Abstract
Macroautophagy (hereafter autophagy), or ‘self-eating’, is a conserved cellular pathway that controls protein and organelle degradation, and has essential roles in survival, development and homeostasis. Autophagy is also integral to human health and is involved in physiology, development, lifespan and a wide range of diseases, including cancer, neurodegeneration and microbial infection. Although research on this topic began in the late 1950s, substantial progress in the molecular study of autophagy has taken place during only the past 15 years. This review traces the key findings that led to our current molecular understanding of this complex process.
The term ‘autophagy’ comes from the Greek words ‘phagy’ meaning eat, and ‘auto’ meaning self. Autophagy is an evolutionarily conserved process in eukaryotes by which cytoplasmic cargo sequestered inside double-membrane vesicles are delivered to the lysosome for degradation. When autophagy was initially discovered more than 40 years ago, it was perplexing as to why the cell would self-digest its own components. The simplest hypothesis was that autophagy serves as a cellular rubbish-disposal mechanism. However, we have since learnt that this ‘self-eating’ process not only rids the cell of intracellular misfolded or long-lived proteins, superfluous or damaged organelles, and invading microorganisms, but also is an adaptive response to provide nutrients and energy on exposure to various stresses. Autophagy has been connected to human pathophysiology, and continued expansion of our knowledge about autophagy has had implications for fields as wide-ranging as cancer, neurodegeneration, immune response, development and ageing. This timeline reviews the history of autophagy research with a focus on the key events that occurred over the past 15 years, when our molecular understanding of this process first began.
The development of the autophagy concept
More than four decades ago, Clark and Novikoff observed mitochondria from mouse kidneys within membrane-bound compartments termed ‘dense bodies’, which were subsequently shown to include lysosomal enzymes1,2. Ashford and Porter later observed membrane-bound vesicles containing semi-digested mitochondria and endoplasmic reticulum in the hepatocytes of rats that had been exposed to glucagon3, and Novikoff and Essner observed that the same bodies contained lysosomal hydrolases4. One year later, in 1963, at the Ciba Foundation symposium on lysosomes, de Duve founded the field when he coined the term ‘autophagy’ to describe the presence of single- or double-membrane vesicles that contain parts of the cytoplasm and organelles in various states of disintegration. He pointed out that these sequestering vesicles, or ‘autophagosomes’, were related to lysosomes and occurred in normal cells. The origin of the membrane surrounding the autophagosome is still controversial; de Duve suggested that the sequestering membranes are derived from preformed membranes, such as smooth endoplasmic reticulum5.
Cellular autophagy is observed in normal rat liver cells, but is enhanced in the livers of starved animals6, and in 1967 de Duve and Deter confirmed that glucagon induces autophagy7. Ten years later, Pfeifer demonstrated the converse — that insulin inhibits autophagy8. Pioneering work by Mortimore and Schworer further demonstrated that amino acids, which are the end products of autophagic degradation, have an inhibitory effect on autophagy in rat liver cells9. These early lines of evidence are consistent with our current understanding of autophagy as an adaptive catabolic and energy-generating process. Subsequently, Seglen and Gordon carried out the first biochemical analysis of autophagy and identified the pharmacological reagent 3-methyladenine as an autophagy inhibitor10; they also provided the first evidence that protein kinases and phosphatases can regulate autophagy11.
These early studies of autophagy from the 1950s to the early 1980s were based on morphological analyses. de Duve and others primarily examined the terminal stages of the process, the steps just before or after fusion with the lysosome. Subsequent studies by Seglen's laboratory began to use electro-injected radioactive probes to examine the early and intermediate steps of autophagy, leading to the identification of the phagophore (the initial sequestering vesicle that develops into the autophagosome; Fig. 1), as well as the amphisome (a non-lysosomal vesicle formed by the fusion of autophagosomes and endosomes12).
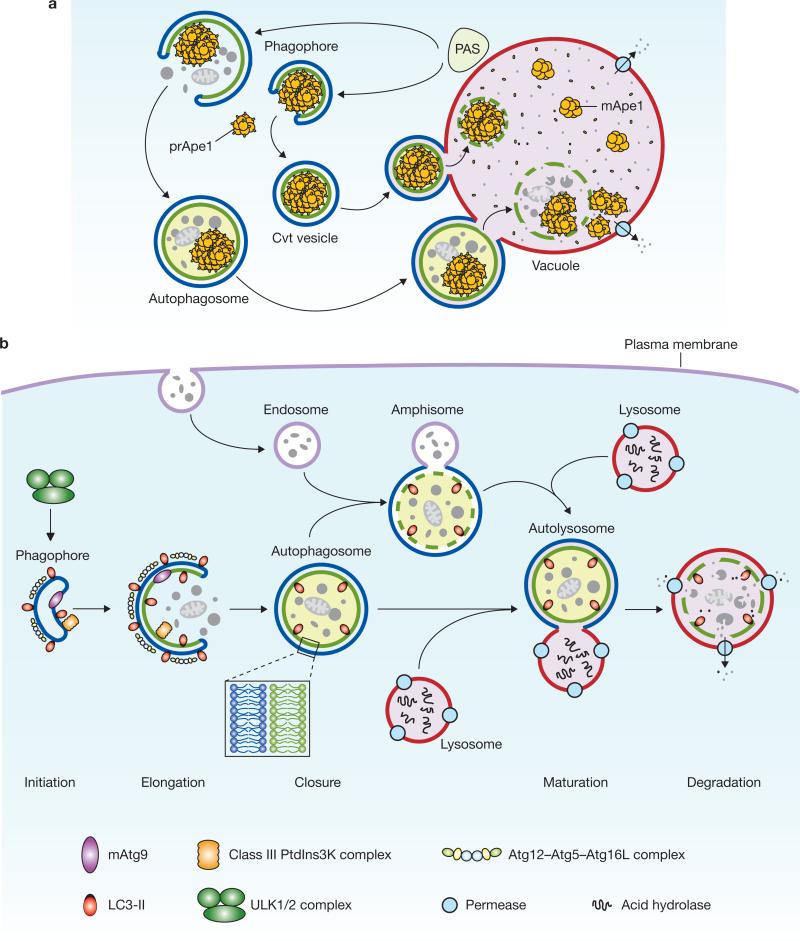
Figure 1.
