Abstract
Recent advances in microbiology implicate the cytoskeleton in the life cycle of some pathogens, such as intracellular bacteria, Rickettsia and viruses. The cellular cytoskeleton provides the basis for intracellular movements such as those that transport the pathogen to and from the cell surface to the nuclear region, or those that produce cortical protrusions that project the pathogen outwards from the cell surface towards an adjacent cell.
Transport in both directions within the neuron is required for pathogens such as the herpesviruses to travel to and from the nucleus and perinuclear region where replication takes place. This trafficking is likely to depend on cellular motors moving on a combination of microtubule and actin filament tracks. Recently, Bearer et al. reconstituted retrograde transport of herpes simplex virus (HSV) in the giant axon of the squid. These studies identified the tegument proteins as the viral proteins most likely to recruit retrograde motors for the transport of HSV to the neuronal nucleus. Similar microtubule-based intracellular movements are part of the biological behavior of vaccinia, a poxvirus, and of adenovirus.
Pathogen-induced surface projections and motility within the cortical cytoplasm also play a role in the life cycle of intracellular pathogens. Such motility is driven by pathogen-mediated actin polymerization. Virulence depends on this actin-based motility, since virulence is reduced in Listeria ActA mutants that lack the ability to recruit Arp2/3 and polymerize actin and in vaccinia virus mutants that cannot stimulate actin polymerization.
Inhibition of intracellular movements provides a potential strategy to limit pathogenicity. The host cell motors and tracks, as well as the pathogen factors that interact with them, are potential targets for novel antimicrobial therapy.
Keywords: Herpes Simplex Virus, Microtubules, Molecular Motors, Myosin, Listeria monocytogenes, vaccinia virus, Actin, Arp2/3 complex, VASP
INTRODUCTION
Intracellular pathogens are well known to co-opt cellular machinery to accomplish their life cycle. Pathogen utilization of cellular machinery is best exemplified by the dependence of some viruses on host cell nuclear factors for replication. Such recruitment potentially extends to virtually every cellular process. Recently, it has been recognized that intracellular pathogens also recruit cytoskeletal proteins from the host cell for movements within and between cells. By recruiting the cellular machinery that speeds intracellular trafficking, pathogens can move about in the cytoplasm much more quickly than could be accomplished by diffusion alone. Movement into the cell from the peripheral site of infection to the nucleus or perinuclear region where most replication takes place is thus accomplished more quickly. Newly replicated pathogens traffic from the nuclear region to the cell surface more quickly when powered by cellular motors. Some pathogens also recruit the cell-surface actin-based machinery in the cortical cytoplasm to generate pedestals and filopodia-like structures that jettison the pathogen towards an adjacent cell which it then infects. In this review, we will examine recent discoveries concerning the interaction between intracellular pathogens and the cytoplasmic proteins of the host involved in cytoskeletal dynamics.
The active movement of cellular components is mediated by two complementary and independent mechanisms: 1) motor protein complexes that transport large aggregates along cytoskeletal “tracks,” and 2) polymerizers, that shove particles forward by intercalating protein monomers into filaments extending behind the particle. Many different intracellular pathogens have now been recognized to utilize one or both of these processes.
Most DNA viruses as well as many RNA viruses move to the nucleus where they can obtain the necessary cellular components for replication. Other viruses replicate exclusively in the perinuclear region. For the virus, movement to and from the nucleus and subsequent escape from the cell after replication require cytoskeletal elements. While replication of intracellular bacteria is not known to be dependent on cellular nuclear factors, intracellular bacteria do need to defend themselves against cytoplasmic host cell factors and to escape the host cell after replication. Both defense and escape involve interaction between the pathogen and the host cell cytoskeleton.
Targets for therapeutic intervention include either the molecules of the pathogen or the host cell factors. In this review, we discuss the cytoskeletal elements upon which the life cycle of specific pathogens depends. These elements are potential targets for drug therapy of infectious disease. We will focus on herpes simplex virus for motor-driven transport and discuss other selected pathogens for actin polymerization-based motility.
THE CELL BIOLOGY OF TRANSPORT
Because the neuron produces a long axonal process, it provides a good model system in which intracellular transport can be observed and experimentally manipulated. Axoplasm is actively transported out neuronal processes. Axoplasm accumulates on the proximal side of a constriction placed around the peripheral nerve in the rat, as demonstrated in the landmark discoveries of Weiss and Hiscoe [1]. Axoplasmic material moves out from the cell body, termed “anterograde transport,” at two distinct rates, termed fast and slow transport, as demonstrated by radioactive labeling [2]. By morphological analysis, the components in the fast transport compartment are composed of the membranous cytoplasmic organelles such as mitochondria, endoplasmic reticulum, and secretory-like vesicles [3,4]. Transport back from synapse to cell body is termed “retrograde transport.”
In the neuron, both fast and slow anterograde transport appear to be necessary for proteins synthesized in the cell body to be delivered out neuronal processes to distant synapses. Retrograde transport carries growth factors, such as nerve growth factor (NGF) [5], and organelles, including lysosomes, back to the cell body [6]. Retrograde transport of NGF may not be required for it to affect neuronal survival [7]. Transport similar to that discovered in neurons occurs in most other cells as well. Thus, studies in the axon have identified molecules and paradigms applicable to most other cells and across species from yeast to human.
Slow transport is less well understood than fast axonal transport [8-10]. Proteins trafficked at slow transport speeds include actin and tubulin, monomers for replenishment of the tracks [11,12]. Prion disease may be transmitted throughout the CNS by slow transport mechanisms [13].
