Abstract
AMP-activated protein kinase (AMPK) is a metabolic fuel gauge conserved along the evolutionary scale in eukaryotes that senses changes in the intracellular AMP/ATP ratio1. The interest in AMPK has recently been raised by evidence showing that AMPK plays an important role to explain the therapeutic benefits of metformin2, 3, thiazolidinediones4 and exercise5, which form the cornerstones of the clinical management of type 2 diabetes and associated metabolic disorders. In general, activation of AMPK acts to maintain cellular energy stores, switching on catabolic pathways that produce ATP, mostly by enhancing oxidative metabolism and mitochondrial biogenesis, while switching off anabolic pathways that consume ATP1. This regulation can take place acutely, through the regulation of fast post-translational events, but also by transcriptionally reprogramming the cell in order to meet energetic needs. Our study demonstrates that AMPK controls the expression of genes involved in energy metabolism in skeletal muscle by acting in coordination with another metabolic sensor, the NAD+-dependent type III deacetylase SIRT1. AMPK enhances SIRT1 activity by increasing cellular NAD+ levels, resulting in the deacetylation and modulation of the activity of downstream SIRT1 targets that include the peroxisome proliferator-activated receptor-γ coactivator 1α and the forkhead transcription factors FOXO1 and FOXO3a. The AMPK-induced SIRT1-mediated deacetylation of these targets explains many of the convergent biological effects of AMPK and SIRT1 on energy metabolism.
Keywords: AMPK, SIRT1, PGC-1α, energy expenditure, FOXO3a, FOXO1
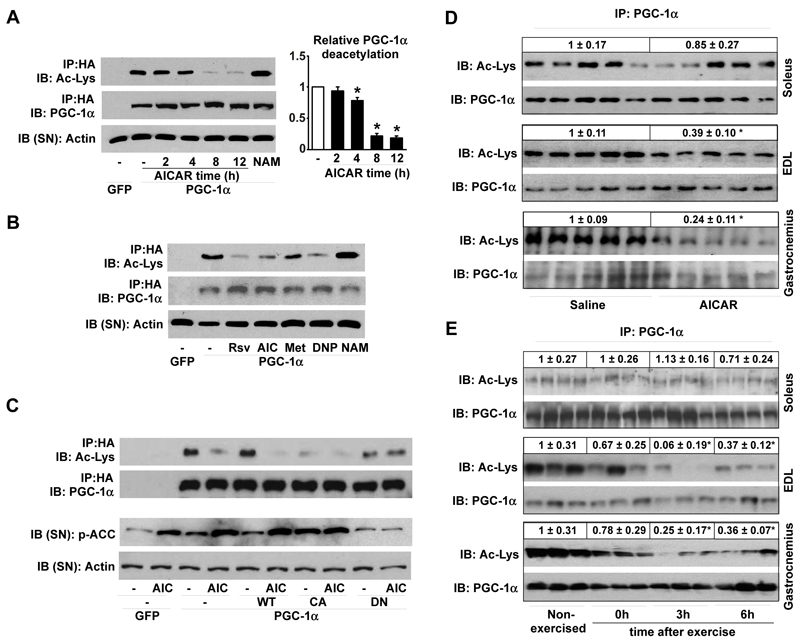
AMPK is a critical regulator of mitochondrial biogenesis in response to energy deprivation6. While the mechanisms by which AMPK modulates mitochondrial gene expression are not entirely elucidated, they seem to require the peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α), either by increasing its expression7 or direct phosphorylation8. Since PGC-1α is also activated by SIRT1-mediated deacetylation9-12, we speculated that AMPK alters PGC-1α activity by changing its acetylation status. Treatment of C2C12 myotubes with the AMPK activator 5-aminoimidazole-4-carboxamide-1-beta-D-riboside (AICAR) decreased PGC-1α acetylation after 4hrs of treatment (Fig.1A). Unrelated AMPK activators (Fig.S1A-B), such as resveratrol13, 14, metformin3, dinitrophenol (DNP)15 and A-76966216, also decreased PGC-1α acetylation (Fig.1B, Fig.S1C). Furthermore, overexpression of a constitutively active form of AMPKα1 led to a robust deacetylation of PGC-1α that could not be further enhanced by AICAR (Fig.1C). In contrast, when a dominant negative form of AMPKα1 was overexpressed, AICAR was unable to deacetylate PGC-1α (Fig.1C). The activation of AMPK hence triggers PGC-1α deacetylation in C2C12 myotubes.
Figure 1. Activation of AMPK triggers PGC-1α deacetylation in C2C12 myotubes and skeletal muscle.
(A-B) C2C12 myotubes infected with adenoviruses for GFP or FLAG-HA-tagged PGC-1α (PGC-1α) were treated with vehicle (−), AICAR (0.5mM, 2-8hrs) or nicotinamide (NAM; 5mM; 12hrs). Then, acetyl-lysine levels were checked on PGC-1α immunoprecipitates (IP). The supernatant (SN) was blotted against actin as input control. Relative quantification of PGC-1α acetylation is shown on the right (B) As in (A), but myotubes were treated for 8hrs with vehicle (−), Resveratrol (Rsv; 50μM), AICAR (AIC), Metformin (Metf; 1mM), DNP (0.5mM) or NAM. (C) C2C12 were infected with adenoviruses encoding GFP, PGC-1α, and wild-type (WT), constitutively active (CA) or dominant negative (DN) forms of AMPKα1. After AICAR treatment, total lysates were analyzed as in (A). (D) PGC-1α acetylation was measured on total protein (soleus and EDL) or nuclear extracts (gastrocnemius) from muscles of mice treated with AICAR or saline. Relative acetylation levels are shown on top of the panels (E) Soleus, EDL and gastrocnemius were obtained from non-exercised or exercised mice at 0, 3 or 6hrs after cessation of exercise, and analysed as in (D). Values are expressed as mean +/− S.E.M. * indicates statistical difference vs. corresponding vehicle, saline or non-exercised group at P < 0.05.
