Abstract
GW182 family proteins interact with Argonaute proteins and are required for the translational repression, deadenylation and decay of miRNA targets. To elicit these effects, GW182 proteins interact with poly(A)-binding protein (PABP) and the CCR4–NOT deadenylase complex. Although the mechanism of miRNA target deadenylation is relatively well understood, how GW182 proteins repress translation is not known. Here, we demonstrate that GW182 proteins decrease the association of eIF4E, eIF4G and PABP with miRNA targets. eIF4E association is restored in cells in which miRNA targets are deadenylated, but decapping is inhibited. In these cells, eIF4G binding is not restored, indicating that eIF4G dissociates as a consequence of deadenylation. In contrast, PABP dissociates from silenced targets in the absence of deadenylation. PABP dissociation requires the interaction of GW182 proteins with the CCR4–NOT complex. Accordingly, NOT1 and POP2 cause dissociation of PABP from bound mRNAs in the absence of deadenylation. Our findings indicate that the recruitment of the CCR4–NOT complex by GW182 proteins releases PABP from the mRNA poly(A) tail, thereby disrupting mRNA circularization and facilitating translational repression and deadenylation.
Keywords: CCR4–NOT, decapping, miRNAs, mRNA decay, NOT1, TNRC6
Introduction
GW182 family proteins play an essential role in miRNA-mediated gene silencing in animal cells (Huntzinger and Izaurralde, 2011; Fabian and Sonenberg, 2012). Several eukaryotic species including humans possess three GW182 paralogs (known as TNRC6A, B and C), whereas there is only one family member in Drosophila melanogaster (Dm GW182). GW182 proteins function as scaffold proteins for the assembly of silencing complexes on mRNA targets. Accordingly, they interact with Argonaute proteins (AGOs) through an N-terminal Argonaute binding domain and recruit additional effector complexes through a C-terminal silencing domain (SD), which is required for silencing in human cells (Figure 1A) (Huntzinger and Izaurralde, 2011; Fabian and Sonenberg, 2012). In particular, the SDs of the human TNRC6A, B and C directly interact with the cytoplasmic poly(A)-binding protein (PABP) and with the PAN3 and NOT1 subunits of the PAN2–PAN3 and CCR4–NOT deadenylase complexes, respectively (Fabian et al, 2009, 2011; Zekri et al, 2009; Huntzinger et al, 2010, 2013; Jínek et al, 2010; Kozlov et al, 2010; Braun et al, 2011; Chekulaeva et al, 2011).
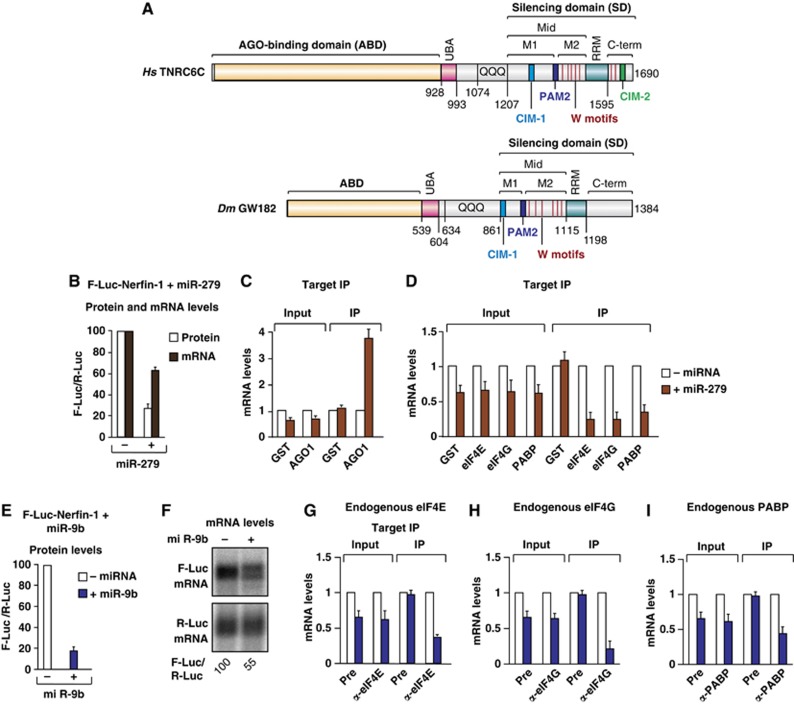
Figure 1.
The association of Dm eIF4E, eIF4G and PABP with silenced miRNA targets. (A) The domain organization of human TNRC6C and Dm GW182. ABD, AGO-binding domain; UBA, ubiquitin associated-like domain; QQQ, region rich in glutamine; Mid, middle region containing the PAM2 motif, which divides the Mid region into the M1 and M2 regions; RRM, RNA recognition motif; C-term, C-terminal region. The positions of the CIM-1 and CIM-2 motifs are indicated. Vertical red lines indicate the positions of tryptophans in the M2 and C-term regions that contribute to NOT1 binding. Amino-acid positions at domain boundaries are indicated below the protein outlines. (B–D) S2 cells were transfected with a mixture of three plasmids: one expressing an F-Luc-Nerfin-1 reporter, another expressing the miR-279 primary transcript or the corresponding empty vector (–), and a third expressing Renilla luciferase (R-Luc). The transfection mixtures contained plasmids expressing the indicated HA-tagged proteins. (B) Firefly luciferase activity (white bars) and mRNA levels (black bars) were normalized to those of the Renilla luciferase and set at 100 in the absence of miR-279 (–). (C, D) HA-tagged proteins were immunoprecipitated using anti-HA antibodies. HA–GST served as a negative control. The levels of F-Luc-Nerfin-1 mRNA (normalized to R-Luc mRNA) in the inputs and IPs were analysed by RT–qPCR. For each HA-tagged protein, the normalized values of F-Luc-Nerfin-1 mRNA were set at 1 in the absence of miR-279 (white bars). (E–I) S2 cells were transfected with a mixture of three plasmids as described above except that miR-9b was used instead of miR-279. (E) Firefly luciferase activity was normalized to that of the Renilla luciferase and set at 100 in the absence of miR-9b. (F) Northern blot of representative RNA samples. Numbers below the panel indicate F-Luc mRNA levels normalized to the R-Luc transfection control and set at 100 in the absence of miR-9b. (G–I) Endogenous eIF4E, eIF4G and PABP were immunoprecipitated using polyclonal antibodies. The corresponding preimmune (Pre) sera served as negative controls. The association of F-Luc-Nerfin-1 mRNA with the endogenous proteins was analysed using RT–qPCR as described above. In all figures shown in this manuscript, bars represent mean values and error bars standard deviations from three independent experiments.
Source data for this figure is available on the online supplementary information page.
The SD is a bipartite region consisting of the middle (Mid) and C-terminal (C-term) regions of the GW182 proteins. The middle region is further divided into the M1 and M2 regions (Figure 1A), which together with the C-term region contribute to the interactions with deadenylases and PABP in an additive manner (Fabian et al, 2009, 2011; Zekri et al, 2009; Huntzinger et al, 2010, 2013; Braun et al, 2011; Chekulaeva et al, 2011). For example, PAN3 interacts with both the M2 and C-term regions of human TNRC6-SDs, whereas NOT1 interacts with tryptophan-containing sequences in the M1, M2 and C-term regions (Figure 1A) (Braun et al, 2011; Chekulaeva et al, 2011; Fabian et al, 2011; Huntzinger et al, 2013). The NOT1-binding motifs in the M1 and C-term regions were termed CCR4–NOT-interacting motifs 1 and 2 (CIM-1 and CIM-2), respectively (Figure 1A) (Fabian et al, 2011). In addition to the CIM-1 and CIM-2 motifs, tryptophan residues in the M2 region of the SD contribute to the interaction with NOT1 (Chekulaeva et al, 2011). Finally, binding to PABP is mediated by a conserved PAM2 motif (PABP-interacting motif 2) located between the M1 and M2 regions of the SD (Figure 1A) (Fabian et al, 2009; Huntzinger et al, 2010; Jínek et al, 2010; Kozlov et al, 2010). This motif was originally identified in the translational regulators Paip1 and Paip2 (PABP-interacting proteins 1 and 2) (Kahvejian et al, 2001), and it confers direct binding to the C-terminal MLLE domain of PABP (Fabian et al, 2009; Huntzinger et al, 2010; Jínek et al, 2010; Kozlov et al, 2010).
Remarkably, although the interaction of GW182 proteins with PABP and deadenylase complexes is conserved in D. melanogaster, the mode of interaction differs (Huntzinger et al, 2010, 2013; Braun et al, 2011). For example, the Dm SD lacks the CIM-2 motif, and in contrast to the human SDs, its deletion from Dm GW182 reduces but does not abolish binding to deadenylases (Huntzinger et al, 2010, 2013; Braun et al, 2011). In agreement with these observations, sequences upstream of the SD contribute to deadenylase binding in D. melanogaster (Chekulaeva et al, 2011; Huntzinger et al, 2013). Moreover, in contrast to the human proteins, Dm GW182 also interacts with PABP indirectly through the M2 and C-term regions in cultured cells (Zekri et al, 2009; Huntzinger et al, 2010, 2013). Therefore, the Dm GW182 PAM2 motif is dispensable for PABP binding and silencing in D. melanogaster (Chekulaeva et al, 2009, 2010, 2011, 9; Zekri et al, 2009; Eulalio et al, 2009a; Huntzinger et al, 2010, 2013).
