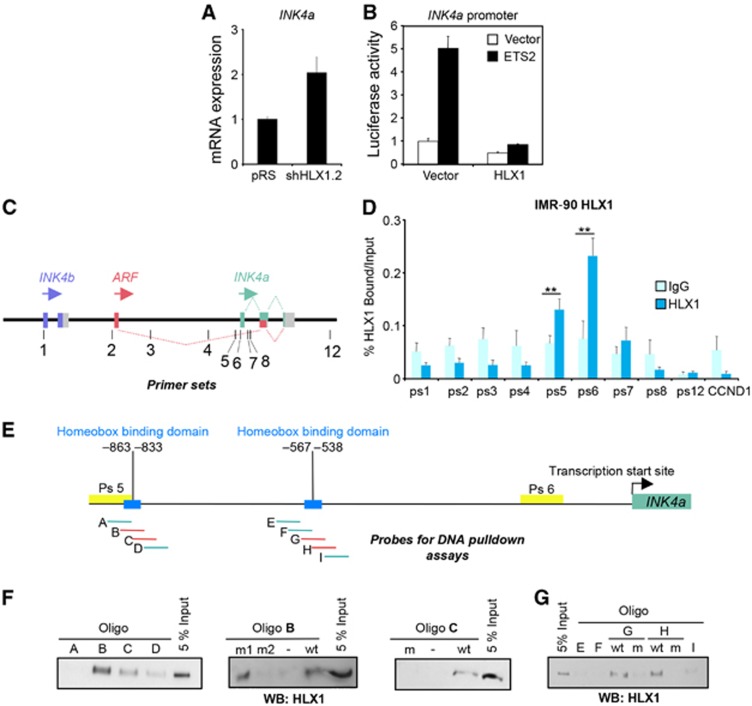
We next concentrated on studying how HLX1 controls p16INK4a expression. Using qRT–PCR, we observed that the regulation of p16INK4a by HLX1 occurred at the mRNA level (Figure 4A; Supplementary Figure S3B). To investigate whether HLX1 directly controlled INK4a transcription, we first conducted a reporter assay using an INK4a promoter reporter activated by expression of ETS2 (Ohtani et al, 2001). Consistent with a direct role of HLX1 in repressing INK4a, co-expression of HLX1 prevented this induction (Figure 4B, compare black bars). To confirm the direct regulation of INK4a by HLX1, we carried out chromatin immunoprecipitation (ChIP) in cells overexpressing HLX1. Analysis of HLX1 binding on the INK4/ARF locus showed a significant association in the proximity of the INK4a promoter (Figure 4C and D). Sequence analysis identified two putative Homeobox binding motifs in this region, centred at ∼−550 and ∼−850 relative to the INK4a TSS (Figure 4E). Using DNA pulldown, we showed that HLX1 can bind to both motifs in a sequence-dependent manner (Figure 4F and G). Overall, these results suggest that HLX1 regulates senescence through direct repression of p16INK4a.
Figure 4.
HLX1 binds to the INK4a promoter. (A) HLX1 regulates INK4a mRNA levels. IMR-90 cells were infected with pRS-shHLX1.2 or a control pRS vector and the levels of INK4a mRNA were evaluated by qRT–PCR. (B) Luciferase reporter assay showing that HLX1 expression prevents the activation of the INK4a promoter by ETS2. (C) Cartoon depicting the INK4/ARF locus and the primer sets used for ChIP. Primer sequences are in Supplementary Table S3. (D) HLX1 binds in the proximity to the INK4a promoter, as assessed by ChIP from IMR-90 cells infected with pBABE-HLX1. Chromatin from IMR-90 cells at passages 24–25 was used for these experiments. Binding of HLX1 to primer sets 5 and 6 was significant. (E) Cartoon depicting the INK4a/ARF locus showing the Homeobox binding domains (blue), the regions amplified by the ChIP primer sets 5 and 6, and the probes used for the DNA pulldown experiments. (F, G) DNA pulldown assays showing HLX1 binding to probes corresponding to the ∼−850 (F) and∼−550 (G) Homeobox binding domain regions. Sequences of oligos used for the probes are included in Supplementary Table S4. wt, wild type; m, mutant. **P<0.01.
Transcriptional repression by HLX1 involves PRCs and HDAC1
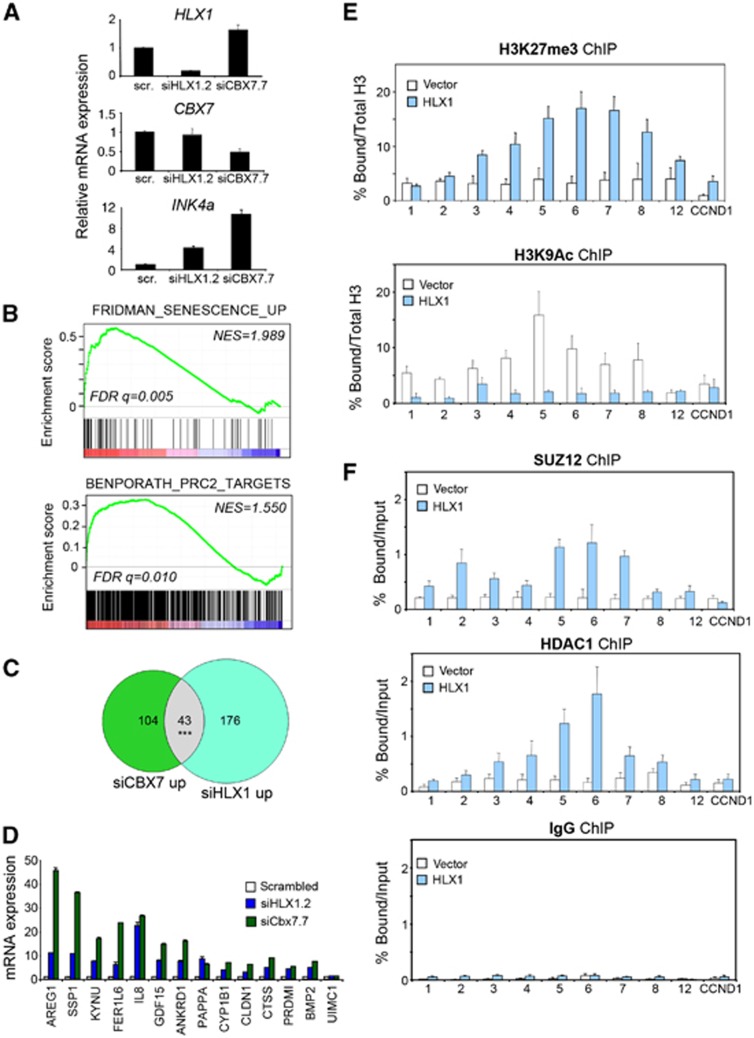
To further investigate the mechanism used by HLX1 to repress p16INK4a and regulate senescence, we conducted mRNA expression profiling of IMR-90 cells with depleted HLX1 expression (Figure 5A). HLX1 knockdown resulted in the identification of a senescence signature (Fridman and Tainsky, 2008) by gene set enrichment analysis (GSEA, Figure 5B). Interestingly, HLX1 depletion also caused the upregulation of HDAC and Polycomb target genes (Figure 5B; Supplementary Figure S7A and B), which would suggest a role for HDAC and PRCs in repressing HLX1 targets. Gene sets of PRC targets described in two independent studies (Bracken et al, 2006; Ben-Porath et al, 2008), and defined by either the depletion of specific PRC2 components or by its combination, were all found to be associated with HLX1 depletion (Supplementary Figure S7C and D). Similar results were observed analysing an independent data set derived from mouse leukaemic cells (Kawahara et al, 2012; Supplementary Figure S7E). Moreover, the expression of a set of genes decorated by H3K27me3 marks was also enriched in IMR-90 cells transfected with siHLX1.2 (Supplementary Figure S7C, right). As these results suggested that a subset of Polycomb targets might be co-regulated by