To validate these observations in vivo, we examined PGC-1α acetylation in hindlimb muscles after a single AICAR injection. PGC-1α acetylation was markedly reduced by AICAR in extensor digitorum longus (EDL) and gastrocnemius, but not in soleus (Fig.1D). The soleus is an oxidative muscle, where basal PGC-1α activity is presumably higher than in glycolytic muscles such as the EDL. Supporting this hypothesis, basal PGC-1α acetylation levels were lower in soleus than EDL (Fig.S2A), explaining why the soleus is refractory to AICAR. AICAR-induced PGC-1α deacetylation in muscle correlated with an increase of PGC-1α target genes (Fig.S2B). Consistent with PGC-1α acetylation levels, AICAR had, however, minor effects on mitochondrial gene expression in the soleus (Fig.S2C).
We then tested whether AMPK activation through exercise decreases PGC-1α acetylation. An exhaustive single-bout of treadmill running transiently activated AMPK (Fig.S3A) and induced PGC-1α deacetylation with a maximal effect 3hrs after ending exercise (Fig.1E). As observed with AICAR infusion, the soleus was refractory to exercise-induced PGC-1α deacetylation (Fig.1E) despite being effectively recruited, as indicated by the diminished glycogen levels and increased AMPK activity (Fig.S3B-C). The decrease in PGC-1α acetylation in EDL and gastrocnemius, but not in soleus, translated in a marked induction of PGC-1α target genes, such as carnitine palmitoyltransferase 1b (CPT-1b), pyruvate dehydrogenase kinase 4 (PDK4) or GLUT4 (Fig.S3D), while these genes only minimally responded in soleus (Fig.S3E). Together, these results physiologically correlate AMPK activation, PGC-1α deacetylation and PGC-1α activity
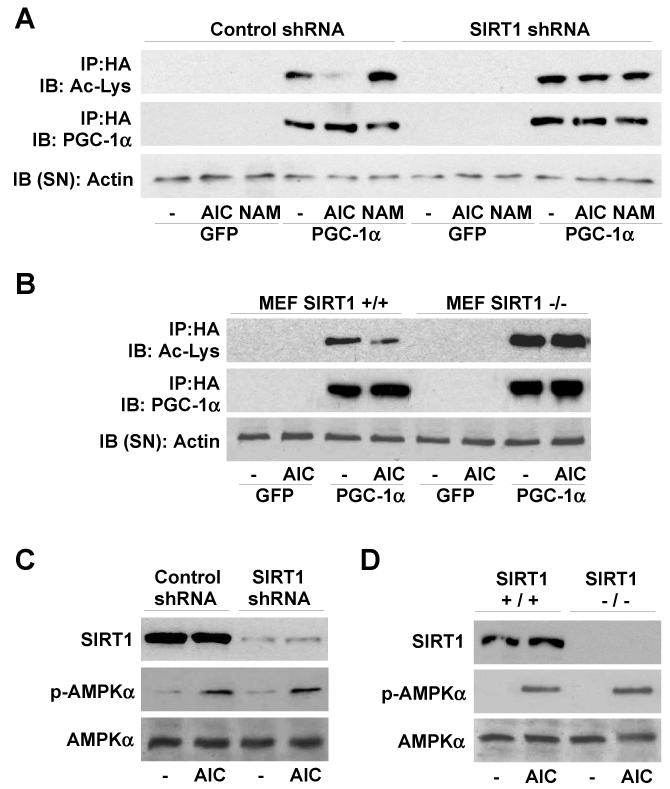
Since SIRT1 interacts with and deacetylates PGC-1α9, 11, we next evaluated whether SIRT1 mediates the AICAR-induced deacetylation of PGC-1α. Pre-treatment of C2C12 myotubes with nicotinamide (NAM), a type III histone deacetylase inhibitor17, blocked AICAR-induced PGC-1α deacetylation (Fig.S4). Furthermore, AICAR failed to decrease PGC-1α acetylation when SIRT1 expression was knocked-down with an shRNA9 (Fig.2A) or genetically ablated, as in SIRT1−/− mouse embryonic fibroblasts18 (Fig.2B). In none of these models the lack of SIRT1 affected AICAR-induced AMPK phosphorylation (Fig.2C-D). The protein levels of SIRT2 and SIRT3, the closest homologs of SIRT1, were not affected by reduced SIRT1 expression (Fig.S5), ruling out their implication in the blunted response to AICAR.
Figure 2. SIRT1 mediates AMPK-induced PGC-1α deacetylation.
(A and C) C2C12 myocytes were infected with adenoviruses encoding GFP, PGC-1α, and either control or SIRT1 shRNAs. After 8hrs (A) or 1hr (C) of AICAR treatment, PGC-1α acetylation and AMPK phosphorylation were analyzed (D-E) SIRT1+/+ and SIRT1−/− MEFs were infected with GFP and FLAG-HA-PGC-1α and treated for 8hrs (D) or 1hr (E) with AICAR to test PGC-1α acetylation and AMPK phosphorylation.