Our previous studies showed that interaction of human GW182 proteins with PABP is important for mediating the silencing of miRNA targets in human cells (Huntzinger et al, 2010; Braun et al, 2011). However, how PABP contributes to silencing is unclear, and several non-mutually exclusive models have been proposed (Huntzinger and Izaurralde, 2011; Fabian and Sonenberg, 2012). One model proposes that GW182 proteins compete with eIF4G for binding to PABP, thereby preventing mRNA circularization and consequently inhibiting translation (Fabian et al, 2009; Zekri et al, 2009). A second model suggests by analogy with Paip2 that the GW182–PABP interaction may reduce the affinity of PABP for the poly(A) tail, thereby repressing translation (Fabian et al, 2009; Zekri et al, 2009; Huntzinger et al, 2010). A third model suggests that the PABP–GW182 interaction may accelerate miRNA-mediated deadenylation (Fabian et al, 2009; Jínek et al, 2010). Finally, a recent study in D. melanogaster cell-free extracts indicates that PABP stimulates silencing by facilitating the association of miRISC complexes with mRNA targets (Moretti et al, 2012). This study also shows that, upon miRISC binding, PABP progressively dissociates from the target. PABP dissociation was independent of deadenylation at early time points and was more pronounced at later time points due to target deadenylation (Moretti et al, 2012).
In contrast to the studies mentioned above, studies in zebrafish embryos and in D. melanogaster cell-free extracts in which silencing is mediated by exogenously supplemented miRNAs indicate that PABP is dispensable for silencing (Fukaya and Tomari, 2011; Mishima et al, 2012). Paradoxically, the GW182 PAM2 motif contributed to silencing efficiency in zebrafish embryos (Mishima et al, 2012). The conflicting results regarding the role of PABP in silencing suggest that PABP may be required for the silencing of specific targets and/or under specific cellular conditions.
To gain further insight into the role of PABP in silencing, we investigated the association of eIF4E, eIF4G and PABP with silenced versus translationally active miRNA targets. We show that silencing causes dissociation of these proteins from the mRNA target. eIF4E association is restored in cells depleted of decapping factors; in these cells, miRNA targets are deadenylated but are not further degraded. In contrast, eIF4G binding is not restored in these cells, suggesting that the association of eIF4G with miRNA targets is destabilized upon deadenylation. In contrast, PABP dissociates from miRNA targets in the absence of deadenylation. PABP dissociation is mediated by the GW182 proteins. However, contrary to expectations, the interaction of GW182 proteins with PABP is dispensable for PABP dissociation. Instead, the interaction between GW182 proteins and the CCR4–NOT complex is required to lower PABP association with the mRNA target. Accordingly, we show that tethered NOT1 and a catalytically inactive POP2 mutant are sufficient to cause PABP dissociation from bound mRNAs in the absence of deadenylation. Our results indicate that the recruitment of the CCR4–NOT complex to miRNA targets by GW182 proteins releases PABP from the mRNA poly(A) tail, thereby facilitating translational repression and increasing the accessibility of the poly(A) tail for deadenylases.
Results
Association of eIF4E, eIF4G and PABP with silenced miRNA targets
To monitor the association of eIF4E, eIF4G and PABP with silenced miRNA targets, we performed coimmunoprecipitation assays with endogenous or HA-tagged versions of these proteins and determined the levels of an actively translated or silenced miRNA reporter in the immunoprecipitates (IP) using real-time quantitative reverse transcription PCR (RT–qPCR). We used an miRNA reporter consisting of the firefly luciferase (F-Luc) open reading frame followed by the 3′ UTR of the D. melanogaster gene nerfin-1 (silenced by miR-9b and miR-279). This reporter is silenced predominantly at the translational level, although a 1.5- to 2-fold reduction in mRNA levels was consistently observed (Figure 1B, E and F). As a positive control, we analysed the association of HA–AGO1 with the reporter.
We observed that HA–AGO1 coimmunoprecipitated the F-Luc-Nerfin-1 reporter in the presence of miR-279 (Figure 1C), thus demonstrating the specificity of the assay. In contrast to AGO1, the association of HA-tagged eIF4E, eIF4G and PABP with the F-Luc-Nerfin-1 reporter decreased in the presence of miR-279 (Figure 1D). This effect could be explained by the degradation of the F-Luc-Nerfin-1 mRNA, although a greater reduction of reporter levels was observed in the IP compared with the input (Figure 1D, compare IP versus Input). Similar results were obtained when silencing of the F-Luc-Nerfin-1 reporter was mediated by miR-9b (Figure 2A–E, below). Western blot analyses indicated that the HA-tagged proteins were expressed at levels comparable to the endogenous proteins and were precipitated with similar efficiencies in the presence or absence of miRNAs (Supplementary Figure S1A and B).
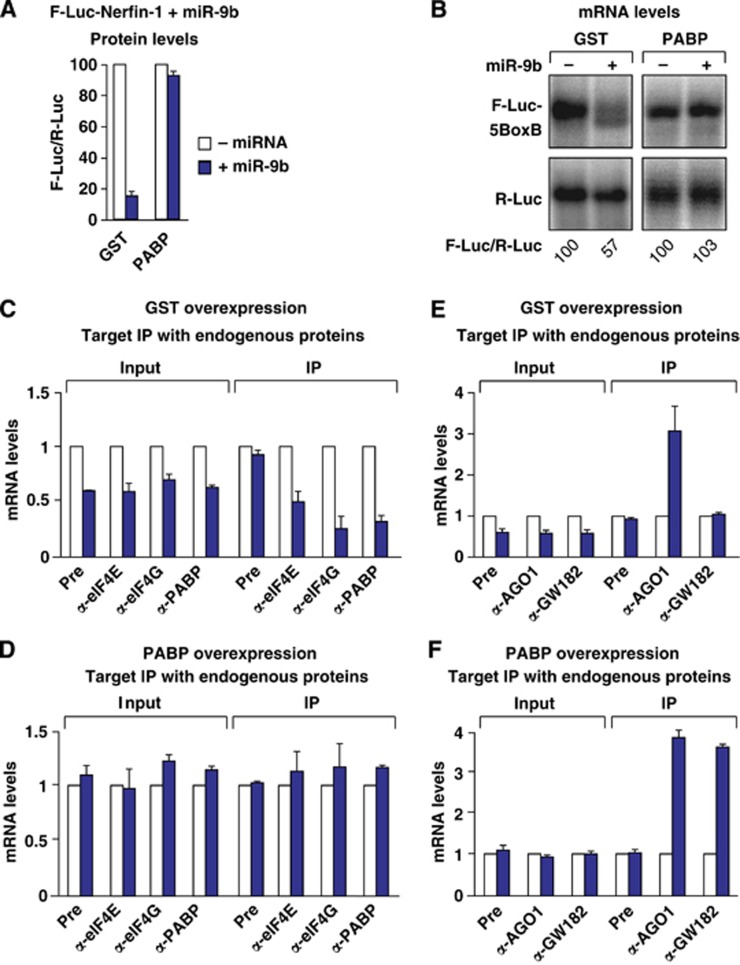
Figure 2.
PABP overexpression suppresses silencing downstream of miRISC binding. (A–F) S2 cells were transfected with a mixture of plasmids as described in Figure 1. The transfection mixtures additionally contained plasmids expressing PABP or GST as indicated. (A) For each condition, firefly luciferase activity was normalized to that of the Renilla luciferase and set at 100 in the absence of miR-9b (white bars). (B) Northern blot of representative RNA samples analysed as described in Figure 1. (C–F) Endogenous eIF4E, eIF4G, PABP, AGO1 and GW182 were immunoprecipitated using polyclonal antibodies, both in GST (C, E) and in PABP (D, F) expressing cells. The levels of F-Luc-Nerfin-1 mRNA in the inputs and IPs were analysed by RT–qPCR as described in Figure 1.
Source data for this figure is available on the online supplementary information page.
To validate these observations, we performed coimmunoprecipitations using antibodies specific to eIF4E, eIF4G and PABP. Similar to the results obtained with the HA-tagged proteins, we observed a decrease in the association of the F-Luc-Nerfin-1 reporter with the endogenous proteins in the presence of miR-9b (Figure 1E–I; Supplementary Figure S1C–E).
Importantly, GW182 depletion suppressed the silencing of the F-Luc-Nerfin reporter by miR-9b and restored its association with eIF4E, eIF4G and PABP to the levels observed in the absence of miR-9b (Supplementary Figure S2A–E). Western blot analysis indicated that GW182 levels were reduced to <10% of control levels in the depleted cells (Supplementary Figure S2C). Nevertheless, in GW182-depleted cells, AGO1 associated with the reporter in an miRNA-dependent manner (Supplementary Figure S2E), indicating that GW182 is not required for target recognition and binding by AGO1. However, although AGO1 was bound to the target, the target was not silenced in GW182-depleted cells, as reported previously (Behm-Ansmant et al, 2006; Eulalio et al, 2008; Zekri et al, 2009; Huntzinger et al, 2013). These results demonstrate that, on its own, the binding of AGO1 to the target is not sufficient for full silencing and does not cause the dissociation of eIF4E, eIF4G or PABP.
To further validate our results, we inhibit silencing using an independent approach. Specifically, it has been reported that PABP overexpression suppresses silencing (Zekri et al, 2009; Walters et al, 2010). We therefore asked whether PABP release from miRNA targets was counteracted in cells overexpressing PABP. We observed that PABP overexpression suppressed silencing of the F-Luc-Nerfin-1 reporter by miR-9b and restored mRNA levels, as previously shown (Figure 2A and B) (Zekri et al, 2009; Walters et al, 2010). In cells overexpressing PABP, endogenous eIF4E, eIF4G and PABP remained bound to the target (Figure 2D), whereas they dissociated from the target in control cells expressing glutathione S-transferase, wherein the target is partially degraded (GST; Figure 2C). Importantly, both endogenous AGO1 and GW182 remained bound to the miRNA reporter (Figure 2F), indicating that PABP overexpression suppresses silencing downstream of miRISC binding.