HLX1, we performed mRNA profiling of IMR-90 cells in which the expression of the Polycomb protein CBX7 was targeted by an siRNA (siCBX7.7; Figure 5A). As expected, depletion of CBX7 resulted in increased expression of Polycomb targets, INK4a among them (Supplementary Figure S7F; Figure 5A). By comparing the genes upregulated upon depletion of CBX7 or HLX1, we observed a significant overlap (43 commonly upregulated genes, P<2.2 × 10−16; Figure 5C). We confirmed the co-regulation of INK4a and 14 of these genes by qRT–PCR (Figure 5A and D). While the overlap of genes downregulated upon CBX7 or HLX1 knockdown was also significant, that was not the case for antiregulated genes (P=0.766).
Figure 5.
Transcriptional repression by HLX1 involves PRCs and HDAC1. (A) qRT-PCR data showing INK4a, HLX1 and CBX7 expression in samples used to conduct mRNA expression analysis. (B) GSEA of gene sets associated with senescence (Fridman and Tainsky, 2008) and PRC2 targets (Ben-Porath et al, 2008) in transcriptional profiles of IMR-90 cells in which HLX1 has been knocked down. NES, normalized enrichment score; FDR, false discovery rate. (C) Venn diagram showing the overlap of genes which increase >2-fold upon CBX7 or HLX1 knockdown. The overlap is significant (P<2.2 × 10−16). The overlap (n=12) of genes antiregulated upon knockdown of CBX7 (n=443) or HLX1 (n=642) is non-significant (P=0.766). (D) qRT–PCR confirms the co-regulation upon depletion of HLX1 or CBX7 of 14 genes among the candidates identified in the mRNA expression study. (E, F) HLX1 expression results in increased binding of SUZ12 and HDAC1 at the INK4/ARF locus. ChIP analyses showing histone modifications (E) and binding of SUZ12 and HDAC1 (F) at the INK4/ARF locus in IMR-90 cells infected with a control vector or expressing HLX1. The experiment was performed with IMR-90 vector and IMR-90 HLX1 cells at passages 24 and 25. ***P<0.001.
To extend these observations to the regulation of the INK4/ARF locus, we analysed the occupancy of SUZ12, HDAC1 and different histone modifications in late passage IMR-90 cells expressing HLX1. A decrease in H3K4me3 and H3K9Ac, concomitant with an increase in H3K27me3, was observed at the INK4/ARF locus upon expression of HLX1 (Figure 5E and data not shown). In addition, we observed an increased binding of PRC2 members (EZH2 and SUZ12) and HDAC1 in the INK4/ARF locus in cells expressing HLX1 (Figure 5F and data not shown).
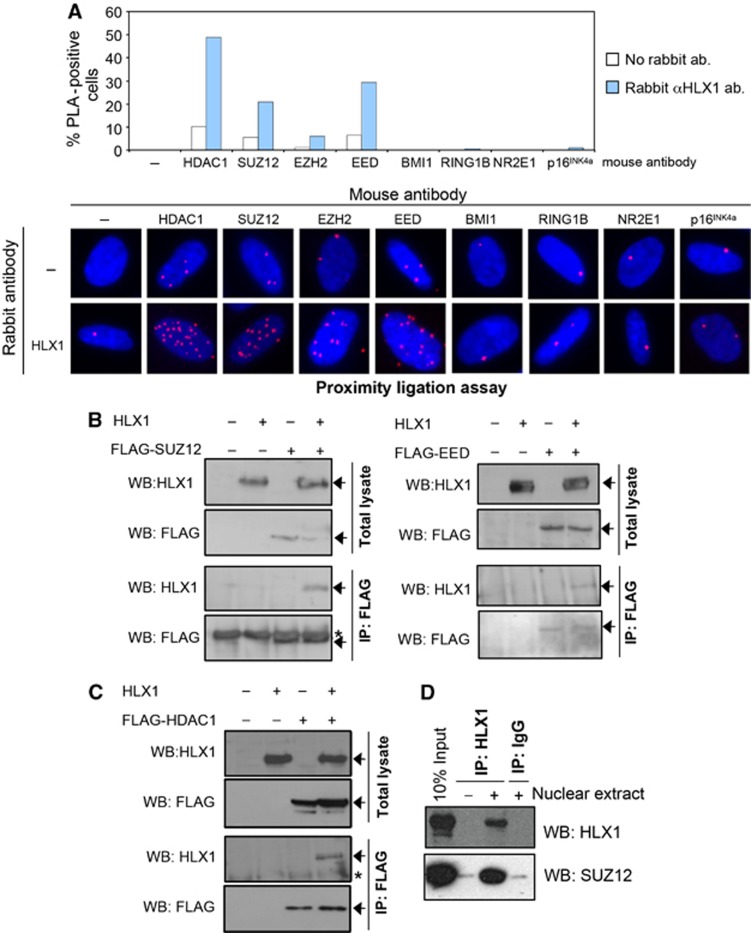
HLX1 associates with HDAC1 and the PRC2
We next investigated whether HLX1 associates with HDAC1 or PRCs. First, we used in situ proximity ligation assays (PLAs) (Fredriksson et al, 2002; Thymiakou and Episkopou, 2011). PLA takes advantage of specific antibodies and DNA amplification to identify co-localizing proteins. Using PLA, we detected that HLX1 associates with HDAC1, SUZ12 and other PRC2 components (Figure 6A). The association was observed between endogenous proteins (Figure 6A) and confirmed by overexpressing one or both interactors (Supplementary Figure S8). The specificity of the PLA assay was established with multiple controls (Figure 6A; Supplementary Figure S8). We next expressed HLX1 and FLAG-tagged versions of components of the PRC2 and PRC1 (Figure 6B; Supplementary Figure S9) or HDAC1 (Figure 6C). By performing co-IP experiments, we observed interactions between HLX1 and PRC2 components (Figure 6B) and HLX1 and HDAC1 (Figure 6C) but not between HLX1 and members of PRC1 (Supplementary Figure S9). To gain additional evidence for an interaction between HLX1 and the PRC2, we immunoprecipitated endogenous HLX1 from HeLa cells and using IB showed its association with SUZ12 (Figure 6D).