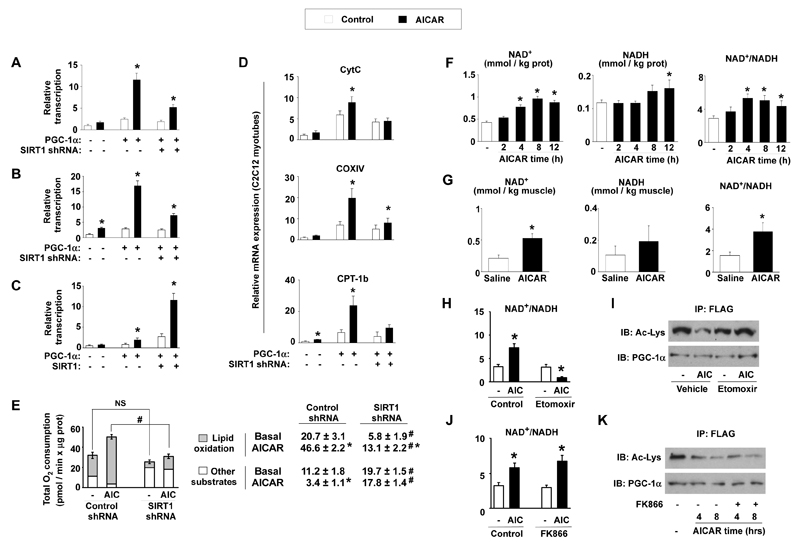
We next explored whether PGC-1α deacetylation influences AMPK-induded PGC-1α transcriptional activity. Since PGC-1α positively autoregulates its own promoter19, we monitored PGC-1α activity by transiently transfecting a luciferase reporter under the control of the PGC-1α promoter, in the absence/presence of PGC-1α and a SIRT1 shRNA. AICAR robustly increased PGC-1α action on its own promoter in both C2C12 myocytes (Fig.3A) and SIRT1+/+ MEFs (Fig.3B). This increase was reduced over 60% when SIRT1 was knocked-down in these cells (Fig.3A-B). Conversely, AICAR only mildly activated PGC-1α in SIRT1−/− MEFs, but full AICAR action was recovered when SIRT1 was reintroduced (Fig.3C). The lack of SIRT1 also compromised AICAR-induced PGC-1α-dependent transcriptional activity on promoters of other target genes, such as PDK420 and MCAD21 (Fig.S6A-B). The overexpression of the constitutively active PGC-1α R13 mutant, where the 13 acetylation sites are mutated, activated the PGC-1α promoter, which could not be further enhanced by AICAR or inhibited by a SIRT1 shRNA (Fig.S6C). This indicates that SIRT1 and the deacetylation of PGC-1α are necessary for AMPK to increase PGC-1α activity.
Figure 3. AICAR modulates PGC-1α-dependent transcriptional activity, mitochondrial gene expression and oxygen consumption through SIRT1 and NAD+ metabolism.
(A) C2C12 myocytes were transfected with a 2-kb mPGC-1α promoter luciferase reporter, a plasmid for mPGC-1α and simultaneously infected with adenovirus encoding control or SIRT1 shRNA. 36hrs later, cells were treated with AICAR (12hrs) and reporter activity was determined. (B) SIRT1+/+ MEFs were analyzed as in (A) (C) SIRT1−/− MEFs were transfected with the 2-kb mPGC-1α reporter and expression plasmids for PGC-1α, SIRT1 or the corresponding empty vectors. Then, cells were treated and analyzed as in (A) (D) C2C12 myocytes were infected with adenoviruses for GFP, PGC-1α and either control or SIRT1 shRNAs. After AICAR treatment, target mRNAs were analyzed by Q-RT-PCR. (E) O2 consumption in C2C12 myotubes infected with PGC-1α, and either control or SIRT1 shRNAs. Total length of the bar equals total O2 consumption. The white part of the bar is the O2 consumption in each group when treated with etomoxir (1mM). Therefore, the grey part represents lipid oxidation-derived O2 consumption. Values for O2 consumption due to the oxidation of lipids and other substrates are indicated on the right. (F) NAD+ and NADH content in C2C12 myotubes treated with AICAR for the times indicated. (G) Whole tibialis anterior muscles from mice treated with saline or AICAR were used for the measurement of NAD+ and NADH. (H-I) C2C12 myotubes preincubated with vehicle or etomoxir (50μM) for 1hr were treated with either vehicle (−) or AICAR (AIC). Then, NAD+ and NADH (H) or PGC-1α acetylation levels (I) were measured. (J-K) As in (H-I), but using FK866 (10nM) instead of etomoxir. All values are expressed as mean +/− S.E.M. * indicates statistical difference vs. vehicle/saline group at p < 0.05. # indicates statistical difference vs. respective control shRNA group at p < 0.05.
We then tested whether the lack of SIRT1 affects AICAR-induced expression of genes related to mitochondrial metabolism and fatty acid utilization, which are under the control of PGC-1α. AICAR-induced expression of genes involved in mitochondrial gene expression, such as estrogen-related receptor-α (ERRα) and PGC-1α, in mitochondrial architecture and electron transport, such as cytochrome C (CytC), cytochrome C oxidase IV (COX IV) and mitofusin-2, and in fatty acid utilization, such as CPT-1b and PDK4, was either prevented or robustly attenuated by knocking down SIRT1 expression in C2C12 myotubes (Fig.3D and Fig.S7) or deletion of the SIRT1 gene in MEFs (Fig.S8). The lack of SIRT1 also impaired the action of metformin on the expression of this gene set (Fig.S9). In contrast, AICAR was unable to further increase the expression of PGC-1α targets in C2C12 myotubes overexpressing the PGC-1α R13 mutant (Fig.S10). Together, these data indicate that, to a large extent, AMPK regulates the expression of mitochondrial and lipid metabolism genes through the modulation of PGC-1α activity by SIRT1.