Schematic depiction of autophagy. (a) In yeast, both autophagy and the Cvt pathway engulf cargoes within distinct double-membrane vesicles, which are thought to originate from the phagophore assembly site (PAS). The PAS is defined as the initial site for autophagy-related (Atg) protein recruitment. The Cvt pathway is one example of selective autophagy, and the only example of a biosynthetic autophagy-related pathway. The Cvt vesicle (140–160 nm in diameter) appears to closely enwrap the specific cargo — the Cvt complex (consisting of the precursor form of aminopeptidase I — prApe1 — and the Atg19 receptor), and exclude bulk cytoplasm. The autophagosome (300–900 nm in diameter) engulfs cytoplasm, including organelles, and also the Cvt complex. The completed vesicles then fuse with the vacuole, the yeast analogue of the mammalian lysosome, and release the inner single-membrane vesicle (autophagic or Cvt body) into the lumen. Subsequent breakdown of the inner vesicles allows the maturation of prApe1 and the degradation of cytoplasm, and hence the recycling of the resulting macromolecules through vacuolar permeases. (b) Mammalian autophagy is initiated by the formation of the phagophore, followed by a series of steps, including the elongation and expansion of the phagophore, closure and completion of a double-membrane autophagosome (which surrounds a portion of the cytoplasm), autophagosome maturation through docking and fusion with an endosome (the product of fusion is known as an amphisome) and/or lysosome (the product of fusion is known as an autolysosome), breakdown and degradation of the autophagosome inner membrane and cargo through acid hydrolases inside the autolysosome, and recycling of the resulting macromolecules through permeases. So far, there is no evidence for a PAS that exists in mammalian cells, and so the mammalian phagophore could be equivalent to the yeast PAS, or derived from the PAS. The core molecular machinery is also depicted, such as the ULK1 and ULK2 complexes that are required for autophagy induction, class III PtdIns3K complexes that are involved in autophagosome formation, mammalian Atg9 (mAtg9) that potentially contributes to the delivery of membrane to the forming autophagosome and two conjugation systems, the LC3-II and Atg12–Atg5–Atg16L complex, which are proposed to function during elongation and expansion of the phagophore membrane.
As early as the 1960s, de Duve suggested that most, if not all, living cells must employ a mechanism for nonspecific bulk segregation and digestion of portions of their own cytoplasm in the lysosome5, but also hinted at the need of a selective proteolytic mechanism acting on abnormal cellular proteins or organelles. In 1973, Bolender and Weibel provided some of the first evidence that a specific organelle (the smooth endoplasmic reticulum) can be engulfed by autophagy13. Four years later, Beaulaton and Lockshin suggested that mitochondria are selectively cleared during insect metamorphosis14. In 1983, Veenhuis demonstrated that superfluous peroxisomes are selectively degraded by autophagy in the yeast Hansenula polymorpha15, and five years later Lemasters and colleagues showed that changes in mitochondrial membrane potential lead to the onset of autophagy16. Further evidence that autophagy can be selective was provided by subsequent studies in yeast and higher eukaryotes.
The molecular era
Insights into the molecular control of autophagy, starting in the late 1990s, revolutionized the ability to detect and genetically manipulate this process, which allowed the field to grow at an extraordinarily fast pace and uncovered the importance of autophagy in human health and disease.
Although autophagy was initially identified in mammals, a significant breakthrough in our understanding of how autophagy is controlled came from analysis in the genetically tractable yeast system. Pioneering work from Ohsumi's group showed that the morphology of autophagy in yeast was similar to that documented in mammals17. They then carried out the first genetic screen for yeast mutants that affected protein turnover (nonspecific macroautophagy)18. This work was followed by similar screens for mutants that affected peroxisome degradation (pexophagy)19 and delivery of a resident vacuolar hydrolase (the cytoplasm to vacuole targeting (Cvt) pathway20, as reviewed in ref. 21). The identification of the first autophagy-related (Atg) gene, ATG1, was published in 1997 (ref. 22). The recent genetic screens for mutants that affect selective mitochondrial degradation (mitophagy), led to the identification of ATG32 and ATG33 (refs 23 and 24).