Figure 6.
HLX1 associates with HDAC1 and the PRC2. (A) In situ PLA of endogenous proteins in IMR-90 shows an association between HLX1 and the PRC2 and HLX1 and HDAC1. A rabbit antibody against HLX1 and the indicated mouse antibodies were used to detect the protein complexes. Representative images (bottom) and a quantification of the percentage of cells displaying an interaction (top) are shown. Cells presenting 10 or more nuclear spots were scored as positive. Blue: DAPI; Red: PLA signal. (B, C) HLX1 co-immunoprecipitates with the PRC2 and HDAC1. HEK 293T cells were transfected with plasmids expressing HLX1 and a FLAG-tagged version of SUZ12 or EED (B) or HDAC1 (C) as indicated. Total lysates or FLAG immunoprecipitates were prepared and immunoblots conducted using antibodies recognizing HLX1 or the FLAG epitope. Specific bands are noted by an arrow, while non-specific bands are denoted by *. WB, western blot; IP, immunoprecipitation. (D) Co-immunoprecipitation between endogenous HLX1 and SUZ12. HeLa nuclear extracts were used to conduct IPs using antibodies against HLX1 or IgG controls. Proteins were resolved by SDS–PAGE and HLX1 or SUZ12 detected by immunoblotting.
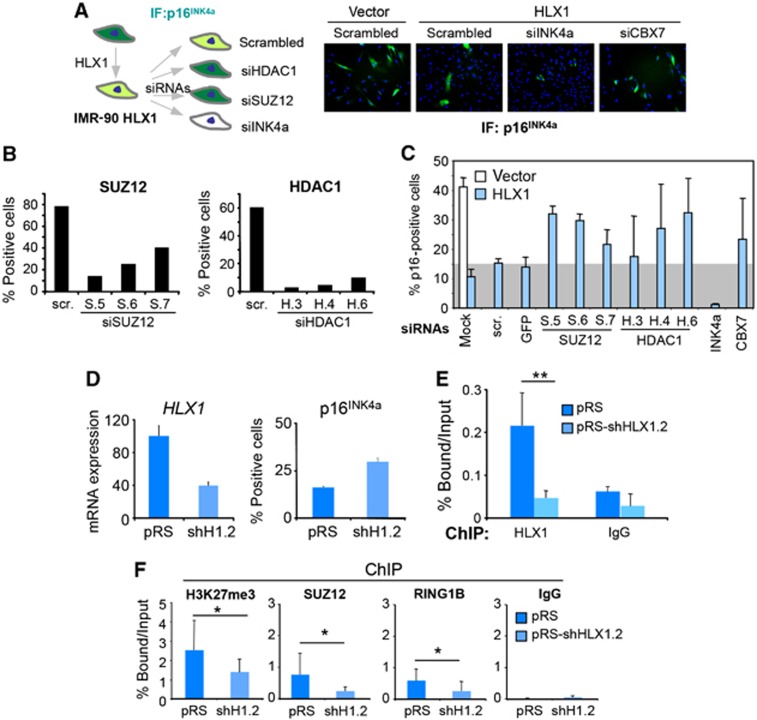
HLX1 repression of p16INK4a depends on PRC2 and HDAC1
In view of the physical association between HLX1 and the PRC2 and HDAC1, we investigated whether repression of p16INK4a by HLX1 depends on PRCs and HDAC1. To this end, IMR-90 cells expressing HLX1 were transfected with multiple siRNAs targeting HDAC1 or SUZ12 to analyse their effect on p16INK4a expression (summarized in Figure 7A). As previously noted, the expression of HLX1 caused a decrease in the percentage of p16INK4a-positive IMR-90 cells and in their nuclear levels of p16INK4a expression (Figure 7A). The ability of the siRNAs used to knock down SUZ12 or HDAC1 was confirmed by quantitative IF (Figure 7B; Supplementary Figure S10A and B). Depletion of SUZ12 or HDAC1 caused a partial reversion of the p16INK4a repression achieved by HLX1 (Figure 7C; Supplementary Figure S10C), suggesting that p16INK4a repression depends on PRCs and HDAC1. To confirm this, we knocked down HLX1 and observed that upregulation of p16INK4a (Figure 7D) correlated with a significant loss not only of HLX1 (Figure 7E) but also of H3K27me3 marks and binding of Polycomb proteins at the INK4/ARF locus (Figure 7F). These changes on INK4a promoter occupancy are reminiscent of what is observed upon depletion of CBX7 (Supplementary Figure S11). Similar results were also observed when we expressed a HLX1 homeodomain mutant (HLX1T328M), which retains its ability to interact with PRC2 but could not be recruited to the INK4a promoter (Supplementary Figure S12). These results suggest that HLX1 recruits PRCs and HDAC1 to repress INK4a expression.