SIRT1 had also a major role in the ability of AMPK to increase mitochondrial respiration, as the long-term effects of AICAR on cellular O2 consumption, a readout of oxidative metabolism, were severely blunted by knocking-down SIRT1 (Fig.3E). We hence determined the contribution of lipid oxidation on total O2 consumption by blocking mitochondrial fatty acid uptake with etomoxir. Confirming previous observations10, knocking down SIRT1 severely decreased lipid oxidation and increased the oxidation of alternative substrates (Fig.3E). In control cells, AICAR markedly increased lipid oxidation-driven O2 consumption, whereas it almost completely blocked the oxidation of other substrates, such as glucose. When SIRT1 expression was knocked-down, AICAR action on lipid oxidation was blunted and, additionally, AICAR was unable to decrease O2 consumption derived from the other substrates (Fig.3E). In line with this, direct measurement of oleate oxidation confirmed that the chronic effects of AICAR on lipid oxidation were blunted in myotubes where SIRT1 expression was knocked down (Fig.S11). The lack of SIRT1 hence alters the long-term actions of AICAR on lipid oxidation and global cellular O2 consumption.
As AICAR or metfomin cannot directly activate SIRT1 (Fig.S12), our data suggests that AMPK signaling modulates SIRT1 activity indirectly. No changes in SIRT1 protein levels were detected after 8hrs of AICAR (Fig.S13A), when PGC-1α is already maximally deacetylated, indicating that changes in activity where not due to increased SIRT1 abundance. We could neither observe interaction of AMPK with SIRT1 up to 8hrs after AICAR, either in the presence or absence of PGC-1α (Fig.S13B). Finally, AMPK could not phosphorylate SIRT1 in vitro either on the full-length protein (Fig.S13C) or on different GST-fragments (Fig.S13D). These results suggest that AMPK regulates SIRT1 action by indirect mechanisms.
Since SIRT1 deacetylase activity is driven by NAD+ levels22, we examined whether AMPK indirectly activates SIRT1 by altering the intracellular NAD+/NADH ratio. Supporting this hypothesis, AICAR increased the NAD+/NADH ratio in C2C12 myotubes and in skeletal muscle (Fig.3F-3G, respectively). The increase in NAD+/NADH ratio was evident 4hrs after AICAR in C2C12 myotubes, and remained elevated after 12hrs (Fig.3F), a timing that perfectly correlates with PGC-1α deacetylation (Fig.1A). Activation of AMPK by metformin, DNP, or overexpression of a constitutively active form of AMPKα1 also increased the NAD+/NADH ratio (Fig.S14A-B). A significant increase in NAD+ was also evident 3hrs after exercise in tibialis anterior muscle, further supporting the hypothesis that changes in NAD+ levels translate AMPK effects onto SIRT1 activity (Fig.S14C).
To determine how AMPK acutely increases the NAD+/NADH ratio, we pharmacologically targeted different possible sources of cellular NAD+ production. Inhibition of the glycolytic enzyme lactate dehydrogenase with oxamic acid did not affect the ability of AICAR to increase NAD+ levels and the NAD+/NADH ratio (Fig.S15A). In contrast, inhibition of mitochondrial fatty acid oxidation with etomoxir was enough to hamper the increase in NAD+/NADH induced by AMPK (Fig.3H and Fig.S15B), indicating that an increase in mitochondrial β-oxidation is required for AMPK to increase the NAD+/NADH ratio. Supporting the role of NAD+/NADH ratio on SIRT1 activity, etomoxir also abolished AMPK-induced PGC-1α deacetylation (Fig.3I),
SIRT1 activity is inhibited by NAM, a product of the deacetylation reaction catalyzed by the sirtuins17. NAM can be cleared and enzymatically reconverted into NAD+ through the NAD+ salvage pathway, whose initial and rate-limiting step is catalyzed in mammals by nicotinamide phosphoribosyltransferase (Nampt)23. Acute blockage of Nampt activity with the specific inhibitor FK86624, however, did not affect AICAR’s capacity to modulate PGC-1α acetylation or NAD+/NADH ratio up to 8hrs after treatment (Fig.3J-K and Fig.S15C). These results apparently collide with observations indicating that Nampt can link AMPK and SIRT1 activities25. These differences may be explained by the fact that a chronic knock-down of Nampt may constitutively inhibit SIRT1 due to reduced intracellular NAM clearance17. Supporting this speculation, knock-down of Nampt for 48hrs promotes PGC-1α hyperacetylation (Fig.S16). The use of FK866 dissociates the direct effects of acute Nampt inhibition from those of indirect SIRT1 inhibition and indicates that AMPK initially regulates the NAD+/NADH ratio and SIRT1 activity independently of Nampt. These results, however, do not rule out that Nampt could participate to sustain the actions of AMPK on SIRT1.