Although the Cvt pathway, pexophagy, mitophagy and macroautophagy are morphologically and mechanistically similar and require most of the Atg components, they are different in important ways. Macroautophagy is generally considered to be nonselective, whereas the Cvt pathway, pexophagy and mitophagy are highly selective. Pexophagy, mitophagy and nonspecific macroautophagy are degradative, whereas the Cvt pathway is biosynthetic, delivering at least two resident hydrolases to the vacuole25,26 (Fig. 1a). Overall, they share one subset of the Atg proteins that are essential for autophagosome formation and referred to as the ‘core’ molecular machinery (reviewed in ref. 27). The core machinery includes four major functional groups: (1) the Atg1–Atg13–Atg17 kinase complex, (2) the class III phosphatidylinositol 3-kinase (PtdIns3K) complex I, consisting of Vps34, Vps15, Atg6 and Atg14, (3) two ubiquitin-like protein conjugation systems (Atg12 and Atg8) and (4) Atg9 and its cycling system. Furthermore, in yeast the autophagy machinery is concentrated at a perivacuolar (the vacuole is the yeast equivalent of the lysosome) site termed the phagophore assembly site (PAS), and the concerted action of the autophagy machinery at the PAS leads to phagophore expansion and autophagosome formation28,29. A fifth set of core components includes proteins needed for the last steps of autophagy when the single-membrane intravacuolar vesicles (that result from fusion of the autophagosome or other sequestering vesicles with the vacuole limiting membrane) and their cargo break down, and permeases release these degradation products back into the cytosol for re-use30–32.
The identification of the ATG genes in yeast led to molecular analysis of autophagy in higher eukaryotes. Mizushima, while in Ohsumi's laboratory, identified the first mammalian autophagy genes, ATG5 and ATG12, and demonstrated that the Atg12–Atg5 conjugation system is conserved33. Perhaps the most critical finding in higher eukaryotes was the identification of the mammalian Atg8 homologue, MAP1LC3 (also known as LC3), by Yoshimori and colleagues, followed by the development of LC3-based assays for monitoring autophagy in mammals and other higher eukaryotes34. However, the increased synthesis or lipidation of LC3 are not sufficient for evaluating autophagy, and it is also critical to follow flux through the entire pathway, including in lysosomes35. In addition to the two conjugation systems, other mammalian Atg homologues have been identified and investigated (Fig. 1b, reviewed in ref. 36). Two Atg1 homologues, ULK1 and ULK2, are essential for autophagy induction and are found in a large complex that includes a mammalian homologue of Atg13 (mAtg13) and the scaffold protein, FIP200 (an orthologue of yeast Atg17). Formation of the human class III PtdIns3K complex, including human Vps34 (hVps34), Beclin 1 (a homologue of Atg6), Atg14L (an orthologue of Atg14) and p150 (a homologue of Vps15), is also conserved. Mizushima et al. used a green fluorescent protein-tagged Atg5 (ref. 37) to follow autophagosome formation, indicating that it proceeds in a step-wise manner, marked by the expansion of the sequestering vesicle (Fig. 1b).
The complexity of autophagy regulation in multicellular eukaryotes is becoming apparent from recent molecular analyses. For example, using Caenorhabditis elegans, Tian et al. identified four autophagy genes that are specific to multicellular animals, named epg-2, epg-3/VMP1, epg-4/EI24 and epg-5: epg-2 mediates cargo recognition and is specific to C. elegans, whereas the other three genes are conserved from worms to mammals38. Finally, two large-scale screens with human cells have identified numerous additional components that may interact with the known autophagy-related proteins, or participate in the signal transduction pathways that control this process39,40.
The origin of the autophagosome membrane is still under considerable debate. For example, recent studies have suggested that the endoplasmic reticulum membrane41–43, the mitochondrial outer membrane44 and the plasma membrane45 can contribute to autophagosome formation, suggesting that a range of organelles can provide the required membrane components (for details, see page 831 of this issue)46.
Structural analysis of the Atg proteins should reveal the mechanism of autophagy; the structure of the mammalian Atg8 homologues, including the γ-aminobutyric acid receptor-associated protein (GABARAP)47 and LC3 (ref. 48) were the first to be reported. Recently, Miller et al. reported the structure of Drosophila melanogaster Vps34 in a complex with PtdIns3K inhibitors, which may help in the design of new drugs that specifically target this kinase49.
Signalling regulation of autophagy
The key breakthrough in our understanding of the signalling pathways that regulate autophagy occurred following the identification of the target of rapamycin kinase (TOR)50,51, which modulates cell growth, cell-cycle progression and protein synthesis. In 1995, Meijer's group showed that rapamycin, an inhibitor of TOR, induces autophagy in rat hepatocytes, and relieves the inhibitory effect of amino acids on autophagy52. They also demonstrated that amino acids stimulate ribosomal protein S6 phosphorylation, an effect inhibited by rapamycin, providing a connection between amino acid-dependent and TOR-dependent regulation (Fig. 2). The TOR signalling pathway is critical because of its ability to integrate the information from nutrient, metabolic and hormonal signals. Research in the yeast system initially lagged behind the mammalian field, but in 1998 Ohsumi's laboratory reported that rapamycin also induces autophagy in yeast53.