Figure 7.
HLX1 repression of p16INK4a is dependent on PRC2 and HDAC1. (A–C) Repression of p16INK4a expression by HLX1 depends on HDAC1 and SUZ12. (A) Cartoon outlining the experiment presented in (A–C) (left). Quantification is provided in (C). (B) The efficiency of siRNAs targeting SUZ12 (left) or HDAC1 (right) in knocking down their respective targets was evaluated by quantitative IF. (C) HLX1 repression of p16INK4a is dependent on PRCs and HDAC1. Quantification of p16INK4a IF in IMR-90 (white bars) and IMR-90 HLX1 (blue bars) transfected with the indicated siRNAs. As a reference, a grey background represents the p16INK4a levels in HLX1 cells transfected with control siRNAs. (D–F) HLX1 knockdown results in loss of H3K27me3 and binding of PcG proteins at the INK4a promoter. (D) IMR-90 cells were infected (at passages 12 and 13) with a vector knocking down HLX1 (pRS-shHLX1.2). qRT–PCR showing HLX1 knockdown (left) and percentage of p16INK4a-positive cells quantified from IF (right) are shown. (E, F) Chromatin of these cells (at passages 14 and 15) was used to conduct ChIP, showing a decrease in HLX1 occupancy (E), H3K27me3 and binding of SUZ12 and RING1B (F) upon HLX1 knockdown at the INK4/ARF locus (using primer set 6). ChIP signal for HLX1, H3K27me3, SUZ12 and RING1B was significantly lower in cells with depleted HLX1. All ChIP signals were significantly higher (P<0.001) than their corresponding IgG signals. *P<0.05, **P<0.01.
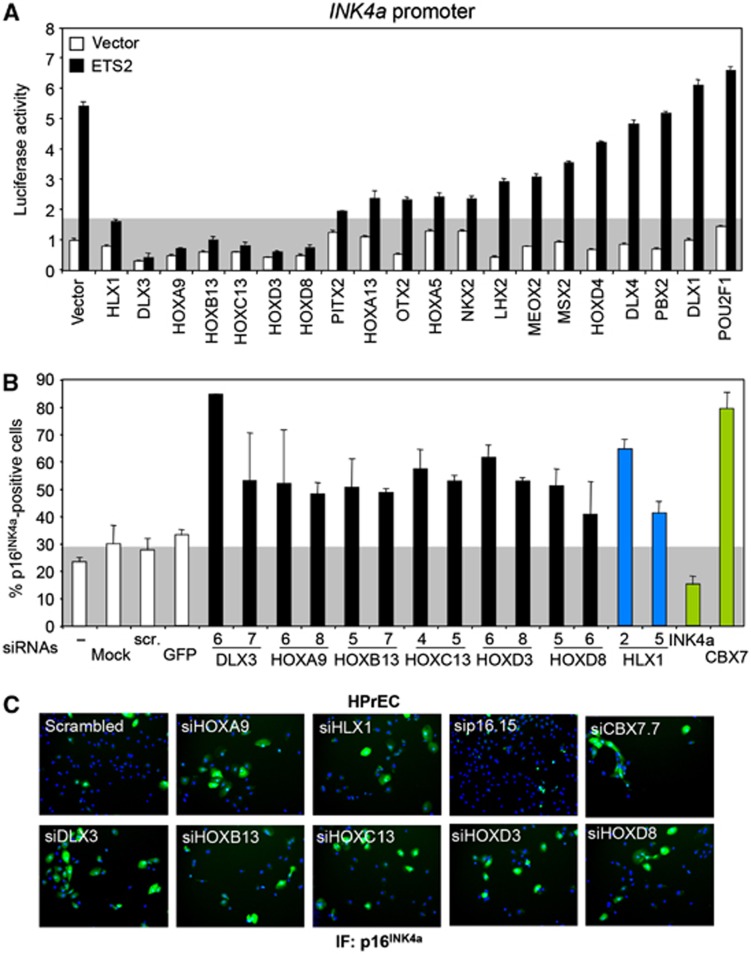
Several homeodomain-containing proteins also regulate p16INK4a expression
The mammalian complement of homeodomain-containing proteins exceeds 260 (Berger et al, 2008). To understand whether the regulation of p16INK4a by HLX1 is a special case or a more general phenomenon, we tested 19 additional Homeobox genes for their ability to regulate INK4a. Using the INK4a promoter reporter assay described earlier (Figure 4B), we identified six additional INK4a repressors (DLX3, HOXA9, HOXB13, HOXC13, HOXD3 and HOXD8; Figure 8A). We next knocked down the expression of these genes using two siRNAs for each (Supplementary Figure S13A) and analysed the effects on p16INK4a expression by IF. We performed these experiments in HPrEC, as they are a more sensitive system for measuring changes in p16INK4a expression (Supplementary Figure S4). Knockdown of any of the six homeodomain-containing proteins caused decreased cell growth and p16INK4a upregulation (Figure 8B and C; Supplementary Figure S13B). Interestingly, we analysed the similarity between the Homeobox DNA-binding profiles (Berger et al, 2008) of the seven homeodomain-containing proteins regulating p16INK4a (HLX1 and the six identified here) and found a significantly higher correlation between them (r=0.82, P=0.038) than between random combinations of any other seven homeodomain-containing proteins (Supplementary Figure S13C).
Figure 8.
Several homeodomain-containing proteins also regulate p16INK4a expression. (A) A luciferase screening identifies multiple Homeobox proteins preventing the activation of an INK4a promoter reporter by ETS2. A grey background represents the luciferase activity in HLX1/ETS2 transfected cells. (B, C) Knockdown of Homeobox genes results in p16INK4a induction. HPrEC were transfected with independent siRNAs targeting the genes identified in (A) as putative INK4a repressors. p16INK4a IF was carried out 5 days after transfection. Quantification of the percentage of p16INK4a-positive cells is shown (B), together with representative images (C). As a reference, a grey background represents the p16INK4a levels in cells transfected with control siRNAs. Controls are shown as white bars, HLX1 siRNAs as dark blue bars and siRNAs targeting INK4a (sip16.15) or CBX7 (siCBX7.7) as green bars.