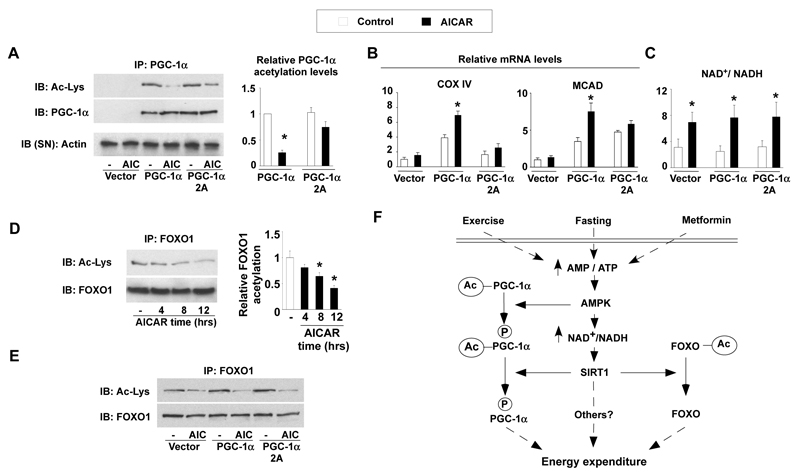
Whereas it has been reported that AMPK activates PGC-1α through direct phosphorylation8, our data show that deacetylation of PGC-1α upon AMPK activation is also required to activate PGC-1α. To understand how both posttranslational modifications intertwine, we used the PGC-1α 2A mutant lacking the two AMPK-phosphorylation sites8. The mutation of these sites markedly reduced deacetylation of PGC-1α by AICAR (Fig.4A), and, consistently, blunted AICAR action on the expression of mitochondrial genes (Fig 4B). The expression of the PGC-1α 2A mutant, however, did not alter AICAR action on the NAD+/NADH ratio, which increased to a similar extent as in myocytes expressing wild-type PGC-1α (Fig.4C and S17). This suggested that the activation of SIRT1 by AMPK should theoretically be unaffected by the PGC-1α 2A mutant and, consequently, still impact on other SIRT1 substrates beyond PGC-1α, like the FOXO transcription factors26. AICAR treatment triggered the deacetylation of endogenous FOXO1 (Fig.4D), as well as other members of the FOXO family, such as FOXO3a (Fig.S18), in C2C12 myotubes. The deacetylation of FOXO1 in response to AICAR was similar in myocytes expressing the wild-type or the 2A mutant of PGC-1α (Fig.4E), demonstrating that the PGC-1α 2A mutation only alters PGC-1α deacetylation but not general SIRT1 activation in response to AICAR. Hence, the data suggest a scenario where phosphorylation of PGC-1α constitutes a priming signal for subsequent deacetylation by SIRT1 (Fig.4F). Interestingly, AMPK can also phosphorylate the FOXO transcription factors27, which are also targets for SIRT1 deacetylation26. It is therefore tempting to speculate that the coordinated sequential actions of AMPK and SIRT1 could be a conserved mechanism for AMPK to modulate the specificity amongst SIRT1 targets, with phosphorylation discriminating which substrates should be deacetylated and preventing random deacetylation.
Figure 4. The PGC-1α phosphorylation mutant is resistant to deacetylation.
(A) C2C12 myocytes were transfected with the wild-type or the 2A mutant form of PGC-1α, using empty vector as control. After 36hrs, cells were treated with AICAR and total lysates were used to test PGC-1α acetylation. Relative acetylation levels of PGC-1α are shown on the right. (B) Cells were treated as in (A), and, after AICAR treatment, target mRNA levels were analyzed by RT-Q-PCR. (C) Cells were treated as in (A), and acidic or alkali lysates were obtained to measure NAD+ and NADH. (D) C2C12 myotubes were treated with AICAR for the times indicated. Then, total protein lysates were used for immunoprecipitation of FOXO1. Relative FOXO1 acetylation is shown on the right (E) As in (A), but immunoprecipitations were performed against FOXO1. (F) Scheme illustrating the convergent actions of AMPK and SIRT1 on PGC-1α. Pharmacological (metformin) and physiological (fasting or exercise) activation of AMPK in muscle triggers an increase in the NAD+/NADH ratio, which activate SIRT1. AMPK also induces the phosphorylation of PGC-1α and primes it for subsequent deacetylation by SIRT1. The impact of AMPK and SIRT1 on the acetylation status of PGC-1α and other transcriptional regulators, such as the FOXO family of transcription factors, will then modulate mitochondrial function and lipid metabolism. All values are presented as mean+/-SE. * indicates statistical difference vs. corresponding vehicle group at P<0.05.
This work demonstrates that deacetylation of PGC-1α is a key mechanism by which AMPK triggers PGC-1α activity in cultured myotubes and in skeletal muscle. We also unveil SIRT1 as a key, albeit not the sole, mediator of AMPK action on PGC-1α transcriptional activity, the expression of mitochondrial and lipid metabolism genes and O2 consumption (Fig.3). The acute actions of AMPK on lipid oxidation alter the balance between cellular NAD+ and NADH, which acts as a messenger to activate SIRT1. This study, hence, constitutes a step forward in the understanding of the mechanisms by which AMPK transcriptionally regulates energy expenditure. The implication of SIRT1 in the transcriptional actions of AMPK provides a possible explanation to the overlapping metabolic effects of SIRT1 and AMPK activators7, 12, 16, 28, 29. Furthermore, the interplay between SIRT1 and AMPK might be reciprocal as specific SIRT1 agonists promote fatty acid oxidation and indirectly activate AMPK through metabolic adaptations29. Hence, the inter-dependent regulation of SIRT1 and AMPK provides a finely tuned amplification mechanism for energy homeostasis under low nutrient availability. Altogether, these findings constitute a conciliatory view of the possible implications of AMPK and SIRT1 on the pleiotropic beneficial effects of calorie restriction on metabolic homeostasis and lifespan, where both enzymes were known to participate, but never linked.
METHODS
Exercise protocol
8-week old non-fasted C57BL6/J male mice were subjected to a resistance running test, using a variable speed belt treadmill enclosed in a plexiglass chamber with a stimulus device consisting of a shock grid attached to the rear of the belt (Panlab, Barcelona, Spain). Animals were acclimatized to the chamber the day preceding the running test. For the habituation, mice run at 21 cm/s for 10 minutes with a 5° incline. For the actual test, we used a protocol at 5° incline where, beginning at 18 cm/s, speed increased gradually by 3 cm/s every 5 minutes. The distance run and the number of shocks were monitored during the test, and exhaustion was assumed when mice received more than 50 shocks in a 2.5 minutes interval. Mice were removed from the treadmill upon exhaustion.