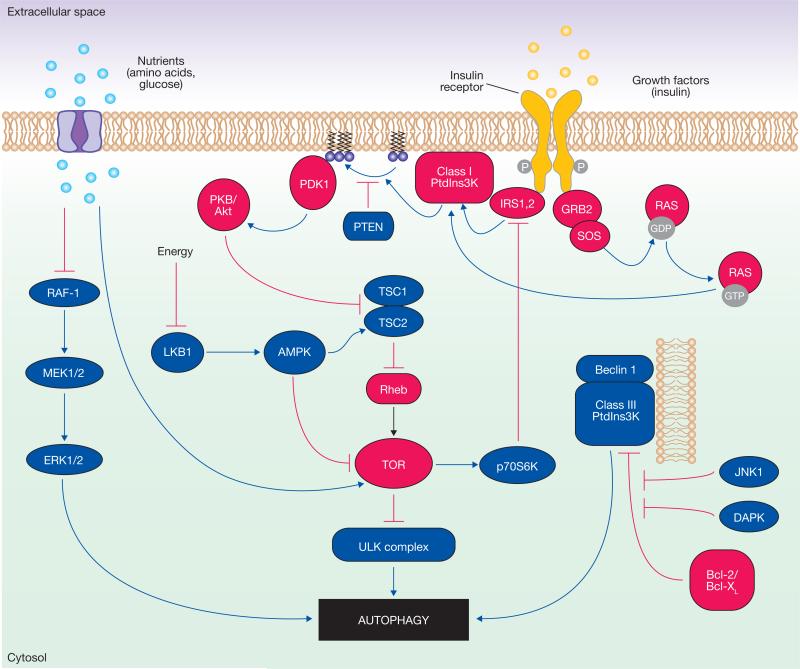
Figure 2.
Signalling regulation of mammalian autophagy. In the figure, the blue components represent the factors that stimulate autophagy, whereas the red ones correspond to inhibitory factors. Autophagy is regulated by a complex signalling network of various stimulatory (blue arrows) and inhibitory (red bars) inputs. TOR plays a central role in autophagy by integrating the class I PtdIns3K signalling and amino acid-dependent signalling pathways. Activation of insulin receptors stimulates the class I PtdIns3K complex and small GTPase Ras, leading to activation of the PtdIns3K–PKB–TOR pathway. PKB phosphorylates and inhibits the tuberous sclerosis complex 1/2 (TSC1–TSC2), leading to the stabilization of Rheb GTPase, which in turn activates TOR, causing inhibition of autophagy. Amino acids inhibit the Raf-1–MEK1/2–ERK1/2 signalling cascade, leading to inhibition of autophagy. Energy depletion causes the AMP-activated protein kinase (AMPK) to be phosphorylated and activated by LKB1. AMPK phosphorylates and activates TSC1–TSC2, leading to inactivation of TOR and autophagy induction. p70S6K kinase is a substrate of TOR that may negatively feed back on TOR activity, ensuring basal levels of autophagy that are important for homeostasis. JNK1 and DAPK phosphorylate and disrupt the association of anti-apoptotic proteins, Bcl-2 and Bcl-XL, with Beclin 1, leading to the activation of the Beclin 1-associated class III PtdIns3K complex and stimulation of autophagy. Beclin 1 is shown bound to the phagophore membrane.
In 1997, Meijer's group found that amino acid-induced S6 phosphorylation was prevented by the PtdIns3K inhibitors wortmannin and LY294002 in rat hepatocytes54 and thus by analogy with rapamycin, should induce autophagy. Unexpectedly, these PtdIns3K inhibitors (and 3-methyladenine) blocked autophagy in the absence of amino acids. One explanation for this apparent contradiction was the presence of two classes of phosphoinositides and phosphatidylinositol kinases. Indeed, Codogno's group, in collaboration with Meijer's laboratory, showed that the class III PtdIns3K product, phosphatidylinositol 3-phosphate (PtdIns(3)P), is essential for autophagy, whereas the class I PtdIns3K products, phosphatidylinositol (3,4)-bisphosphate (PtdIns(3,4)P2) and phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3), have inhibitory effects55. In agreement with these results, overexpression of PTEN, which hydrolyzes PtdIns(3,4)P2 and PtdIns(3,4,5)P3, stimulates autophagy56. The PtdIns3K inhibitors inhibit both classes of PtdIns3K enzymes, and thus downregulate both autophagy and S6 phosphorylation.
Insulin had been shown to inhibit autophagy and we now know that the initial steps in insulin signal transduction occur at the plasma membrane and lead to the activation of the class I PtdIns3K and the production of PtdIns(3,4,5)P3 to promote the membrane recruitment and activation of protein kinase B (PKB; also known as AKT) through 3-phosphoinositide-dependent protein kinase 1 (PDK1; Fig. 2). Subsequent studies demonstrated that activation of this pathway, by expressing an active form of PKB, or expressing a constitutively active form of PDK1, has an inhibitory effect on autophagy56,57. Moreover, TOR is a downstream target: rapamycin reverses the inhibition of autophagy that results from activation of the class I PtdIns3K pathway.