HOXA9 also represses p16INK4a transcription in a PRC2- and HDAC1-dependent manner
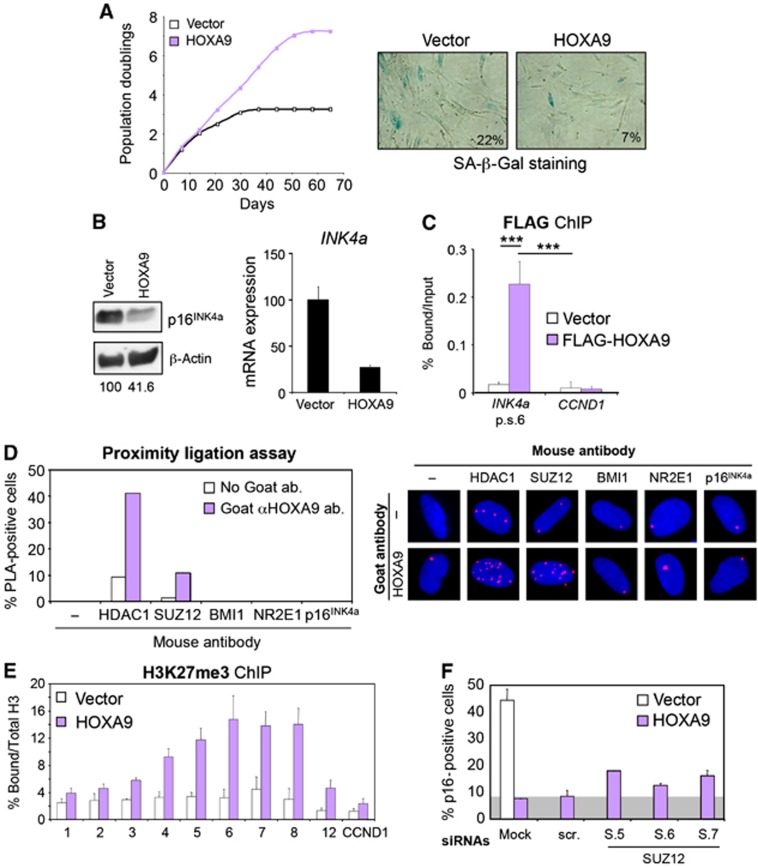
Among the Homeobox proteins with ability to regulate p16INK4a, we identified HOXA9, a known oncogene in leukaemia, and HOXB13, linked to prostate cancer progression (Kim et al, 2010), suggesting the potential significance of p16INK4a repression by Homeobox proteins in cancer. Furthermore, we recently showed that HOXA9 contributes in part to leukaemogenesis by negating senescence through p16INK4a repression (Smith et al, 2011). Expression of HOXA9 extends lifespan of IMR-90 cells (Figure 9A, left; Supplementary Figure S14A) accompanied with increased numbers of BrdU-positive cells and a decreased percentage of SA-β-Gal-positive cells (Figure 9A, right and data not shown). This correlated with a decrease in INK4a expression in IMR-90 cells expressing HOXA9 (Figure 9B). The repression of p16INK4a by HOXA9 was probably direct as HOXA9 was detected bound in the proximity to the INK4a promoter similar to HLX1 (Figure 9C).
Figure 9.
HOXA9 also represses p16INK4a transcription in a PRC2- and HDAC1-dependent manner. (A) Expression of HOXA9 delays senescence of IMR-90 cells. Growth curve showing the extension of lifespan on IMR-90 cells expressing HOXA9 (left). Representative pictures showing less SA-β-Gal-positive cells in IMR-90 cells expressing HOXA9 at passage 22 (right). (B) HOXA9 expression results in decreased p16INK4a levels. Immunoblot (left) and qRT–PCR (right) showing reduced p16INK4a levels in IMR-90 cells expressing HOXA9. The numbers under the blots correspond to quantification of HOXA9 signal normalized to β-actin. (C) HOXA9 binds to the INK4/ARF locus. Chromatin from IMR-90 cells expressing Flag-HOXA9 was used to perform ChIP showing that HOXA9 binds in the proximity to the INK4a promoter. (D) In situ PLA showing association between HOXA9 and HDAC1 and HOXA9 and SUZ12 in IMR-90 cells. Representative pictures (right) and quantification (left) are shown. (E) Expression of HOXA9 results in an increase in H3K27me3 marks at the INK4/ARF locus. (F) HOXA9 repression of p16INK4a is dependent on SUZ12. Quantification of p16INK4a IF in IMR-90 (white bars) and IMR-90 HOXA9 (purple bars) transfected with the indicated siRNAs. Grey background represents the p16INK4a levels in HOXA9-expressing cells transfected with control siRNAs (right). ***P<0.001.
We next wondered whether HOXA9, as a paradigm for the other identified Homeobox genes, represses p16INK4a in a way dependent on PRCs and HDAC1, as HLX1 does. To investigate this, we first analysed the ability of HOXA9 to associate with PRC2 and HDAC1. We observed an association between HOXA9 and HDAC1 and SUZ12 (Figure 9D; Supplementary Figure S14B and C). Similarly to what we observed for HLX1, HOXA9 expression also resulted in increased H3K27me3 marks at the INK4/ARF locus (Figure 9E). Finally, depletion of SUZ12 resulted in a partial loss of HOXA9-mediated p16INK4a repression (Figure 9F), further suggesting that HOXA9 regulates p16INK4a in a PRC2-dependent mechanism as HLX1.
Discussion
Cells undergo profound transcriptional changes when they reach senescence. So far, several transcription factors have been shown to play critical roles in the regulation and implementation of senescence (Lanigan et al, 2011). Having that into consideration, we designed a screen to identify novel transcriptional regulators controlling senescence. As a result, we identified the homeodomain-containing protein HLX1. HLX1 expression extends cellular lifespan and blunts OIS. Although an effect of HLX1 on regulating alternative pathways cannot be discarded, our evidence points to the repression of p16INK4a by HLX1 as a key mechanism explaining the effects of HLX1 on senescence. ChIP, transcriptome analysis and interaction studies suggest that the ability of HLX1 to repress transcription is mediated at least partially by PRCs.