It is important to note that GW182 association with miRNA targets is observed when mRNA degradation is inhibited by either depletion of deadenylation factors (Zekri et al, 2009), decapping factors (Figure 3G) or overexpression of PABP (Figure 2F). Indeed, in contrast to AGO1, GW182 did not preferentially associate with silenced targets in control cells, as reported previously (Zekri et al, 2009) (Figure 2E; Supplementary Figure S3D). These observations suggest that GW182 dissociates from miRNA targets after degradation. Alternatively, GW182 may not be accessible to antibodies when bound to a target mRNA in control cells. However, we consider this last possibility unlikely because, in previous studies, we could not coimmunoprecipitate silenced targets with GW182 proteins fused N-terminally to HA or GFP (green fluorescent protein) tags or C-terminally to a V5 tag (Zekri et al, 2009). Moreover, as shown in Figure 2E and F, we obtained similar results when we analysed the association of endogenous GW182 using polyclonal antibodies.
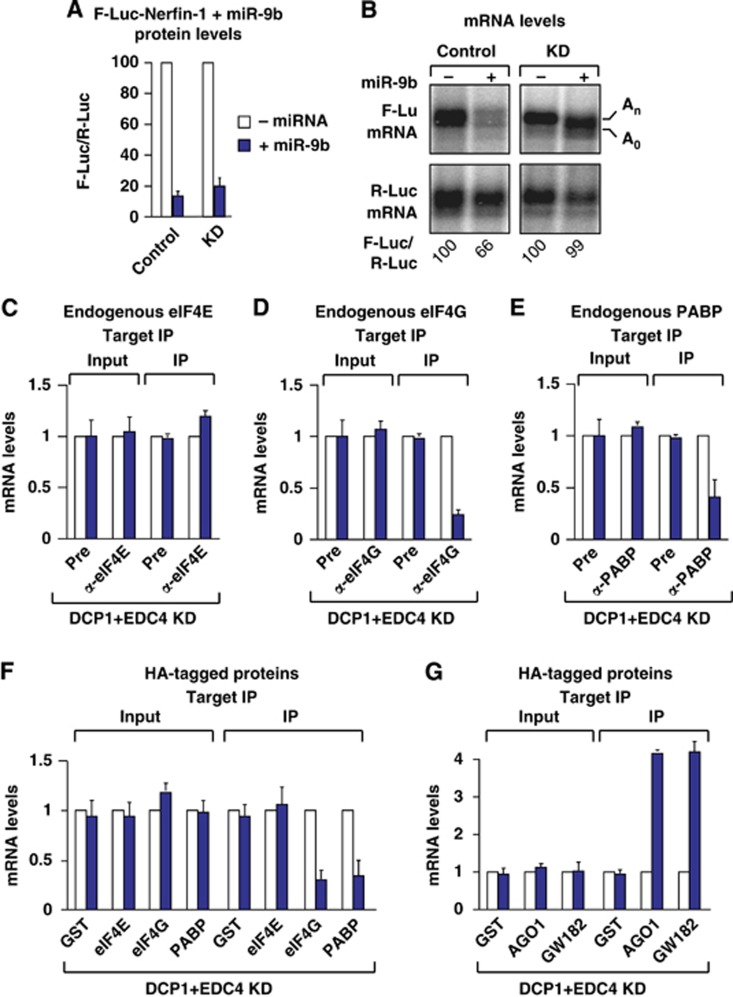
Figure 3.
miRNA target association with eIF4E is rescued in cells depleted of decapping factors. (A–G) Control S2 cells (treated with GFP dsRNA) or cells depleted of DCP1 and EDC4 were transfected with a mixture of three plasmids as described in Figure 1. (F, G) The transfection mixtures additionally contained plasmids for the expression of the indicated HA-tagged proteins. (A) Firefly luciferase activity was normalized to that of the Renilla luciferase. For each condition (control and knockdown cells), the normalized values of F-Luc activity were set at 100 in the absence of miR-9b (white bars). (B) Northern blot of representative RNA samples analysed as described in Figure 1. The positions of the polyadenylated (An) and deadenylated (A0) F-Luc-Nerfin-1 mRNA are indicated. (C–E) Coimmunoprecipitation of the F-Luc-Nerfin-1 reporter with the endogenous indicated proteins in the absence (white bars) or presence of miR-9b (blue bars). (F, G) Coimmunoprecipitation of the F-Luc-Nerfin-1 reporter with HA-tagged proteins in the absence (white bars) or presence of miR-9b, analysed as described in Figure 1.
Source data for this figure is available on the online supplementary information page.
eIF4E association is restored in cells depleted of decapping factors
It is known that miRNAs trigger target mRNA degradation (Huntzinger and Izaurralde, 2011). Therefore, we could not completely exclude the possibility that the decreased association of eIF4E, eIF4G and PABP with silenced targets was an indirect effect of mRNA degradation. We therefore performed immunoprecipitation assays in cells in which the decapping and decay of miRNA targets were blocked by the codepletion of DCP1 and EDC4. In a previous study, we showed that at least two decapping activators must be codepleted to efficiently block decapping in S2 cells (Eulalio et al, 2007). In cells codepleted of DCP1 and EDC4, the degradation of the F-Luc-Nerfin reporter by miR-9b was nearly brought to a halt and mRNA levels were fully restored. The reporter accumulated in the deadenylated form, because deadenylation precedes decapping (Eulalio et al, 2007), and consequently remained translationally repressed (Figure 3A and B). Thus, the reporter accumulating in depleted cells was expected to be less efficiently coimmunoprecipitated with PABP. Consistent with this expectation, we observed a reduction in the association of the silenced F-Luc-Nerfin-1 mRNA with both endogenous and HA-tagged PABP in depleted cells (Figure 3E and F).
Remarkably, eIF4E association was rescued in depleted cells, despite the fact that the reporter was silenced (Figure 3C and F). These results indicate that eIF4E dissociation from silenced miRNA reporters is caused by decapping and/or the activity of decapping factors. Surprisingly, despite the recovery of eIF4E binding, the association of the F-Luc-Nerfin-1 mRNA with endogenous or HA-tagged eIF4G was not restored (Figure 3D and F), suggesting that eIF4G dissociates from repressed miRNA targets following deadenylation. In contrast, AGO1 and GW182 remained bound to the target in depleted cells (Figure 3G). Western blot analyses indicated that the levels of DCP1 and EDC4 in the depleted cells were reduced to 10% of the control levels and that the expression of endogenous eIF4E, eIF4G or PABP was not affected in these cells (Supplementary Figure S3A and B). Similar results were obtained in cells codepleted of Me31B and EDC4 (Supplementary Figure S4).
The dissociation of eIF4G despite normal levels of eIF4E is surprising. However, previous studies have suggested that eIF4G binding to mRNAs may be stabilized through interactions with PABP (Kahvejian et al, 2005; Derry et al, 2006), and deadenylation may thus decrease eIF4G association with mRNA.
Tethered Dm GW182 is sufficient to reduce eIF4E, eIF4G and PABP association with mRNA targets
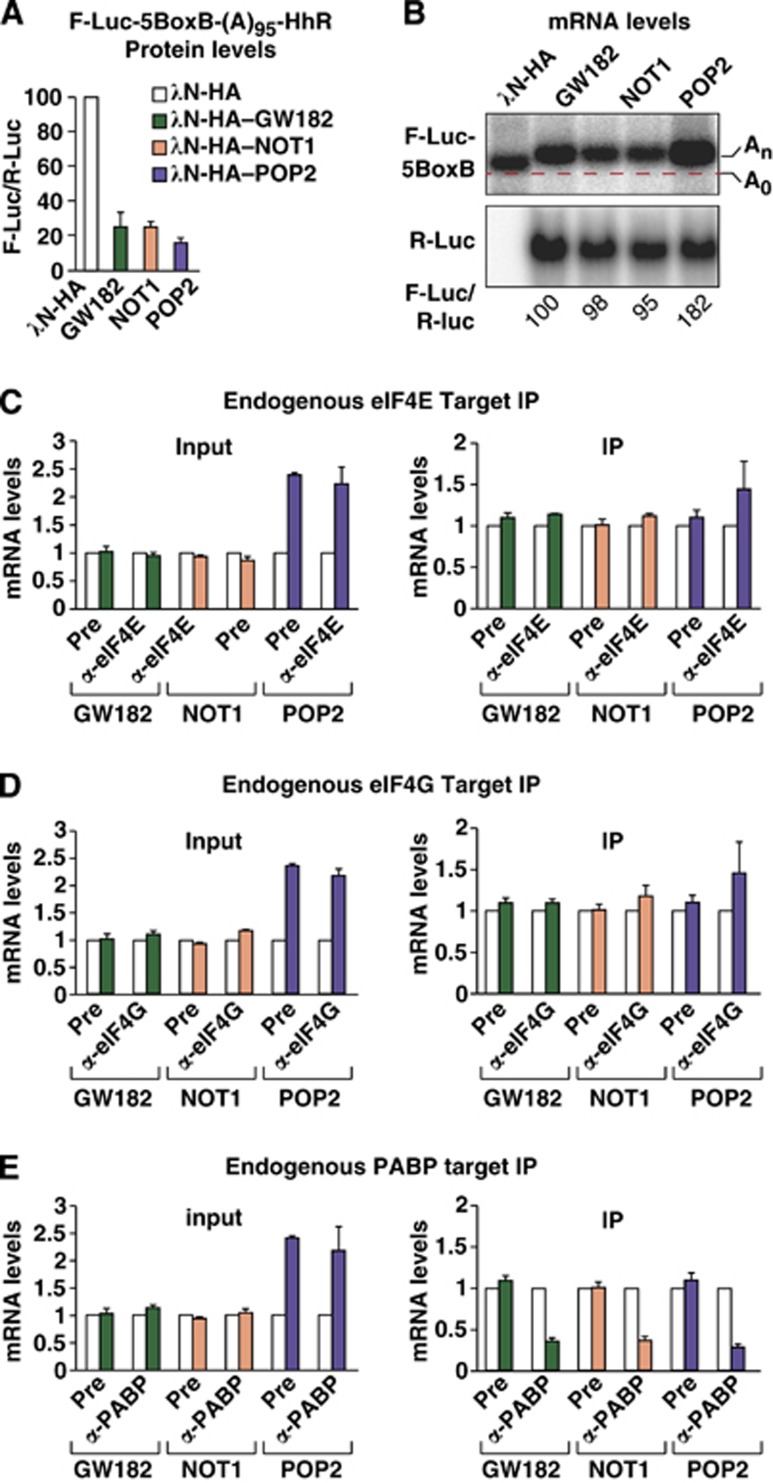
The results obtained in GW182-depleted cells indicate that GW182 is responsible for the reduced association of the F-Luc-Nerfin-1 reporter with eIF4E, eIF4G and PABP. We therefore tested whether silencing triggered by GW182 tethering resulted in a similar dissociation of the endogenous proteins from the mRNA. GW182 was expressed with a tag derived from the N protein of the bacteriophage λ (λN tag) to enable tethering to a firefly luciferase (F-Luc) reporter containing five Box B hairpins (5BoxB) inserted into the 3′ UTR. Dm GW182 tethering promotes the translational repression and degradation of the mRNA reporter (Figure 4A and B, control cells) (Huntzinger and Izaurralde, 2011). To prevent extensive mRNA degradation, we expressed GW182 at low levels, which resulted in a two-fold reduction of reporter abundance and a corresponding decrease in Firefly luciferase activity (Figure 4A and B, control cells). We observed that in cells expressing GW182, the F-Luc-5BoxB reporter associated less efficiently with endogenous eIF4E, eIF4G and PABP (Figure 4C–E), in accordance with the partial degradation of the mRNA.