Although TOR was considered central to autophagy regulation, TOR-independent pathways have been recently reported (Fig. 2). For example, Beclin 1 can be activated by the stress-responsive c-Jun amino-terminal kinase 1 (JNK1) and death-associated protein kinase (DAPK)58,59.
Health and disease
Cancer
Accumulating evidence reveals that alterations in autophagy occur in various human diseases. Cancer was one of the first diseases genetically linked to impaired autophagy: a landmark discovery by Levine's laboratory found that Beclin 1, a phylogenetically conserved protein essential for autophagy, is also a haploinsufficient tumour suppressor60. Beclin 1 was originally isolated as a Bcl-2 (B-cell lymphoma 2)-interacting protein. Binding of Beclin 1 to the anti-apoptotic protein Bcl-2 decreases Beclin 1-associated hVps34 PtdIns3K activity and thereby inhibits autophagy61. beclin 1 monoallelic deletion on chromosome locus 17q21 occurs in 40–75% of human ovarian, breast and prostate cancers62. Mice with heterozygous loss of beclin 1 show an accelerated rate of spontaneous tumour development 63,64, and Atg4C- deficient mice display a similar propensity65. These observations suggest that autophagy is important for tumour suppression.
White and colleagues have provided evidence to explain the apparent paradox as to why autophagy, which functions primarily as a cell-survival pathway, also functions in tumour suppression (Fig. 3). First, in apoptosis-defective cells, autophagy prevents death from necrosis, a process that might exacerbate local inflammation and promote tumour growth66. Second, stressed autophagy-defective tumour cells accumulate p62 (also known as sequestosome 1), damaged mitochondria, reactive oxygen species (ROS) and protein aggregates, which might cause DNA damage, oncogene activation and tumorigenesis67,68. However, autophagy may also promote tumour cell survival during metabolic stress in the tumour microenvironment, under conditions of hypoxia and low nutrients. Accordingly, genetic or pharmacological inhibition of autophagy was shown to enhance the cytotoxicity of cancer chemotherapy agents and to promote tumour regression69,70. Thus, autophagy can act both positively and negatively with regard to cancer cell survival; autophagy probably functions to prevent cancer initially, but once a tumour develops, the cancer cells utilize autophagy for their own cytoprotection.
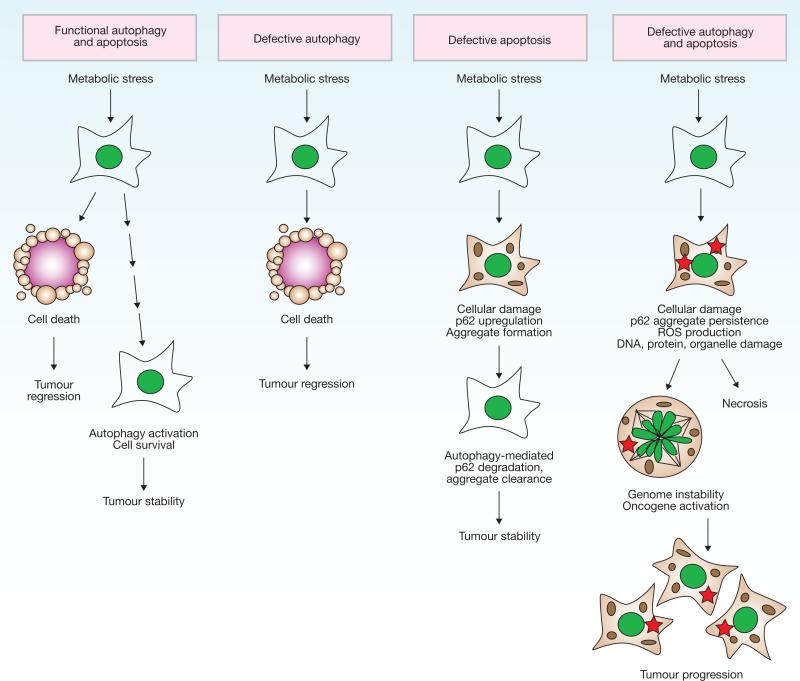
Figure 3.
A model for the roles of apoptosis and autophagy in tumorigenesis. A common cellular response to metabolic stress is cell death mediated by apoptosis, which limits tumour growth. Tumours may trigger autophagy-mediated cell survival in certain metabolic-stressed tumour regions. In apoptotic-defective, metabolic-stressed tumour cells, activation of autophagy prevents death from necrosis, whereas defects in autophagy lead to accumulation of p62, damaged mitochondria, ROS and protein aggregates, resulting in genome damage and tumorigenesis. For additional information, see refs 68 and 119.
Neurodegeneration
Early studies by Rubinsztein's laboratory demonstrated that autophagy affects the degradation of certain aggregate-prone proteins, such as those involved in Huntington's disease71. Induction of autophagy by inhibition of TOR attenuates the accumulation of mutant huntingtin aggregates and protects against neurodegeneration in fly and mouse models of Huntington's disease72. Subsequent studies provided compelling evidence that activation of autophagy is a beneficial physiological response in nearly all neurodegenerative diseases73,74. In addition, two parallel mouse studies using neuronal-specific knockouts of Atg5 or Atg7 demonstrate that basal autophagy controls the constitutive turnover of soluble, cytosolic proteins to prevent the accumulation of abnormal neuroproteins that may cause symptoms of neurodegeneration75,76.