The question of how PRCs are recruited to their target genes in mammalian cells is still unresolved. The proposed model is that interaction with multiple transcription factors and lincRNAs can dictate this specificity (Bracken and Helin, 2009). lincRNAs, such as ANRIL, and transcription factors, such as TWIST1 and Zfp277, recruit PRCs to repress INK4a (Negishi et al, 2010; Yang et al, 2010; Yap et al, 2010). Here, we add Homeobox proteins (such as HLX1 or HOXA9) to the list of transcription factors that rely on PRCs to repress INK4a. There seems to be redundancy in the ability of homeodomain-containing proteins to repress p16INK4a. To test this, we knocked down the expression of HLX1 in cells expressing HOXA9 and vice versa (Supplementary Figure S15A). Although the results obtained are compatible with Homeobox redundancy in INK4a regulation, the issue might be more complex as homeobox proteins can form heterodimers (Shah and Sukumar, 2010) (and in particular HLX1 and HOXA9; Supplementary Figure S15B and C). Therefore, further work is needed to unravel the significance of multiple homeobox protein regulating INK4a. In addition, Homeobox proteins are often expressed in a tissue-specific manner and we believe that specific Homeobox proteins can recruit PRC2 to repress p16INK4a expression in different tissues. For example, Hlx1−/− mice have defects in the expansion of embryonic liver and gut, caused by restricted proliferation in the absence of apoptosis in those organs (Hentsch et al, 1996). These are two of the tissues in which HLX1 is highly expressed. Whether this phenotype is driven by abnormal expression of p16INK4a or it is unrelated remains to be determined. Interestingly, a recent study identified HLX1 as a gene frequently overexpressed in patients with acute myeloid leukaemia (AML) that correlates with poor survival (Kawahara et al, 2012). Although the authors suggest regulation of PAK1 and BTG1 as mechanisms explaining the prevalence of HLX1 in AML, the ability of HLX1 to regulate senescence may be an additional trait contributing to its role in AML.
On a more general note, it is interesting to speculate whether a downregulation of any of these homeodomain proteins could explain the increase in p16INK4a expression during senescence. However, HLX1 levels did not change significantly during replicative senescence or OIS (Supplementary Figure S16A), and those of the other homeodomain proteins analysed were, in general, very low and either did not change significantly or were upregulated during replicative senescence (Supplementary Figure S16B) or OIS (data not shown). Therefore, although homeodomain-containing proteins regulate the steady-state levels of p16INK4a, their expression during senescence does not explain p16INK4a upregulation. The expression of EZH2 and other PRC2 components decreases during senescence (Bracken et al, 2007; Supplementary Figure S16C). Those changes together with JMJD3 upregulation (Agger et al, 2009; Barradas et al, 2009) explain better the increase in p16INK4a levels during senescence. The fact that many Homeobox genes are PRC targets might also explain why they are found upregulated during senescence (Supplementary Figure S16B).
Interestingly, OCT4 and NANOG, two core components of the pluripotency network of transcription factors responsible for recruiting PRCs to targets in embryonic stem (ES) cells, are homeodomain-containing proteins and PRC members form part of the OCT4 interactome (van den Berg et al, 2010). An examination of significantly enriched sequences present in PcG target genes in ES cells revealed multiple homeodomain binding motifs (Ku et al, 2008), further suggesting that certain homeodomain-containing proteins could recruit PRC to a subset of targets.
Given that the expression of p16INK4a is kept at bay not only at embryonic stages, but also later during normal development and that PRCs are critical for this process, it is not far-fetched to speculate that Homeobox genes, which are expressed in specific cell types, could have a role in recruiting PRCs to silence INK4a. In summary, the aforementioned results suggest that multiple Homeobox proteins can regulate p16INK4a in a similar way to how HLX1 and HOXA9 do. Therefore, we propose that recruitment of PRCs and HDAC1 by Homeobox proteins to repress p16INK4a and other target genes could be a conserved mechanism with potential implications in development and cancer.
Materials and methods
Antibodies
Primary antibodies used were β-actin (sc-47778, Santa Cruz), BrdU (A21303, Invitrogen), HDAC1 (ab19845, Abcam; ab46985, Abcam), HLX1 (09-084, Millipore; HPA005968, Sigma; Abnova, H00003142-B01P), p16INK4a (JC-8, CRUK; sc-468, Santa Cruz; sc-56330, Santa Cruz), p21CIP1 (P1484, Sigma), CBX7 (Abcam, ab21873), p53 (sc-126, Santa Cruz), RB (BD Parmingen, 554136), SUZ12 (ab12073, Abcam; ab107253, Abcam), FLAG (F3165, Sigma), HA (MMS-101P, Covance), EZH2 (ab3748, Abcam; AC22, 3147, Cell Signaling), EED (342-441, H00008726-M05, Abnova), α-tubulin (T6074, Sigma), HOXA9 (sc-17155, Santa Cruz), BMI1 (ab14389, Abcam), RING1A (2820, Cell Signaling), RING1B (mouse monoclonal, kind gift of H Koseki), NR2E1 (sc-100905, Santa Cruz), H3K9ac (07–352, Millipore), Histone H3 (ab1791, Abcam) and H3K27me3 (CS-069-100, Diagenode).
Plasmids
Plasmids encoding FLAG-tagged versions of EED, BMI1, RING1A, HPH2, ETS2, HDAC1, SUZ12 and HOXA9 and pLXSN-E7 have been described before. MSCV retroviral vectors expressing HEY1, MSX2, NAB2, HLX1, SPDEF, TGIF1 and TSC22D4 were generated by cloning the cDNAs from pCMV6-based plasmids (Origene). The pGL2-INK4a promoter reporter has been described previously (Hara et al, 1996). pcDNA3-HLX1 and pBABE-HLX1 plasmids were generated by cloning HLX1 (untagged, or FLAG-tagged) in the corresponding vectors. A322T and T328M mutations were introduced in HLX1 by site-directed mutagenesis. pCMV6 plasmids expressing Homeobox proteins were obtained from Origene. shRNAs were cloned in pRetroSUPER (pRS), as previously described (Brummelkamp et al, 2002). The shRNA sequence targeting HLX1 incorporated to the pRS retroviral vector was shHLX1.2 (TTGTTTGGTGCAGAGAACGCT). Lentiviral pLKO-based shRNA targeting HLX1 (pLKO-shHLX1.2) was obtained from Sigma (clone ID TRCN0000014821). Lentiviral pLKO-based shRNA targeting CBX7 (pLKO-shCBX7.5) was obtained from Sigma (clone ID TRCN0000019144). Retroviral knockdown of p16INK4a (GAGGAGGTGCGGGCGCTGC) (Voorhoeve and Agami, 2003), p21CIP1 (TCCCACAATGCTGAATATACA) (Banito et al, 2009) and p53 (GTAGATTACCACTGGAGTC) (Brummelkamp et al, 2002) was performed using previously described shRNA constructs.