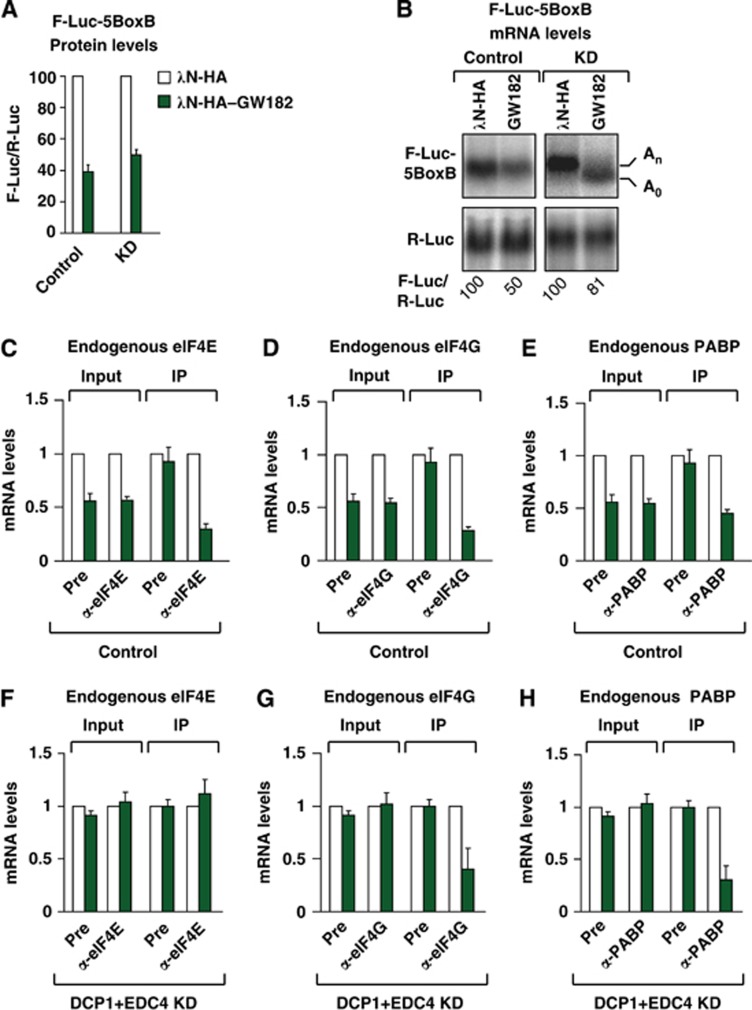
Figure 4.
GW182 tethering reduces the association of eIF4E, eIF4G and PABP with the mRNA target. (A–H) Control S2 cells (treated with GFP dsRNA) or cells codepleted of DCP1 and EDC4 (KD) were cotransfected with a mixture of three plasmids: one expressing the F-Luc-5BoxB reporter, another expressing the λN-HA peptide or λN-HA–GW182, and a third expressing Renilla luciferase (R-Luc). (A) Firefly luciferase activity was normalized to that of the Renilla luciferase and set at 100 in cells expressing the λN-HA peptide (white bars). (B) Northern blot of representative RNA samples. (C–H) Levels of the F-Luc-5BoxB reporter coimmunoprecipitating with endogenous eIF4E, eIF4G or PABP in control (C–F) or knockdown (F–H) cells analysed as described in Figure 1.
Source data for this figure is available on the online supplementary information page.
Importantly, silencing by GW182 tethering recapitulated the observations obtained with the F-Luc-Nerfin-1 reporter; in cells codepleted of DCP1 and EDC4, the F-Luc-5BoxB reporter accumulated in a deadenylated form but was not further degraded (Figure 4B, knockdown cells, KD). eIF4E association with the mRNA reporter was restored (Figure 4F), whereas the association with eIF4G was not (Figure 4G). As expected, PABP was released as the reporter was deadenylated (Figure 4B and H).
PABP dissociates from miRNA targets in the absence of deadenylation
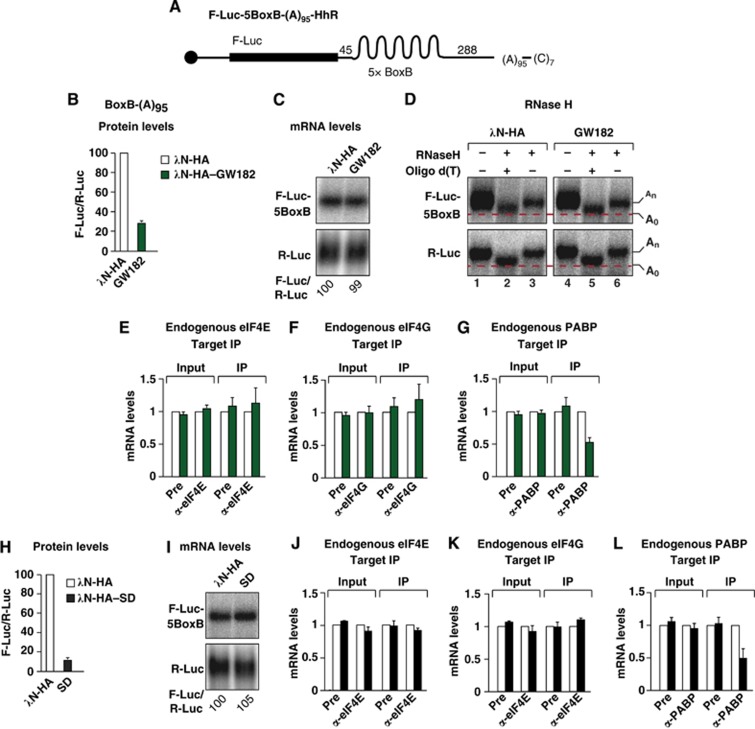
The experiments described above suggest that eIF4G and PABP dissociate from silenced, deadenylated reporters. To determine whether deadenylation per se is sufficient to explain our findings, we investigated the association of eIF4E, eIF4G and PABP with a reporter containing an internal poly(A) tail of 95 residues (F-Luc-5BoxB-(A)95-Hhr; Figure 5A). This reporter is not deadenylated because its 3′ end is generated by a self-cleaving hammerhead ribozyme (HhR) and contains a 3′ poly(C) tail of seven residues (Figure 5A). Consequently, the reporter was silenced by GW182 tethering without undergoing deadenylation and degradation (Figure 5B and C). We confirmed that tethering of GW182 did not cause deadenylation of the reporter using an oligo(dT)-directed ribonuclease H (RNase H) cleavage assay (Figure 5D). In control cells (i.e., cells expressing λN-HA), both the F-Luc-5BoxB-(A)95-Hhr reporter and the R-Luc transfection control mRNA migrated more rapidly after cleavage with oligo(dT)-directed RNase H had removed the internal and natural poly(A)-tails, respectively (Figure 5D, lane 2 versus 3). A similar pattern was observed in cells expressing λN-HA–GW182 (Figure 5D, lane 5 versus 6), indicating that the internal poly(A) tail of 95 nucleotides was still present in the reporter, despite its repression at the translational level.
Figure 5.