Furthermore, autophagy may eliminate protein aggregates through a selective mechanism77,78. One possible autophagy receptor is p62/SQSTM1, a multifunctional adaptor protein that contains an LC3-interacting region (LIR) and a ubiquitin-associated (UBA) domain79. Mounting evidence suggests that p62 and NBR1, another apparent autophagy receptor with similar domains80, serve as cargo receptors to selectively deliver polyubiquitylated, misfolded, aggregated proteins and damaged, potentially deleterious organelles for clearance through autophagy in both mammals and Drosophila79–83 (for more details see also the Perspective by Peter and colleagues on page 836 of this issue)84. Importantly, as an in vivo LC3-interacting protein that is constantly degraded by autophagy, p62 has been widely used as a marker for autophagic flux36.
Innate and adaptive immunity
Although as early as 1984 Rikihisa reported that autophagy is induced during Rickettsia infection85, it was not until the emergence of new tools to detect autophagy in infected cells that it became clear autophagy affects diverse aspects of immunity. In 2004, Yoshimori's group, and simultaneously the laboratories of Deretic and Colombo, provided landmark studies showing autophagy is an important defence mechanism against certain invading bacterial pathogens, such as Mycobacterium tuberculosis and Streptococcus pyogenes86,87. Other studies soon extended the list of invading microorganisms that interact with autophagy88–90. Recently, Kurata's group, in collaboration with Yoshimori's laboratory, provided the first evidence that an intracellular pattern-recognition receptor, PGRP-LE, affects recognition and delivery of invading Listeria monocytogenes to the autophagy-mediated host defence system in Drosophila91. Randow's laboratory provided further evidence that a human autophagy receptor, NDP52 (nuclear dot protein 52 K), detects ubiquitin-coated Salmonella enterica and directs this bacteria into autophagosomes by simultaneously binding to LC3 (ref. 92).
Furthermore, the sequestration of intracellular pathogens during autophagy is not limited to bacteria and parasites. More than a decade ago, Liang et al. demonstrated that enforced neuronal expression of Beclin 1 protected mice against alphavirus replication and encephalitis93. Subsequent studies with herpes simplex virus confirmed the role of autophagy in engulfing newly assembled viruses inside the host cells94, whereas autophagy inhibition is essential for viruses to evade innate immunity and cause disease95. It is worth noting that as with certain bacteria, viruses may have evolved strategies to utilize the autophagic machinery to establish their own replicative niche. Finally, in addition to a role in innate immunity, autophagy also promotes the adaptive immune response. In particular, Münz's laboratory provided the first demonstration that autophagy is involved in efficient MHC class II presentation of an endogenously synthesized viral protein (Epstein-Barr virus nuclear antigen 1; EBNA1)96; the involvement of autophagy in facilitating the processing and presentation of MHC class I antigen was recently demonstrated by Desjardins's laboratory97.
Ageing and longevity
A common feature of all ageing cells is a progressive accumulation of damaged proteins and organelles (such as defective mitochondria) and decreased autophagic activity could be important for this. Early studies from Bergamini's laboratory showed that autophagy function declines with age in vivo in rodents and in vitro in isolated hepatocytes98,99. They also carried out the first critical analysis showing that caloric restriction, the only intervention known to effectively slow down ageing, prevents the decline of autophagic activity with age99,100. The first experimental study implicating autophagy genes in ageing was performed by Levine's group, which showed that knockdown of bec-1 (C. elegans Beclin 1 orthologue-1), inhibits a lifespan-extending phenotype in mutants lacking the insulin signalling gene, daf-2 (ref. 101). Subsequent studies in Drosophila confirm a critical role for autophagy in promoting longevity, based on the observation that Atg7-deficient flies are short-lived102, whereas promoting basal levels of autophagy enhances longevity in adult flies103. Recent advances in understanding the molecular links between autophagy and ageing control are reviewed on page 842 of this issue104 and suggest that various signalling pathways and environmental factors may converge on autophagy to regulate ageing.
Current efforts to avoid the decline of autophagy function with age include the practice of using an anti-lipolytic drug that mimics the beneficial effect of caloric restriction on autophagy105. Spermidine, a naturally occurring polyamine, also promotes longevity by inducing autophagy, although its lifespan-extending effect has not been investigated clinically106.
Development and cell death
Since the discovery of the Atg machinery in yeast, Tsukada and Ohsumi noted that yeast autophagy mutants cannot sporulate during starvation18. Many subsequent studies in various organisms confirmed the role of autophagy in development. For example, autophagy mutants of Dictyostelium discoideum are defective in multicellular development107, inactivation of C. elegans autophagy genes disrupts normal dauer formation101, mutation of Drosophila Atg1 or Atg3 results in premature death from the larval to the pupal stage107 and loss of beclin 1 in mice results in early embryonic lethality63,64. Given this, it has long been presumed that autophagy supplies nutrients during developmental remodelling processes that occur during starvation. However, caution must be exercised in drawing conclusions about these phenotypes. For example, Atg7–/– Drosophila show normal metamorphosis102, and Atg5–/– and Atg7–/– mice (generated by mating heterozygotes) survive embryogenesis and appear normal at birth109,110. These studies, however, might overlook the role of autophagy during very early development. Indeed, Mizushima's laboratory provided compelling evidence using oocyte-specific Atg5–/– mice that autophagy is induced shortly after fertilization and is essential during a short period of early embryogenesis, the oocyte-to-embryo transition, but not for later embryo development111 (see also the review by Mizushima and Levine on the role of autophagy in mammalian cell differentiation and development on page 823 of this issue)112.