 Preceding the running test, we randomly subdivided mice into 3 different groups (8 mice/group): one group that would be sacrificed immediately after the exercise test, another which would be sacrificed 3-hrs after the exercise test, and, finally, a group that would be sacrified 6-hrs after the cessation of exercise. The time and distance run before exhaustion was similar in the three groups (data not shown), around 600 meters after 40 minutes of exercise. Mice sacrificed 3 and 6 hrs after exercise had free acces to food and water once the running protocol was finished.
Preceding the running test, we randomly subdivided mice into 3 different groups (8 mice/group): one group that would be sacrificed immediately after the exercise test, another which would be sacrificed 3-hrs after the exercise test, and, finally, a group that would be sacrified 6-hrs after the cessation of exercise. The time and distance run before exhaustion was similar in the three groups (data not shown), around 600 meters after 40 minutes of exercise. Mice sacrificed 3 and 6 hrs after exercise had free acces to food and water once the running protocol was finished.
Reagents and Materials
AICAR was purchased from Toronto Research Chemicals (Toronto, ON, Canada). Anti-PGC-1α (H300) and anti-actin goat antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-PGC-1α, anti-Acetyl-Lysine, anti-AMPKα anti-phospho-AMPKα (Thr172) and anti-FOXO1 polyclonal antibodies were purchased from Cell Signalling (Beverly, MA). Anti-Sir2 and anti-phospho AcetylCoA Carboxylase (ACC) (Ser79) were purchased from Upstate Biotechnology Inc. (Lake Placid, NY). Anti-Nampt antibody was purchased from Bethyl laboratories (Montgomery, TX). Anti-FLAG (M2) and anti-HA monoclonal antibodies as well as most commonly used chemicals were purchased from Sigma Aldrich. Nampt siRNAs were purchased from Dharmacon Inc. (Lafayette, CO). The A-769662 compound was a kind gift from Grahame Hardie (University of Dundee, Dundee, UK). Claude Ammann from TopoTarget Switzerland SA provided the FK866 compound.
Plasmids and adenoviral vectors
Adenoviruses encoding for GFP, FLAG-HA-PGC-1α, FLAG-R13-PGC-1α, control and SIRT1 shRNAs were described previously1. Adenoviruses encoding for the different forms of AMPKα1 subunit were a kind gift from Pascal Ferré and Fabienne Foufelle (INSERM Unit 671, Paris, France)2. The plasmids encoding for the mouse PDK4 gene promoter3, mouse MCAD gene promoter4, and the 2A-PGC-1α mutant5 have all been described. The plasmid encoding for FLAG-tagged FOXO3a was purchased from Addgene (Cambridge, MA).
Cell culture, adenoviral infection and treatments
C2C12 skeletal muscle cells were grown and differentiated as described6. Unless otherwise stated, C2C12 were considered as myotubes after 96 hrs of differentiation. Differentiation medium was supplemented with 0.1 mM oleic acid. Adenoviral infections of C2C12 myocytes were performed after 48 hrs of differentiation. Cells were washed with PBS and left for 1 hr in serum-free DMEM 4.5 g/l glucose containing the appropiate amount of viral particles (MOI = 100 per each virus used, using GFP as control to make even the final viral amount). Then, the media was replaced with fresh differentiation media for additional 47 hrs before any treatment took place. MEFs were cultured in DMEM 4.5 g/l glucose supplemented with 10% fetal calf serum (FCS). Adenoviral infection of MEFs was performed when cells reached 70% of confluency, and processed as with C2C12 myocytes but using 10% FCS DMEM medium instead of differentiation medium at the end. DMSO was used as vehicle for the different treatements. Plasmids and siRNAs were transfected using Lipofectamine 2000 (Invitrogen) and following the manufacturer’s instructions. SIRT1+/+ and SIRT1−/− cells were a kind gift of Fred Alt (Harvard Medical School, Boston, Massachusetts, USA).
Total protein extraction
To obtain total protein extracts from cellular samples, cells were rapidly washed with ice-cold PBS before adding cold lysis buffer (25 mM Tris HCl pH = 7.9, 5 mM MgCl2, 10% Glycerol, 100 mM KCl; 0,1% NP40; 0.3 mM dithrioteitol, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 50 mM sodium fluoride, containing freshly added protease inhibitor cocktail (Calbiochem)). For acetylation studies, 5 mM nicotinamide and 1 mM sodium butyrate were added to the buffer. After 1 minute, cells were scraped, transferred into an Eppendorf tube and left on ice for 5 more minutes. Then cells were homogenized with a 25-gauge needle, left for 5 more minutes on ice and centrifuged at 13000 rpms for 10 minutes. The supernatant was collected and kept at −80 °C.
Nuclear extracts
Nuclear extracts from gastrocnemius muscles were obtained as described previously6.
Immunoprecipitation and western blot
Routinely, 500 micrograms of protein from cultured cells or 2 milligram of protein from muscle sampes (total lysates or nuclear extracts) were used for immunoprecipitation. 40 microliters of Protein G-sepharose resuspended in Lysis Buffer were used for pleclearing the sample and immunoprecipitation after conjugating the beads with 3-5 mg of antibody. The resulting immunoprecipitate was boiled with 50 microliters of Laemmli Sample Buffer (LSB) and used for Western Blot applications. For immunoprecipitation using rabbit polyclonal antibodies, Protein-A-sepharose beads were used instead of Protein-G-conjugated beads. Western blot and protein detection was performed as described previously6.
Gene expression analysis
RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). Complementary DNA was generated using Superscript II enzyme (Invitrogen) and quantitative real-time PCR was performed as described previously6 using acidic ribosomal protein (ARP) to normalize the expression. The oligonucleotides primers used for PCR analysis are provided at the end of the section.