Cell culture
IMR-90 and WI-38 human diploid fibroblasts, HEK 293T and HeLa cells were obtained from the ATCC. Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 10% fetal bovine serum. Primary human keratinocytes (NHK) and prostate epithelial cells (HPrEC) were obtained from Lonza and maintained in keratinocyte growth medium (KGM-2, Lonza) and prostate epithelial cell growth medium (PrEGM, Lonza), respectively. IMR-90 ER:RAS cells have been previously described (Barradas et al, 2009). Passage numbers are indicated in experiments involving IMR-90 cells. For experiments involving siRNAs or shRNAs, IMR-90 cells were infected or transfected at passages 12 and 13 and used for different assays at passages 14 and 15. IMR-90 cells at passages 17–19 were infected with vectors expressing HLX1 or HOXA9. Growth curves were performed and cells expanded. The different assays were performed between passages 22 and 28.
Retroviral and lentiviral infection
Methods, used for retrovirus and lentivirus production and infection, are detailed in Gil et al (2004).
SA-β-Gal, BrdU incorporation and crystal violet staining
These assays were performed as described previously (Gil et al, 2004). For BrdU incorporation, IMR-90 cells were incubated with 50 μM BrdU for 16 h. Upon IF, images were acquired with an IN Cell Analyzer 1000 and >1000 cells per condition were analysed. For the SA-β-Gal, at least 100 cells per condition were counted.
Transfection of siRNAs
IMR-90 fibroblasts, NHK or HPrEC, were reverse transfected with 30 nM siRNA in 96-well or 6-well plates, using a 3.5% solution of HiPerFect transfection reagent (QIAGEN). The Cy3-labelled siGLO cyclophilin B siRNA (Dharmacon) was used to monitor transfection efficiency and the AllStars scrambled siRNA or siRNA targeting GFP (QIAGEN) served as negative controls. For a list of siRNAs, see Supplementary Table S2.
Analysis of global changes in protein levels
SILAC in combination with Orbitrap mass spectrometry was used to evaluate differential protein levels in IMR-90 cells infected with a control vector (pRS) versus a vector knocking down HLX1 (pRS-shHLX1.2), as described in detail in Supplementary data.
Immunofluorescence
IF was performed using an IN Cell Analyzer 1000 (GE Healthcare). Image processing and quantification was performed using the IN Cell Investigator software (v1.7; GE Healthcare), as described in Supplementary data.
Immunoblotting
IB was performed following standard procedures and quantified using ImageJ software.
Quantitative RT–PCR analysis
Total RNA was extracted using the RNeasy Mini kit (QIAGEN) and reverse transcription was carried out using SuperScript II (Invitrogen). PCR was performed on an Opticon 2 (Bio-Rad) using SYBR Green PCR Master Mix (Applied Biosystems) or an ABI7900HT Fast Real-Time PCR System (Applied Biosystems) using TaqMan Universal PCR Master Mix (Applied Biosystems). Gene expression was normalized to RPS14, GAPDH or TBP. Primer sets and TaqMan Gene Expression Assays (Applied Biosystems) used are listed in Supplementary Table S3.
Luciferase reporter assays
HEK 293T cells were reverse transfected in 96-well plates with 150 ng firefly luciferase reporter plasmid, 10 ng TK-Renilla luciferase and 60 ng of the different transcription factors, unless otherwise indicated, using TransIT-LT1 (Mirus). The cells were lysed 48 h post transfection and processed using the Dual-Glo Luciferase Assay System (Promega). The expression of the firefly luciferase reporter plasmid was normalized to the expression of the Renilla luciferase reporter.
Gene expression profiling
For microarray experiments, cRNA was hybridized to Human Gene 1.0 ST arrays (Affymetrix) following manufacturer’s recommendations. Three biological replicates were performed for each condition. Microarray analysis was carried out at EMBL. Microarray data were normalized using Robust Multichip Average (RMA) and differentially expressed genes were identified using Linear Models for MicroArrays (LIMMA). A cutoff of a Benjamini-Hochberg (BH) false discovery rate <0.05 was used to identify significant genes. All analyses were carried out in R (v2.13.0). As a quality-control check, a hierarchical clustering of the samples was used to visualize whether samples grouped by treatment.
Gene set enrichment analysis
We used GSEA (v2.07) to examine the association between gene sets and gene expression upon siHLX1 or siCBX7 treatment. GSEA is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states. We ranked the genes by log2 fold change obtained from LIMMA contrasts. Pre-ranked GSEA was performed using curated Molecular Signatures Database (v3.0) with a size of 5–1200 genes ( http://www.broadinstitute.org/gsea/msigdb/index.jsp). A gene set was considered significantly enriched when the nominal P-value was <0.05 and the false discovery rate q-value was <0.25. To define the ‘Bracken’ Polycomb target gene sets, 341 potential PcG target genes were taken from Supplementary Table 1 of (Bracken et al, 2006). Gene symbols were examined and updated to ensure they contained only official HUGO symbols. For each knockdown, we identified the upregulated genes and created GSEA-specific genesets in gmx format.
DNA pulldown assay
Lysate from 293T cells overexpressing HLX1 was prepared in HKMG buffer (10 mM Hepes pH 7.9, 100 mM KCl, 5 mM MgCl2, 10% glycerol, 0.5% Nonidet P-40 (NP-40), 1 mM DTT and protease inhibitors (Complete EDTA-free, Roche)) and pre-cleared with pre-equilibrated streptavidin-coupled Dynabeads (Invitrogen). Pairs of complementary oligonucleotides (40-bp long, with the sense oligonucleotide biotinylated at the 5′ end, Sigma, sequences described in Supplementary Table S4) were annealed and incubated overnight at 4°C with the cell lysate. DNA-bound proteins were collected by incubation for 1 h at 4°C with streptavidin-coupled Dynabeads (Invitrogen), washed four times in HKMG buffer, separated by SDS–PAGE and HLX1 detected by IB.