In addition to the well-documented role of autophagy in cell survival, a function for autophagy in cell death has long been proposed. Autophagic cell death was originally described in tissues undergoing active development. In the early 1960s and 1970s, ultrastructural studies revealed that in Drosophila autophagic vacuoles accumulate during an early stage in the destruction of most larval tissues113,114. Autophagy is often called ‘type II programmed cell death', in contrast to apoptosis, or ‘type I programmed cell death’. Yu et al. and Shimizu et al. provided the first evidence that when apoptosis is compromised, activation of autophagy leads to cell death115,116. Notably, the complex crosstalk between ‘self-digestion’ by autophagy and ‘self-killing’ by apoptosis may be key in diverse aspects of development and disease pathogenesis. Autophagic cell death is especially important for development because certain developmental programmes require massive cell elimination. Although there is no definitive evidence that autophagy is necessary for developmental cell death in mammals, Berry et al. have provided compelling evidence that in Drosophila autophagy is indeed required for developmental degradation of salivary gland cells117. McPhee et al. provided further evidence that an engulfment receptor, Draper, is required for the induction of autophagy during degradation of salivary glands, but not starvation-induced autophagy in the fat body, which is associated with survival118. This suggests that Draper functions to separate autophagy associated with cell death from autophagy leading to cell survival118. However, since it is difficult to separate the independent roles of autophagy and apoptosis during the rapid destruction that occurs in the salivary gland, the physiological role of autophagy in developmental cell death is rather complicated, and the observations of ‘autophagic cell death’ may not be correct, even if autophagy occurs in dying cells. Thus, there is little direct evidence that autophagy drives physiological cell death, and most researchers now refer to cell death ‘with autophagic features’, reflecting the fact that autophagy is primarily a cell survival mechanism.
Finally, it is worth noting that the putative function of autophagy in cell death is not restricted to developmental programmed cell death but also extends to cell death that occurs during various pathological conditions, such as cancer, neurodegeneration, immunity and ageing. There is no doubt that the process of autophagy, which has the capacity to degrade entire organelles, can be extremely detrimental to cellular physiology if not properly regulated. Therefore, a full understanding of the paradoxical roles of autophagy in promoting life and death will be critical for a practical assessment of autophagy and its use as a therapeutic intervention.
CONCLUSIONS
Three critical points emerge from this historical survey. First, our current knowledge of autophagy, especially in human physiology, represents only the tip of the iceberg. Autophagy may function primarily as a cytoprotective mechanism, for example, to maintain nutrient and energy homeostasis during starvation conditions, or to clear defective proteins, damaged organelles and invasive pathogens that cause various diseases. However, activation of autophagy can also be harmful: autophagy might allow cancer cells to become resistant to chemotherapy, or excessive autophagy might cause undesirable cell death. Thus, defining the precise roles of autophagy in specific disease contexts, and determining whether stimulation or inhibition of autophagy is more beneficial are future goals. Second, we need a greater understanding of the regulatory pathways that control autophagy. In particular, how does the cell determine the specificity and magnitude of autophagy based on complex signalling inputs? Finally, there are still many fundamental questions about the molecular actions of the Atg proteins, the membrane source(s) for autophagosome formation, the mechanism of sequestering vesicle formation and the selective nature of autophagy. Our knowledge about autophagy is growing rapidly. Perhaps soon, we will be able to manipulate autophagy to fight disease and promote health.