Oxygen Consumption
C2C12 myotubes or MEF cells were incubated for 5 hrs/day with AICAR during two days, in the presence of 0.1 mM oleic acid. Then oxygen consumption was measured using the Seahorse Biotechnology XF24 equipment (Seahorse Bioscience Inc., North Billerica, MA 01862, USA) as described7.
Oleate oxidation
The estimation of oleate oxidation rates was performed as described previously8.
Reporter Gene Assays
C2C12 myocytes were transfected in 48-well plates at 90% of confluence with Lipofectamine 2000 (Invitrogen) following the manufacturer’s intructions. Cells were left for 5 hrs with the DNA-lipofectamine mix, and the corresponding adenoviruses were added (each adenovirus at MOI 100) for the last hr of transfection. Then, the medium was removed and replaced by differentiation medium supplemented with 0.1 mM oleic acid for 36 hrs before treatment with AICAR or vehicle for 12 hrs. For MEFs, the protocol was similar, but replacing the transfection medium by DMEM 4.5 g/l glucose 10% FCS supplemented with 0.1 mM oleic acid. Firefly luciferase activity was measured and normalized to β-gal activity (always transfected simultaneously). Empty pGL3basic reporter gene vector and pCDNA.3.1 vector were used as control vectors. pGL3-PGC-1α 2 Kb promoter was purchased from Addgene (Cambridge, MA). The pcDNA.3.1 HA-PGC-1α plasmid was generated in Pere Puigserver’s laboratory. pcDNA-FLAG-SIRT1 plasmid was developed in our own laboratory. Reporter contructs for PDK4 and MCAD promoters were produced by Daniel P. Kelly’s laboratory.
Measurement of SIRT1 activity
Experiments testing the direct effects of AICAR and metformin on SIRT1 activity were performed as described9.
NAD+/NADH measurements
NAD+ and NADH nucleotides were directly measured as described before10. In brief, whole tibialis anterior muscles or two 10 cm dishes of C2C12 myotubes were homogenized in 200 microliters of acid extraction buffer to measure the NAD+ concentration, or 200 microliters of alkali extraction to obtain NADH concentration. Then, homogenates were neutralized and the concentration of nucleotides was measured fluorimetrically after an enzymatic cycling reaction using 5 microliters of sample. Values for both nucleotides were detected within the linear range. NAD+/NADH ratios were calculated by comparing the ratios obtained from each animal (randomly, one tibialis was used for NAD+ measurements and the other for NADH) or from parallel cell dishes in each experiment. The ratios obtained from different animals or individual cell culture experiment ratios were then used as individual values to calculate the mean and S.E.
Glycogen measurement
Muscle pieces (15–20 mg) were hydrolyzed in 250 μl of 2 M HCl at 95°C for 2 hrs. The solution was then neutralized with 250 μl 2 M NaOH, and the resulting free glycosyl units were assayed spectrophotometrically using a hexokinase-dependant assay kit (Amresco).
Protein kinase assays
For protein kinase assays on full-length SIRT1 and Nampt, FLAG-tagged proteins were produced using a coupled in vitro transduction and translation system (TNT, Promega Corporation, Madison, USA). Active AMPK and SAMS peptide, as positive control substrate for AMPK, were purchased from Upstate Biotechnology. AMPK was mixed with either FLAG-SIRT1, FLAG-Nampt, control vector or SAMS peptide (200 mM) in a solution containing 30 mM HEPES pH 7.4, 0.65 mM DTT, 0.02% Brij-35, 10 mM MgAc and 0.2 mM AMP. The reaction started by the addition of 0.1 mM ATP (containing [γ32P]ATP at 1000 cpm/pmol), and was stopped after 20 minutes by adding 5 ml of 3% phosphoric acid and 15 ml of the reaction mix were transferred to a piece of P81 phosphocellulose Whatmann paper and washed extensively with ortophosphoric acid solution. Then, the paper was dried with acetone and radioactivity was counted by Cherenkov counting. The rest of the reaction mix was diluted in LSB and boiled for 5 minutes. Western blots were performed to ensure the correct presence of the proteins in the mix (data not shown). For kinase assays on different GST-SIRT1 fragments and GST-PGC-1α 1-400 (as positive control), proteins were expressed in bacteria (BL21 strain; Invitrogen) and purified by using Glutathione-Sepharose 4B beads (GE Healthcare, 17-0756-01). In vitro kinase assays were carried out according to the manufacturer’s specifications (Millipore, 14-305). Briefly, recombinant protein was incubated with 32 mM HEPES pH 7.4, 0.01% Brij-35, 18.75 MgCl2, 150 mM AMP, 125 mM ATP, 2.5 mCi [γ32P] ATP, and 0.65 mM DTT in the presence or absence of 200 ng activated AMPK for 30 min at 30°C. The glutathione beads were then washed twice and eluted protein was analyzed by SDS-PAGE and radiolabeled phosphate incorporation was assessed by autoradiography. Protein levels were determined by Coomassie blue staining.