GW182 tethering causes PABP dissociation in the absence of deadenylation. (A) Schematic representation of the F-Luc-5BoxB-(A)95-HhR reporter carrying 5BoxB hairpins, consecutive poly(A) and poly(C) stretches of 95 and 7 residues, respectively, and a self-cleavable HhR. (B–L) S2 cells were transfected with a mixture of three plasmids: one expressing the F-Luc-5BoxB-(A)95-HhR reporter, another expressing the indicated λN-HA-tagged proteins, and a third expressing Renilla luciferase (R-Luc). (B, H) Firefly luciferase activity was normalized to that of the Renilla luciferase and set at 100 in cells expressing the λN-HA peptide (white bars). (C, I) Northern blot of representative RNA samples analysed as described in Figure 1. (D) RNA samples isolated from the tethering assay, shown in (C), were treated with RNase H in the absence or presence of oligo(dT) and analysed by northern blotting. (E–G, J–L) The association of the F-Luc-5BoxB-(A)95-HhR reporter with the indicated endogenous proteins in the absence (white bars) or presence of GW182 (green bars) or GW182-SD (black bars) was analysed as described in Figure 1.
Source data for this figure is available on the online supplementary information page.
We next analysed the association of eIF4E, eIF4G and PABP with the reporter in the absence or presence of GW182. We observed that the reporter coimmunoprecipitated with both endogenous eIF4E and eIF4G to similar degrees whether it was silenced or not (Figure 5E and F). These results suggest that the dissociation of eIF4G from polyadenylated reporters described above occurs only on mRNAs undergoing deadenylation and may reflect changes in the mRNP composition when deadenylation is coupled to decapping.
Unexpectedly, PABP dissociated from the silenced reporter despite the fact that the poly(A) tail was internal and not subjected to deadenylation (Figure 5G). These results indicate that GW182 triggers PABP dissociation from repressed mRNAs independently of deadenylation. Finally, we observed that tethering of the GW182-SD alone caused a similar reduction in the association of endogenous PABP with the mRNA reporter (Figure 5H–L), indicating that the SD is sufficient to promote PABP dissociation.
The interaction of GW182 proteins with the CCR4–NOT complex is required to promote PABP dissociation
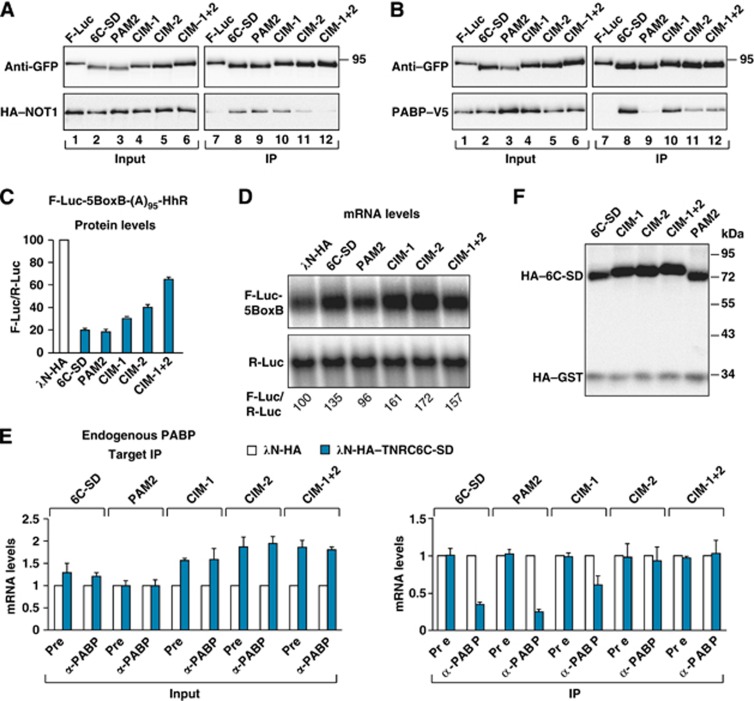
Because the mechanism of silencing is conserved and human TNRC6 proteins complement silencing in D. melanogaster cells (Huntzinger et al, 2010, 2013; Chekulaeva et al, 2011), we next tested whether the human TNRC6C-SD could promote PABP dissociation. This question was particularly interesting for the following reasons. First, a single amino-acid substitution in the PAM2 motif of human TNRC6-SDs abrogates PABP binding, whereas the equivalent mutation in the Dm protein is ineffectual (Fabian et al, 2009; Huntzinger et al, 2010, 2013; Jínek et al, 2010). Second, recent studies have identified tryptophan-containing sequences that are required for human SDs to interact with the CCR4–NOT complex and have described mutations in these sequences that abolish binding (Chekulaeva et al, 2011; Fabian et al, 2011). Thus, human SDs provide the opportunity to specifically disrupt PABP or deadenylase binding and to thereby study the contribution of these interactions to PABP displacement. Specifically, we introduced mutations in the CIM-1 and CIM-2 motifs individually or in combination. In immunoprecipitation assays, the mutation in the CIM-2 motif reduced NOT1 binding (Figure 6A, lane 11). NOT1 binding was further reduced when the CIM-1 and CIM-2 motifs were mutated simultaneously (Figure 6A, lane 12). We also introduced a F1389A substitution in the PAM2 motif that abolishes binding to PABP but not to the CCR4–NOT complex (Figure 6A and B, lane 9) (Fabian et al, 2009; Huntzinger et al, 2010, 2013).
Figure 6.
PABP release requires the interaction of GW182 proteins with the CCR4–NOT complex. (A, B) The interactions of the TNRC6C-SD protein (wild-type or mutant) with NOT1 and PABP were analysed by coimmunoprecipitation using anti-GFP antibodies. GFP-tagged F-Luc served as a negative control. (C–F) S2 cells were transfected with a mixture of three plasmids as described in Figure 5. (C) Firefly luciferase activity was normalized to that of the Renilla luciferase and set at 100 in cells expressing the λN-HA peptide (white bar). (D) Northern blot of representative RNA samples analysed as described in Figure 1. (E) The association of the F-Luc-5BoxB-(A)95-HhR reporter with the endogenous PABP in the absence (white bars) or presence (blue bars) of TNRC6C-SD was analysed as described in Figure 1. (F) Western blot showing comparable expression of TNRC6C-SD wild-type and mutants.
Source data for this figure is available on the online supplementary information page.
We observed that the TNRC6C-SD caused translational repression of the reporter containing an internal poly(A) stretch in the absence of mRNA destabilization (Figure 6C and D). Furthermore, the TNRC6C-SD caused PABP dissociation (Figure 6E). We next examined the contribution of the PAM2, CIM-1 and CIM-2 motifs to PABP dissociation. We observed that, in contrast to wild-type TNRC6C-SD, mutations in the CIM-2 motif alone or in combination with the CIM-1 motif (CIM-1+2 double mutant) abolished the ability of the protein to dissociate PABP (Figure 6E). Mutations in the CIM-1 motif alone impaired but did not abolish PABP release; this result is consistent with the notion that the CIM-1 and CIM-2 motifs are not functionally equivalent (Fabian et al, 2011). Surprisingly, the F1389A mutation in the PAM2 motif did not affect the ability of the TNRC6C-SD to displace PABP. All mutant proteins repressed the translation of the reporter, although the CIM mutants were impaired to different extents (Figure 6C). In particular, the CIM-2 mutant was two-fold less efficient in repressing reporter expression, suggesting that the contribution of PABP displacement to the repression is approximately two-fold. Importantly, the proteins carrying mutations in the CIM-1 and CIM-2 motifs stabilized mRNA reporter levels (Figure 6D and E, inputs), despite inducing translational repression, further confirming that the reporter is not deadenylated under our experimental conditions. All proteins were expressed at comparable levels (Figure 6F). We concluded that the interaction with the CCR4–NOT complex is required for GW182 proteins to promote PABP dissociation.
Tethering of CCR4–NOT complex subunits promotes PABP dissociation
Because the dissociation of PABP requires interaction with the CCR4–NOT complex, and subunits of this complex have been shown to promote translational repression in the absence of deadenylation (Cooke et al, 2010; Chekulaeva et al, 2011; Bawankar et al, 2013), we next sought to determine whether displacement of PABP could be mediated by the CCR4–NOT complex. We observed that tethered NOT1 repressed the translation of the F-Luc-5BoxB-(A)95-Hhr reporter without changing mRNA levels (Figure 7A and B), as previously reported (Chekulaeva et al, 2011; Bawankar et al, 2013). In contrast, tethered PAN3 did not repress reporter expression (Supplementary Figure S5), suggesting that the PAN2–PAN3 complex cannot repress translation in the absence of deadenylation. As observed for GW182, tethered NOT1 caused PABP dissociation (Figure 7E), whereas eIF4E and eIF4G remained bound to the mRNA reporter (Figure 7C and D). These results suggest that the CCR4–NOT complex mediates the release of PABP observed when GW182 is tethered to the reporter.
Figure 7.
Tethered subunits of the CCR4–NOT complex promote PABP dissociation in the absence of deadenylation. (A–E) A tethering assay was performed using the F-Luc-5BoxB-(A)95-HhR reporter and the indicated λN-HA-tagged proteins. (A) Firefly luciferase activity was normalized to that of the Renilla luciferase and set at 100 in cells expressing the λN-HA peptide (white bars). (B) Northern blot of representative RNA samples. The first lane of the upper panel shows the migration of a corresponding reporter lacking the internal poly(A). (C–E) The association of the F-Luc-5BoxB-(A)95-HhR reporter with endogenous eIF4E, eIF4G and PABP in the absence (white bars) or presence of the indicated proteins (coloured bars) was analysed as described in Figure 1.
Source data for this figure is available on the online supplementary information page.
To further confirm that the CCR4–NOT complex is sufficient to promote PABP dissociation, we tethered a catalytically inactive form of POP2. Previous studies indicated that both wild-type POP2 and a catalytically inactive mutant promoted translational repression of a reporter in the absence of mRNA deadenylation (Cooke et al, 2010; Chekulaeva et al, 2011; Bawankar et al, 2013). In accordance with these studies, tethering of the POP2 catalytically inactive mutant repressed translation of the F-Luc reporter containing an internal poly(A) stretch without causing mRNA degradation; rather, the POP2 mutant increased the abundance of the mRNA reporter two-fold (Figure 7B–E, inputs). Despite the increase in mRNA levels, the reporter was repressed at the translational level (Figure 7A), and PABP was displaced from the internal poly(A) tail (Figure 7E).