ACKNOWLEDGEMENTS
This work was supported by NIH grant GM53396 to D.J.K.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Clark SL., Jr. Cellular differentiation in the kidneys of newborn mice studied with the electron microscope. J. Biophys. Biochem. Cytol. 1957;3:349–362. doi: 10.1083/jcb.3.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novikoff AB. The proximal tubule cell in experimental hydronephrosis. J. Biophys. Biochem. Cytol. 1959;6:136–138. doi: 10.1083/jcb.6.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J. Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novikoff AB, Essner E. Cytolysomes and mitochondrial degeneration. J. Cell Biol. 1962;15:140–146. doi: 10.1083/jcb.15.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Duve C, Wattiaux R. Functions of lysosomes. Annu. Rev. Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 6.Novikoff AB, Essner E, Quintana N. Golgi apparatus and lysosomes. Fed. Proc. 1964;23:1010–1022. [PubMed] [Google Scholar]
- 7.Deter RL, Baudhuin P, de Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J. Cell Biol. 1967;35:C11–16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeifer U. Inhibition by insulin of the physiological autophagic breakdown of cell organelles. Acta Biol. Med. Ger. 1977;36:1691–1694. [PubMed] [Google Scholar]
- 9.Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–176. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- 10.Seglen PO, Gordon PB. 3-methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl Acad. Sci. USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holen I, Gordon PB, Seglen PO. Protein kinase-dependent effects of okadaic acid on hepatocytic autophagy and cytoskeletal integrity. Biochem. J. 1992;284:633–636. doi: 10.1042/bj2840633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem. Biophys. Res. Commun. 1988;151:40–47. doi: 10.1016/0006-291x(88)90556-6. [DOI] [PubMed] [Google Scholar]
- 13.Bolender RP, Weibel ER. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of treatment. J. Cell Biol. 1973;56:746–761. doi: 10.1083/jcb.56.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaulaton J, Lockshin RA. Ultrastructural study of the normal degeneration of the intersegmental muscles of Anthereae polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J. Morphol. 1977;154:39–57. doi: 10.1002/jmor.1051540104. [DOI] [PubMed] [Google Scholar]
- 15.Veenhuis M, Douma A, Harder W, Osumi M. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch. Microbiol. 1983;134:193–203. doi: 10.1007/BF00407757. [DOI] [PubMed] [Google Scholar]
- 16.Lemasters JJ, et al. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta. 1998;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 17.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 19.Titorenko VI, Keizer I, Harder W, Veenhuis M. Isolation and characterization of mutants impaired in the selective degradation of peroxisomes in the yeast Hansenula polymorpha. J. Bacteriol. 1995;177:357–363. doi: 10.1128/jb.177.2.357-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 22.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 23.Kanki T, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teter SA, et al. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 2001;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epple UD, Suriapranata I, Eskelinen E-L, Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 1998;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 34.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2009;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Y, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010;141:1042–1055. doi: 10.1016/j.cell.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 39.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipinski MM, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 43.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 44.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tooze S. A & Yoshimori, T. The origin of the autophagosomal membrane. Nat. Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 47.Bavro VN, et al. Crystal structure of the GABAA-receptor-associated protein, GABARAP. EMBO Rep. 2002;3:183–189. doi: 10.1093/embo-reports/kvf026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugawara K, et al. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- 49.Miller S, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 51.Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 52.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 53.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 54.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 55.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 56.Arico S, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 57.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int. J. Biochem. Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 58.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zalckvar E, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 61.Pattingre S, et al. Bcl-2 anti-apoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Aita VM, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 63.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marino G, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J. Biol. Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 66.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mathew R, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carew JS, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr–Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 72.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 73.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. α-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 74.Yu WH, et al. Macroautophagy—a novel β-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurode-generation in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 76.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 77.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell. Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 79.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 80.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 81.Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 83.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl Acad. Sci. USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kraft C, Peter M, Hoffman K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 85.Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced rickettsiae. Anat. Rec. 1984;208:319–327. doi: 10.1002/ar.1092080302. [DOI] [PubMed] [Google Scholar]
- 86.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 87.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 88.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste C. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J. Clin. Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 90.Ogawa M, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 91.Yano T, et al. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 93.Liang XH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tallóczy Z, Virgin HW, IV., Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 95.Orvedahl A, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 96.Paludan C, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 97.English L, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Del Roso A, et al. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp. Gerontol. 2003;38:519–527. doi: 10.1016/s0531-5565(03)00002-0. [DOI] [PubMed] [Google Scholar]
- 99.Donati A, et al. Age-related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J. Gerontol. A Biol. Sci..Med. Sci. 2001;56:B288–293. doi: 10.1093/gerona/56.7.b288. [DOI] [PubMed] [Google Scholar]
- 100.Cavallini G, Donati A, Gori Z, Bergamini E. Towards an understanding of the anti-aging mechanism of caloric restriction. Curr. Aging Sci. 2008;1:4–9. doi: 10.2174/1874609810801010004. [DOI] [PubMed] [Google Scholar]
- 101.Meléndez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 102.Juhasz G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Simonsen A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 104.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat. Cell Biol. 2010;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 105.Bergamini E. Targets for antiageing drugs. Expert Opin. Ther. Targets. 2005;9:77–82. doi: 10.1517/14728222.9.1.77. [DOI] [PubMed] [Google Scholar]
- 106.Eisenberg T, et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 107.Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 108.Juhasz G, Csikos G, Sinka R, Erdelyi M, Sass M. The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 2003;543:154–158. doi: 10.1016/s0014-5793(03)00431-9. [DOI] [PubMed] [Google Scholar]
- 109.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 110.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tsukamoto S, et al. Autophagy is essential for pre-implantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 112.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schin KS, Clever U. Lysosomal and free acid phosphatase in salivary glands of Chironomus tentans. Science. 1965;150:1053–1055. doi: 10.1126/science.150.3699.1053. [DOI] [PubMed] [Google Scholar]
- 114.Nopanitaya W, Misch DW. Developmental cytology of the midgut in the flesh-fly, Sarcophaga bullata (Parker). Tissue Cell. 1974;6:487–502. doi: 10.1016/0040-8166(74)90040-8. [DOI] [PubMed] [Google Scholar]
- 115.Yu L, et al. Regulation of an ATG7–beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 116.Shimizu S, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 117.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McPhee CK, Logan MA, Freeman MR, Baehrecke EH. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465:1093–1096. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]