Oligonucleotide primer list
Acidic Ribosomal Protein:
Reverse: AAAGCCTGGAAGAAGGAGGTC
Forward: AGATTCGGGATATGCTGTTGG
PGC-1α
Reverse: GGGTTATCTTGGTTFFCTTTATG
Forward: AAGTGTFFAACTCTCTGGAACTG
PGC-1β
Reverse: TGGAGACTGCTCTGGAAGGT
Forward: TGCTGCTGTCCTCAAATACG
NRF-1
Reverse: GATGACCACCTCGACCGTTT
Forward: CGGAGTGACCCAAACTGAAC
ERRα
Forward: CACAGCCTCAGCATCTTCAA
Reverse: ACTGCCACTGCAGGATGAG
PPARα
Reverse: TTGAAGGAGCTTTGGGAAGA
Forward: AGGAAGCCGTTCTGTGACAT
PPARβ/δ
Reverse: ACTGGCTGTCAGGGTGGTTG
Forward: AATGCGCTGGAGCTCGATGAC
TFAm
Reverse: ATGTCTCCGGATCGTTTCAC
Forward: CCAAAAAGACCTCGTTCAGC
Cyt C
Reverse: TCCATCAGGGTATCCTCTCC
Forward: GGAGGCAAGCATAAGACTGG
COX IV
Reverse: GCTCGGCTTCCAGTATTGAG
Forward: AGAAGGAAATGGCTGCAGAA
ATP5g1
Reverse: AFTTGGTGTGGCTGGATCA
Forward: GCTGCTTGAGAGATGGGTTC
Mitofusin-2
Reverse: CAATCCCAGATGGCAGAACTT
Forward: ACGTCAAAGGGTACCTGTCCA
mCPT-1b
Reverse: GCACCCAGATGATTGGGATACTGT
Forward: TTGCCCTACAGCTGGCTCATTTCC
PDK4
Reverse: GGAACGTACACAATGTGGATTG
Forward: ATCTAACATCGCCAGAATTAAACC
MCAD
Reverse: AGCTGATTGGCAATGTCTCCAGCAAA
Forward: GATCGCAATGGGTGCTTTTGATAGAA
GLUT4
Reverse: AGGTGAAGATGAAGAAGCCAAGC
Forward: CTTCTTTGAGATTGGCCCTGG
SOD1
Reverse: TTGTTTCTCATGGACCACCA
Forward: AGGCTGTACCAGTGCAGGAC
Nampt
Reverse: AGTGGCCACAAATTCCAGAGA
Forward: CCGCCACAGTATCTGTTCCTT
Supplementary Material
Acknowledgements
This work was supported by grants of CNRS, Ecole Polytechnique Fédérale de Lausanne, INSERM, ULP, NIH (DK59820 and DK069966), EU FP6 (EUGENE2; LSHM-CT-2004-512013), EU Ideas programme (Sirtuins; ERC-2008-AdG-23118). CC is supported by grants of Fondation de la Recherche Medicale (FRM) and EMBO. JNF was supported by a FEBS grant. The authors thank Fabianne Foufelle and Pascal Ferre, Bruce Spiegelman, Daniel P. Kelly, Shin-ichiro Imai, Grahame Hardie, Claude Ammann (Topotarget) and Fred Alt for kindly providing materials and the members of the Auwerx and Puigserver labs for support and discussion.
Footnotes
Additional experimental procedures can be found in the Supplemental Data.
Author contributions. CC designed and executed experiments, interpreted data and wrote the manuscript. ZGH, JCM, JNF, ML and LN performed experiments and JNF helped with writing. PJE and PP provided crucial reagents and helped with data interpretation. JA supervised the design and interpretation of the experiments and participated in the writing of the manuscript.
Conflicts of interest. PP consults for and JCM and PJE are employed by Sirtris, a subsidiary of GSK that develops drugs targeting sirtuins.
REFERENCE LIST
- 1.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–85. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 2.Shaw RJ, et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–6. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–74. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fryer LG, Parbu-Patel A, Carling D. The Anti-diabetic drugs rosiglitazone and metformin stimulate AMP-activated protein kinase through distinct signaling pathways. J Biol Chem. 2002;277:25226–32. doi: 10.1074/jbc.M202489200. [DOI] [PubMed] [Google Scholar]
- 5.Barnes BR, et al. Changes in exercise-induced gene expression in 5′-AMP-activated protein kinase gamma3-null and gamma3 R225Q transgenic mice. Diabetes. 2005;54:3484–9. doi: 10.2337/diabetes.54.12.3484. [DOI] [PubMed] [Google Scholar]
- 6.Zong H, et al. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A. 2002;99:15983–7. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suwa M, Nakano H, Kumagai S. Effects of chronic AICAR treatment on fiber composition, enzyme activity, UCP3, and PGC-1 in rat muscles. J Appl Physiol. 2003;95:960–8. doi: 10.1152/japplphysiol.00349.2003. [DOI] [PubMed] [Google Scholar]
- 8.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–22. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 10.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 12.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A. 2007;104:7217–22. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi T, et al. Metabolic stress and altered glucose transport: activation of AMP-activated protein kinase as a unifying coupling mechanism. Diabetes. 2000;49:527–31. doi: 10.2337/diabetes.49.4.527. [DOI] [PubMed] [Google Scholar]
- 16.Cool B, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3:403–16. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 18.Chua KF, et al. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–6. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wende AR, Huss JM, Schaeffer PJ, Giguere V, Kelly DP. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25:10684–94. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–74. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 22.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 23.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 24.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–42. [PubMed] [Google Scholar]
- 25.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 27.Greer EL, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–19. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 28.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
REFERENCES FOR THE SUPPLEMENTAL PROCEDURES
- 1.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 2.Woods A, et al. Characterization of the role of AMP-activated protein kinase in the regulation of glucose-activated gene expression using constitutively active and dominant negative forms of the kinase. Mol Cell Biol. 2000;20:6704–11. doi: 10.1128/mcb.20.18.6704-6711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wende AR, Huss JM, Schaeffer PJ, Giguere V, Kelly DP. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol Cell Biol. 2005;25:10684–94. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–74. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 5.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–22. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe M, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–9. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 8.Pich S, et al. The Charcot-Marie-Tooth type 2A gene product, Mfn2, up-regulates fuel oxidation through expression of OXPHOS system. Hum Mol Genet. 2005;14:1405–15. doi: 10.1093/hmg/ddi149. [DOI] [PubMed] [Google Scholar]
- 9.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin SS, Manchester JK, Gordon JI. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J Biol Chem. 2001;276:36000–7. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.