To confirm that the reduced association of the mRNA reporter with PABP indeed reflects PABP dissociation rather than reduced accessibility of PABP to the antibodies (e.g., due steric hindrance effects), we tested whether tethering of other large proteins would interfere with the coimmunoprecipitation of the reporter with PABP. In particular, we tethered either PAN3 or an inactive AGO1 mutant, which does not bind GW182 or miRNAs (F2V2 mutant, Eulalio et al, 2008). Tethering of these proteins did not repress the F-Luc-5BoxB-(A)95-Hhr reporter and did not interfere with its coimmunoprecipitation with PABP (Supplementary Figure S5).
Together, our results indicate that recruitment of the CCR4–NOT complex displaces PABP even in the absence of mRNA deadenylation. They further show that in the absence of deadenylation, the CCR4–NOT complex represses translation without affecting eIF4E and eIF4G association with the mRNA cap structure; thus, the repression most likely occurs after recognition of the cap structure by the eIF4F complex (i.e., eIF4E, eIF4G and eIF4A).
Contribution of PABP dissociation to silencing
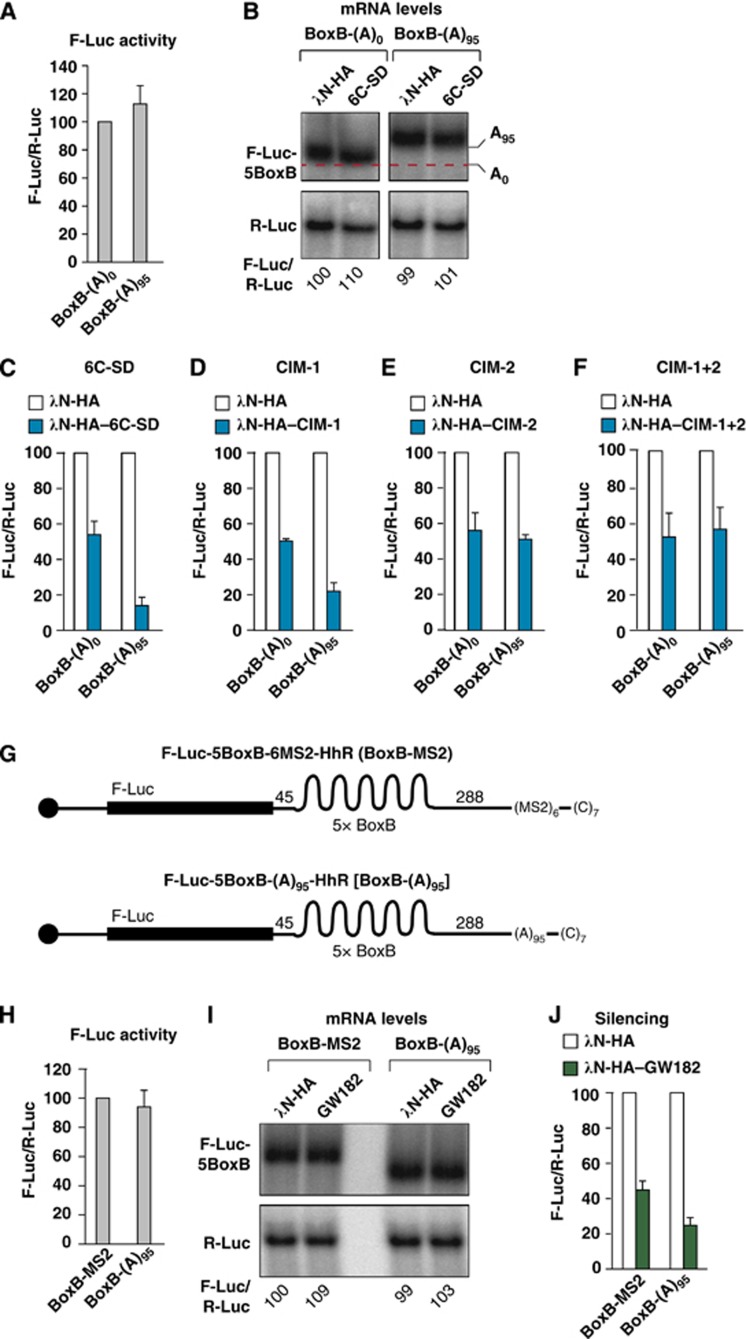
To investigate the contribution of PABP displacement to silencing, we performed three independent experiments. First, we compared the silencing efficiency of the F-Luc-5BoxB-(A)95-HhR with that of the corresponding reporter without an internal poly(A). Because the insertion of an internal poly(A) increases reporter mRNA levels and luciferase activity ∼20-fold (Supplementary Figure S6A and B), we adjusted the amounts of transfected plasmids to obtain similar translation efficiencies (i.e., similar mRNA levels and luciferase activity; Figure 8A and B). Under these conditions, we observed that the reporter containing the internal poly(A) was 3.8-fold more efficiently repressed by tethered TNRC6C-SD relative to the reporter lacking the internal poly(A) (Figure 8C), in agreement with previous studies (Humphreys et al, 2005; Wu et al, 2006; Eulalio et al, 2008, 2009b; Beilharz et al, 2009; Moretti et al, 2012). Similar results were obtained with tethered Dm GW182 (Supplementary Figure S6C and D). The difference in silencing efficiencies between the two reporters was attenuated when the CIM-1 mutant was tethered and abolished when the CIM-2 or the CIM-1+2 mutants were tethered (Figure 8D–F). These results reveal a dual role for PABP and the poly(A) in silencing. First, PABP and the poly(A) tail increase silencing efficiency (Figure 8C). Second, PABP release contributes to the repression because TNRC6C-SD mutants that do not displace PABP are impaired in silencing. Interestingly, the TNRC6C-SD mutants that do not release PABP silence unadenylated and polyadenylated targets to a similar extent, whereas the wild-type SD displaces PABP and silences polyadenylated targets more efficiently (Figure 8C–F).
Figure 8.
Contribution of PABP dissociation to silencing. (A–F) S2 cells were transfected with a mixture of three plasmids: one expressing either the F-Luc-5BoxB-HhR reporter [BoxB-(A)0] or the F-Luc-5BoxB-(A)95-HhR reporter [5BoxB-(A)95], another expressing either λN-HA or λN-HA–TNRC6C-SD (wild-type or mutants), and a third plasmid expressing Renilla luciferase (R-Luc). (A) Relative luciferase activity for each reporter in the absence of λN-HA–TNRC6C-SD. Firefly luciferase activities were normalized to those of the Renilla luciferase for each reporter and set at 100 for the BoxB-(A)0 reporter. (B) Northern blot of representative RNA samples. The levels of the F-Luc reporters were normalized to those of R-Luc mRNA and set at 100 for the BoxB-(A)0 reporter in cells expressing the λN-HA peptide. (C–F) Relative luciferase activity for each reporter in the absence or presence of λN-HA–TNRC6C-SD (wild-type or mutants). Firefly luciferase activities were normalized to those of the Renilla luciferase for each reporter and set at 100 in cells expressing the λN-HA peptide (white bars). (G) Schematic representation of the F-Luc-5BoxB-6MS2-HhR compared with the F-Luc-5BoxB-(A)95-HhR reporter. Labels are as described in Figure 5A. (H–J) S2 cells were transfected with a mixture of three plasmids: one expressing either the F-Luc-5BoxB-6MS2-HhR (BoxB-MS2) or the F-Luc-5BoxB-(A)95-HhR reporter [BoxB-(A)95], another expressing either λN-HA or λN-HA–GW182, and a third plasmid expressing Renilla luciferase (R-Luc). Additionally, a plasmid expressing MS2–PABP was included in the transfection mixtures containing the F-Luc-5BoxB-6MS2-HhR reporter. (H) Relative luciferase activity for each reporter in the absence of λN-HA–GW182. (I) Northern blot of representative RNA samples. The levels of the F-Luc reporters were normalized to those of R-Luc mRNA and set at 100 for the BoxB-MS2 reporter in cells expressing the λN-HA peptide and MS2–PABP. (J) Relative luciferase activity for each reporter in the absence or presence of λN-HA–GW182 analysed as described in (C–F).
Source data for this figure is available on the online supplementary information page.
Second, to evaluate the contribution of PABP dissociation to silencing independently of its stimulatory effect, we sought to generate a reporter to which PABP binds but from which it cannot be released. To this end, we replaced the internal poly(A) stretch in the F-Luc-5BoxB-(A)95-HhR reporter with six binding sites for the bacteriophage MS2 coat protein (F-Luc-5BoxB-6MS2-HhR, Figure 8G) and coexpressed this reporter with PABP fused N-terminally to the MS2 protein. Tethered MS2–PABP led to a two- and six-fold increase in reporter mRNA levels and luciferase activity, respectively, relative to the values obtained with tethered MS2–GST, which served as a negative control (Supplementary Figure S6E and F). Regardless of the presence of MS2–PABP, expression of λN-HA–GW182 silenced the reporter without causing mRNA degradation (Supplementary Figure S6F and G). Next, we compared the silencing efficiency of the reporter to which PABP was tethered relative to the reporter to which PABP binds directly to the internal poly(A). We adjusted reporter levels to obtain similar translation efficiencies (i.e., similar mRNA levels and luciferase activity; Figure 8H and I). We observed that the reporter with an internal poly(A) tail was 1.7-fold more efficiently repressed compared with the reporter to which PABP was tethered through the MS2-binding sites (Figure 8J). These results suggest that PABP dissociation may enhance silencing ∼1.7-fold, in agreement with the results obtained with the CIM-2 mutant. Similar results were obtained with reporter containing an internal poly(A) of 74 residues (corresponding to approximately six PABP-binding sites), which was silenced as efficiently as the F-Luc-5BoxB-(A)95-HhR reporter (Supplementary Figure S6H and I), whereas a reporter with an internal poly(A) of 26 residues was 1.7-fold less efficiently silenced (Supplementary Figure S6H and I).