Chromatin immunoprecipitation
ChIP was performed as previously described (Barradas et al, 2009). Immunoprecipitation of cross-linked chromatin was conducted with the antibodies listed above. After immunoprecipitation, DNA was extracted using the QIAquick PCR purification kit (QIAGEN) and an aliquot amplified by qRT–PCR using oligonucleotide primers described in Supplementary Table S3.
Proximity ligation assay
PLA was performed using a Duolink II kit (Olink Bioscience), as previously described (Thymiakou and Episkopou, 2011).
Immunoprecipitation studies
HEK 293T cells were transfected in 10 cm plates with 20 μg plasmid, using PEI. Forty-eight hours after transfection, cells were washed, scraped in ice-cold PBS and lysed in a buffer containing 50 mM Tris pH 8.0, 0.5% NP-40, 200 mM NaCl, 0.1 mM EDTA, 10% glycerol and protease inhibitors (Complete EDTA-free, Roche). Total cell lysates were then incubated for 2 h at 4°C with the appropriate antibody. Immune complexes were collected by incubation for 1 h at 4°C with Dynabeads protein G (Invitrogen) and washed three times in lysis buffer. Bound proteins were eluted with SDS sample buffer and analysed by gel electrophoresis and IB.
Statistical analysis
Values represent mean±standard error of the mean (s.e.m.). At least three biological replicas were carried out. Statistical analyses were performed using two-tailed Student’s t-test with Prism software. Significance levels were denoted as: *P≤0.05, **P≤0.01 and ***P≤0.001.
Supplementary Material
Acknowledgments
We are grateful to A Fisher, D Landeira, N Brockdorff, H Koseki, J Carroll, H Mohammed, M-M Zhou, MG Rosenfeld, D Beach and B Lenhard for reagents, help and advice and C-H Chen for technical support. M Walsh is supported by the awards SS-AG2482-10 from the Ellison Medical Foundation and 1RO1CA154809 from NCI of the NIH. Core funding from the MRC and grants from MRC Technology, CRUK and AICR funded the research in J Gil’s laboratory. N Popov was funded by an MRC studentship. N Martin was funded by fellowships from the ARC, EMBO and Marie Curie FP7. J Gil is also supported by the EMBO Young Investigator Programme.
Author contributions: JG, NP and NM conceived the project. NP and NM performed most of the experiments and analysed the data. AO and FA performed and analysed the ChIP experiments. NM and ET performed the PLA experiments. GD and TC carried out the bioinformatic analysis. APS carried out the Mass Spectrometry. SR, BBZ and SL generated reagents and performed experiments. JG, CWES, GP, VE and MW designed experiments and analysed the data. JG and VE wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abate-Shen C (2002) Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer 2: 777–785 [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K (2009) The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev 23: 1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J (2009) Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ, Peters G, Gil J (2009) Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev 23: 1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Paro R (2011) Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet 12: 123–135 [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML, Hughes TR (2008) Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA et al. (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Helin K (2009) Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9: 773–784 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, Hansen KH, Helin K (2007) The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M (2007) Cellular senescence in cancer and aging. Cell 130: 223–233 [DOI] [PubMed] [Google Scholar]
- Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U (2002) Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 20: 473–477 [DOI] [PubMed] [Google Scholar]
- Fridman AL, Tainsky MA (2008) Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene 27: 5975–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martinez D, Beach D (2004) Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol 6: 67–72 [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G (2006) Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7: 667–677 [DOI] [PubMed] [Google Scholar]
- Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G (1996) Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol 16: 859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentsch B, Lyons I, Li R, Hartley L, Lints TJ, Adams JM, Harvey RP (1996) Hlx homeo box gene is essential for an inductive tissue interaction that drives expansion of embryonic liver and gut. Genes Dev 10: 70–79 [DOI] [PubMed] [Google Scholar]
- Kawahara M, Pandolfi A, Bartholdy B, Barreyro L, Will B, Roth M, Okoye-Okafor UC, Todorova TI, Figueroa ME, Melnick A, Mitsiades CS, Steidl U (2012) H2.0-like homeobox regulates early hematopoiesis and promotes acute myeloid leukemia. Cancer Cell 22: 194–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK (2009) Polycomb group complexes—many combinations, many functions. Trends Cell Biol 19: 692–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YR, Oh KJ, Park RY, Xuan NT, Kang TW, Kwon DD, Choi C, Kim MS, Nam KI, Ahn KY, Jung C (2010) HOXB13 promotes androgen independent growth of LNCaP prostate cancer cells by the activation of E2F signaling. Mol Cancer 9: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y (2011) Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30: 1956–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS (2010) The essence of senescence. Genes Dev 24: 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Fisher AG (2011) Inactive yet indispensable: the tale of Jarid2. Trends Cell Biol 21: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanigan F, Geraghty JG, Bracken AP (2011) Transcriptional regulation of cellular senescence. Oncogene 30: 2901–2911 [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey L, Helin K (2010) Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 35: 323–332 [DOI] [PubMed] [Google Scholar]
- Negishi M, Saraya A, Mochizuki S, Helin K, Koseki H, Iwama A (2010) A novel zinc finger protein Zfp277 mediates transcriptional repression of the Ink4a/arf locus through polycomb repressive complex 1. PLoS ONE 5: e12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067–1070 [DOI] [PubMed] [Google Scholar]
- Popov N, Gil J (2010) Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics 5: 685–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Sukumar S (2010) The Hox genes and their roles in oncogenesis. Nat Rev Cancer 10: 361–371 [DOI] [PubMed] [Google Scholar]
- Smith LL, Yeung J, Zeisig BB, Popov N, Huijbers I, Barnes J, Wilson AJ, Taskesen E, Delwel R, Gil J, Van Lohuizen M, So CW (2011) Functional crosstalk between Bmi1 and MLL/Hoxa9 axis in establishment of normal hematopoietic and leukemic stem cells. Cell Stem Cell 8: 649–662 [DOI] [PubMed] [Google Scholar]
- Thymiakou E, Episkopou V (2011) Detection of signaling effector-complexes downstream of bmp4 using PLA, a proximity ligation assay. J Vis Exp 49: pii2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP (1999) Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet 23: 474–478 [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, Agami R (2003) The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4: 311–319 [DOI] [PubMed] [Google Scholar]
- Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, Chang SY, Lee OK, Wu KJ (2010) Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol 12: 982–992 [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.