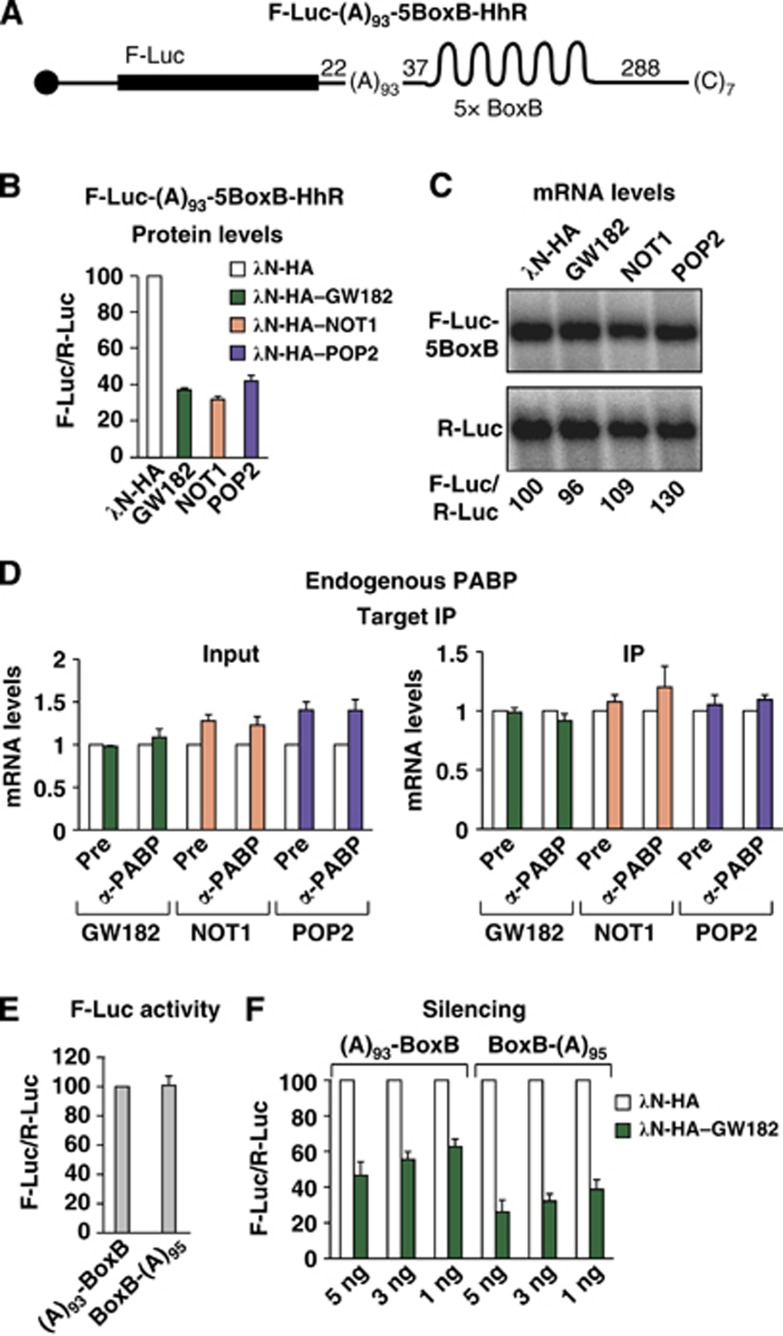
In the third experiment, we generated a reporter with an internal poly(A) stretch of 93 residues upstream of the BoxB hairpins, and thus upstream of the binding site for the GW182 protein (F-Luc-(A)93-5BoxB-HhR; Figure 9A). The internal poly(A) stimulated translation and stabilized mRNA reporter levels relative to the equivalent reporter lacking a poly(A) (Supplementary Figure S6J), but not as efficiently as the reporter containing a distal poly(A) tail. Furthermore, the reporter was silenced by tethered GW182, NOT1 or the catalytically inactive POP2 mutant in the absence of mRNA degradation (Figure 9B and C). Interestingly, tethering of GW182, NOT1 or POP2 did not cause PABP dissociation (Figure 9D), suggesting that the CCR4–NOT complex dissociates PABP molecules bound downstream of its binding site. However, we cannot exclude the possibility that PABP proximity to the 3′ end or the distance to the PABP-binding sites (which is different in the two reporters) impact on the ability of the CCR4–NOT complex to dissociate PABP.
Figure 9.
Contribution of PABP dissociation to silencing. (A) Schematic representation of the F-Luc-(A)93-5BoxB-HhR reporter [(A)93-5BoxB]. (B–D) S2 cells were transfected with a mixture of three plasmids: one expressing the F-Luc-(A)935BoxB-HhR reporter, another expressing the indicated λN-HA-tagged proteins, and a third plasmid expressing Renilla luciferase (R-Luc). (B) Firefly luciferase activities were normalized to those of the Renilla luciferase and set at 100 in cells expressing the λN-HA peptide. (C) Northern blot of representative RNA samples. (D) The association of the F-Luc-(A)93-5BoxB-HhR reporter with endogenous PABP in cells expressing the λN-HA peptide (white bars) or the indicated proteins (coloured bars) was analysed as described in Figure 1. (E, F) S2 cells were transfected with a mixture of two plasmids: one expressing either the (A)93-BoxB or the BoxB-(A)95 reporter and another plasmid expressing Renilla luciferase (R-Luc). (F) Increasing amounts of plasmids expressing either λN-HA or λN-HA–GW182 were included in the transfection mixtures. (E) Relative luciferase activity for each reporter in the absence of λN-HA–GW182. (F) Relative luciferase activity for each reporter in the absence (white bars) or presence of λN-HA–GW182 (green bars).
Source data for this figure is available on the online supplementary information page.
The observation that PABP is not displaced from the F-Luc-(A)93-5BoxB-HhR reporter provided an additional opportunity to test the contribution of PABP displacement to silencing. Again, we adjusted the amounts of plasmids transfected in S2 cells to obtain equivalent translation efficiencies for the reporters containing the poly(A) stretch either upstream or downstream of the BoxB sites (Figure 9E). We observed that GW182 silenced both reporters in a dose-dependent manner (Figure 9F). However, the reporter containing the poly(A) stretch upstream of the tethering site was silenced 1.7-fold less efficiently at each GW182 concentration tested (Figure 9F). These results further confirm that the contribution of PABP dissociation to silencing is ∼1.7- to 2-fold.
Discussion
The data presented in this study provide insights into the mechanism of silencing and the roles of GW182 proteins and the CCR4–NOT complex in this process. We show that the recruitment of the CCR4–NOT complex by GW182 proteins elicits PABP dissociation from silenced miRNA targets. This dissociation occurs in the absence of deadenylation and thus may contribute to the translational repression observed prior to the onset of miRNA target deadenylation and decay.
Contribution of PABP dissociation to miRNA target silencing
Previous studies have provided evidence that the translational repression of miRNA targets occurs before mRNA deadenylation and degradation (Pillai et al, 2005; Mathonnet et al, 2007; Fabian et al, 2009; Zdanowicz et al, 2009; Bazzini et al, 2012; Béthune et al, 2012; Djuranovic et al, 2012; Moretti et al, 2012). However, how animal miRNAs repress translation remains highly controversial, and at least three different mechanisms have been proposed: inhibition of eIF4F binding and/or function in translation initiation, inhibition of initiation at a step downstream of eIF4F binding and inhibition of translation after initiation (reviewed in Djuranovic et al, 2011). We analysed the association of eIF4E with miRNA targets and showed that silencing causes eIF4E dissociation. However, in cells in which miRNA targets undergo deadenylation but decapping and subsequent degradation are inhibited (i.e., in cells depleted of decapping factors), eIF4E remains bound to the targets to which both AGO1 and GW182 are also bound (Figure 3F and G). These results suggest that eIF4E dissociates from miRNA targets as a consequence of decapping (and/or the action of decapping factors), and they rule out models, suggesting that translational repression is achieved by the displacement of eIF4E from the mRNA cap structure by miRISCs. In accordance with this conclusion, eIF4E remains bound to silenced targets containing an internal poly(A) tail, which are not degraded (Figure 5).
We also show that in cells depleted of decapping activators, eIF4G association with silenced mRNAs decreases, despite the fact that eIF4E remains bound (Figure 3). This dissociation is observed for polyadenylated targets undergoing deadenylation and may represent a remodelling step for mRNAs entering the 5′-to-3′ degradation pathway. In contrast, eIF4G remained bound to silenced reporters containing an internal poly(A) tail, which do not undergo deadenylation (Figure 5). Therefore, the dissociation of eIF4G occurs as a consequence of deadenylation and may not represent a direct effect of silencing complexes.
In agreement with studies in a cell-free system that recapitulates silencing (Moretti et al, 2012), we show that PABP has a dual role in silencing. First, PABP and the poly(A) tail stimulate silencing because an mRNA reporter lacking a poly(A) tail is silenced less efficiently than its polyadenylated counterparts (Figure 8C; Supplementary Figure S6I) (Humphreys et al, 2005; Wu et al, 2006; Eulalio et al, 2008, 2009b; Beilharz et al, 2009; Moretti et al, 2012). Second, PABP dissociates from silenced targets and this dissociation contributes to the repression. Moretti et al (2012) demonstrated that PABP stimulates silencing by facilitating miRISC binding to mRNA targets. In our study, we observed this stimulatory effect even when recruitment of silencing complexes was mediated by direct tethering of GW182 proteins (Figure 8C; Supplementary Figure S6I); thus, it is possible that GW182 recruitment was rate limiting in our assay because we expressed the proteins at low levels. Alternatively, PABP may stimulate silencing by another unknown mechanism. Regardless of the precise mechanism by which PABP stimulates silencing, our study shows that after stimulating silencing, PABP dissociates from silenced targets in the absence of deadenylation. These findings, together with the observation that translational repression of miRNA targets precedes deadenylation, suggest that the displacement of PABP from the mRNA poly(A) tail likely contributes to the repression of translation observed before the onset of deadenylation (Pillai et al, 2005; Mathonnet et al, 2007; Fabian et al, 2009; Zdanowicz et al, 2009; Bazzini et al, 2012; Béthune et al, 2012; Djuranovic et al, 2012; Moretti et al, 2012).
The results presented in this and a previous study (Moretti et al, 2012) help to explain why silencing occurs even when PABP is depleted or removed from mRNA poly(A) tails by overexpression or the addition of excess of Paip2 (Fukaya and Tomari, 2011; Mishima et al, 2012). Indeed, if one function of GW182 is to facilitate PABP dissociation from the poly(A) tail, then PABP may become dispensable in extracts in which it has been depleted or displaced from the poly(A) tail by Paip2. Our results are also consistent with the observation that mRNAs lacking a poly(A) tail are nevertheless silenced, although less efficiently than their polyadenylated counterparts (Figure 8; Supplementary Figure S6I) (Humphreys et al, 2005; Wu et al, 2006; Eulalio et al, 2008, 2009b; Beilharz et al, 2009; Braun et al, 2011; Chekulaeva et al, 2011; Fukaya and Tomari, 2011; Bazzini et al, 2012; Mishima et al, 2012).
In summary, PABP displacement represents one mechanism by which GW182 proteins and the CCR4–NOT complex repress translation in the absence of deadenylation. This mechanism contributes approximately two-fold to repression, indicating that additional repressive mechanisms are used by miRISCs to achieve maximal target silencing.
PABP dissociation is mediated by the CCR4–NOT complex
In this study, we demonstrate that recruitment of the CCR4–NOT complex by its interaction with GW182 proteins or through direct tethering of its subunits triggers the release of PABP from mRNA targets. These results suggest that PABP displacement from the mRNA poly(A) tail may be a requirement for deadenylation. Indeed, in addition to reducing translation efficiency, PABP dissociation may increase the accessibility of the poly(A) tail to the CCR4 and POP2 deadenylases of the CCR4–NOT complex. Precisely how the CCR4–NOT complex releases PABP from the poly(A) tail remains unclear. By analogy with Paip2, a specific inhibitor of PABP function (Khaleghpour et al, 2001), it is possible that the CCR4–NOT complex disrupts the PABP–poly(A) interaction. Accordingly, CCR4–NOT complex subunits interact with PABP (Zekri et al, 2009); however, it is currently not known whether these interactions are direct or which subunit(s) of the CCR4–NOT complex contacts PABP. Future studies will investigate the mechanism of PABP release by the CCR4–NOT complex. Given the central role of the CCR4–NOT complex in post-transcriptional mRNA regulation, we anticipate that this novel activity of the complex will contribute to the translational repression of a large variety of different mRNAs.
Materials and methods
Plasmids, antibodies and western blotting
Luciferase reporters and plasmids for the expression of miRNAs and HA-tagged proteins in D. melanogaster cells were described elsewhere (Zekri et al, 2009; Huntzinger et al, 2010; Braun et al, 2011). Additional plasmids and antibodies used in this study are described in the Supplementary data.
RNA interference, transfections and luciferase assays
Protein depletions were performed as described previously (Eulalio et al, 2007). S2 cells were depleted on days 0 and 4, transfected on day 6 and collected on day 9. Information on the amounts of transfected plasmids is provided in the Supplementary data. Firefly and Renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega). Northern blotting was performed as described previously (Behm-Ansmant et al, 2006).
Immunoprecipitation assays
Protein and RNA coimmunoprecipitations were performed as described previously (Zekri et al, 2009; Braun et al, 2011). For RNA coimmunoprecipitations, S2 cells (10–12 × 106 cells) were collected 3 days after transfection, washed with PBS, and lysed in 0.5 ml of NET buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% NP40) supplemented with protease inhibitors. Cells were lysed by three 30-s sonication treatments followed by 15 min of incubation on ice. The lysed cells were then spun at 16 000 g for 15 min at 4 °C. Aliquots (1/5) of the cleared lysate (Input) were set aside for both RNA extraction and western blotting. In parallel, Protein G-sepharose beads (Roche) were pre-incubated with 0.5 mg of yeast RNA and the corresponding antibodies for 1 h at 4°C. The beads were washed three times with NET buffer and incubated with the cleared cell lysates for 1 h at 4°C. After five additional washes, 10–20% of the IP and 1–2% of the input were analysed using western blotting to confirm that similar amounts of protein were immunoprecipitated in the presence or absence of miRNA (or λN-HA-tagged proteins in tethering assays). The remaining IP fraction was used for RNA isolation.
Reverse transcription and quantitative real-time PCR
RNA was isolated from samples corresponding to inputs and IP using TRIzol LS reagent (Invitrogen) in the presence of glycogen as a carrier (20 μg; Roche). DNase treatment was performed using the TURBO DNase-kit (Ambion) for 30 min at 37°C. RNA was detected via cDNA synthesis and qPCR. cDNA was synthesized with M-MuLV reverse transcriptase (Fermentas) and random primers (Sigma) according to the manufacturers’ protocols. For each sample, a mock RT reaction lacking the enzyme was included as a negative control. The comparative Ct method with SYBR Green was used with the Bio-Rad DNA Engine Opticon System and primers, as described previously (Zekri et al, 2009).
Supplementary Material
Acknowledgments
We thank H Budde, S Heimstädt and E Huntzinger for reagents and C Igreja and N Bercovich for helpful discussions. This study was supported by the Max Planck Society and by grants from the Deutsche Forschungsgemeinschaft (DFG, FOR855 and the Gottfried Wilhelm Leibniz Program awarded to EI).
Author contributions: LZ conceived the project, designed, conducted and interpreted the RNA immunoprecipitation assays, and contributed to the writing of the manuscript. DKO performed protein immunoprecipitations and western blotting. EI interpreted the experiments, supervised the project and wrote the manuscript. All authors edited the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E (2013) NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol 10: 226–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ (2012) Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336: 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Humphreys DT, Clancy JL, Thermann R, Martin DI, Hentze MW, Preiss T (2009) microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS One 4: e6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béthune J, Artus-Revel CG, Filipowicz W (2012) Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep 13: 716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E (2011) GW182 proteins recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Filipowicz W, Parker R (2009) Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA 15: 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W (2011) miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Parker R, Filipowicz W (2010) The GW/WG repeats of Drosophila GW182 function as effector motifs for miRNA-mediated repression. Nucleic Acids Res 38: 6673–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A, Prigge A, Wickens M (2010) Translational repression by deadenylases. J Biol Chem 285: 28506–28513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry MC, Yanagiya A, Martineau Y, Sonenberg N (2006) Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol 71: 537–543 [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R (2011) A parsimonious model for gene regulation by miRNAs. Science 331: 550–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green A (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E (2009a) A C-terminal silencing domain in GW182 is essential for miRNA function. RNA 15: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E (2008) GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E (2009b) Deadenylation is a widespread effect of miRNA regulation. RNA 15: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E (2007) Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev 21: 2558–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N (2011) miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nat Struct Mol Biol 18: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates III JR, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N (2009) Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19: 586–593 [DOI] [PubMed] [Google Scholar]
- Fukaya T, Tomari Y (2011) PABP is not essential for microRNA-mediated translational repression and deadenylation in vitro. EMBO J 30: 4998–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T (2005) MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA 102: 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Braun JE, Heimstädt S, Zekri L, Izaurralde E (2010) Two PABP-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J 29: 4146–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kuzuoğlu-Öztürk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E (2013) The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res 41: 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jínek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA (2010) Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol 17: 238–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Roy G, Sonenberg N (2001) The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb Symp Quant Biol 66: 293–300 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N (2005) Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N (2001) Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7: 205–216 [DOI] [PubMed] [Google Scholar]
- Kozlov G, Safaee N, Rosenauer A, Gehring K (2010) Structural basis of binding of P-body associated protein GW182 and Ataxin-2 by the MLLE domain of poly(A)-binding protein. J Biol Chem 285: 13599–13606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N (2007) MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 17: 1764–1767 [DOI] [PubMed] [Google Scholar]
- Mishima Y, Fukao A, Kishimoto T, Sakamoto H, Fujiwara T, Inoue K (2012) Translational inhibition by deadenylation-independent mechanisms is central to microRNA-mediated silencing in zebrafish. Proc Natl Acad Sci USA 109: 1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW (2012) PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol 19: 603–608 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W (2005) Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309: 1573–1576 [DOI] [PubMed] [Google Scholar]
- Walters RW, Bradrick SS, Gromeier M (2010) Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA 16: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103: 4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, Darzynkiewicz E, Hentze MW (2009) Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell 35: 881–888 [DOI] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstädt S, Izaurralde E (2009) The silencing domain of GW182 interacts with PABP to promote translational repression and degradation of miRNA targets and is required for target release. Mol Cell Biol 29: 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.