Both actin filaments and microtubules serve as tracks for transport, either individually or as a functional unit. Actin-based transport of organelles is responsible for cytoplasmic streaming and mitochondrial movements in algae and yeast [14-16]. In neurons, actin-based transport probably acts in synergy with microtubule transport, as either disruption of actin filaments by gelsolin, a filament severing protein, or depolymerization of microtubules stops organelle movements [17 ,18]. In axoplasmic squashes, organelles move on microtubules [3] and on invisible tracks, i.e. not microtubules [19]. Isolated, KI-stripped organelles move on purified actin filaments in vitro [20,21]. Mitochondria move in both directions on both actin and mictotubules in cultured hippocampal neurons [22].
Membrane-bound organelles move in both directions on microtubules in vitro [18,23]. Non-membranous particles also move on microtubules, and these include the intraflagellar transport patiicles (IFT) originally isolated from Chlamydomonas [24] and also found in sensory neurons and kidney tubular epithelium [25,26]. These particles may be analogous to the “packets” observed in other neurons [27], or to the particles containing mRNA encoding a small subset of proteins in axons [28]. Non-membranous particles may be more analogous to microbes than membrane bound organelles.
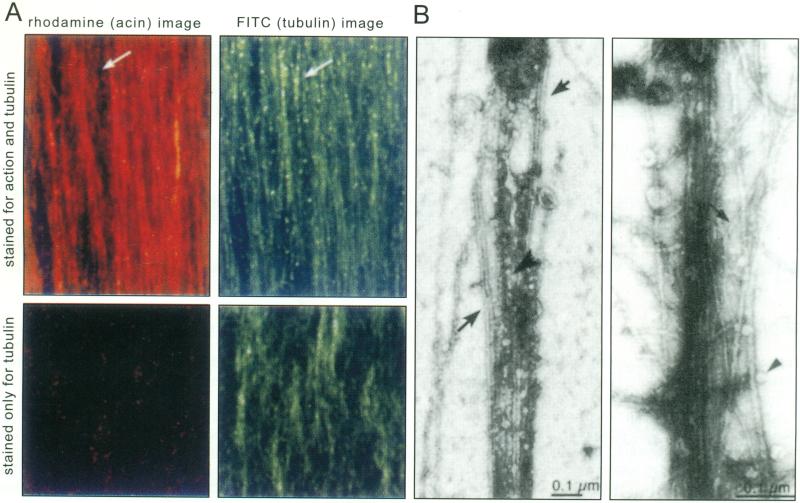
Actin and microtubules are colinear in the axon and therefore are likely to be structurally interdependent [29]. See Fig. (1). In the green alga, Bryopsis, depolymerization of one filament system causes depolymerization of the other and stops motility of intracellular particles [30].
Fig. (1). Actin filaments and microtubules in axoplasm.
A. Actin filaments (red) and microtubules (green) are co-linear in squid axoplasm. Top two panels: extruded axoplasm was fixed and double-labeled with rhodamine phalloidin (left, red) and anti-tubulin (right, green). Note that the two filament systems overlap except in rare areas, such as that indicated by the arrow. Axons stained only for tubulin (bottom panels) do not display any label in the rhodamine channel (bottom left).
B. By electron-microscopy of negatively stained whole mounts of extruded axoplasm, microtubules (large arrows, left panel) and actin filaments (small arrow, right panel) interweave. Membrane-bound vesicles are found attached to either filament type within the plume of interwoven filaments (arrowheads).
(Modified from Bearer and Reese, 1999, J. Neurocytology, by permission).
Many toxins produced by micro-organisms are picked up by these host cell transport pathways. Some specific examples include tetanus toxin, which is trafficked across the full length of the neuron from dendrites to axonal synapse [5]. Lethal factor of anthrax toxin apparently requires some aspect of transport for its toxicity, since inheritance of specific kinesin alleles correlate with resistance to anthrax in inbred mice [31].
BIOCHEMISTRY OF THE FILAMENTOUS PROTEINS THAT SERVE AS TRACKS FOR TRANSPORT
Both microtubules and actin filaments are composed of multiple protein subunits that come together to form filaments in the presence of NTP and salt. These filaments have a fast-growing, “plus” end and a slow growing, “minus” end. Thus, each filament is polarized. Polarity of the filament also influences its properties as a track for transport, since each motor can only go in one direction on any given filament. This holds true for all motors yet identified that move on either microtubule or actin filaments.
Inside cells, the microtubules and actin are in a dynamic state of polymerization and depolymerization. This dynamic activity has been beautifully documented using fluorescently labeled actin or tubulin. Transfection of a plasmid encoding green-fluorescent protein (GFP) fused to either actin [32,33] or tubulin [34] allows the dynamics of each filament system to be observed by video microscopy over time and in response to various stimulants and/or toxins.
Each filament system has a nucleator that stimulates localized formation and growth of polymers. Actin filaments are nucleated by the Arp2/3 protein complex in platelets [35], and probably in most other cells. Formins also nucleate actin (see note). The Arp2 subunit of the seven-protein Arp2/3 complex in platelets was first identified as an actin-binding protein by filamentous actin affinity chromatography [36]. The Arp2/3 complex was identified by its profilin-binding from Acanthamoeba castellani [37] and was found to be the host factor recruited by Listeria monocytogenes to mediate its actin-based motility [38]. The crystal structure of various subunits of the Arp2/3 complex is underway [39,40]. The crystal structure of bovine Arp2/3 complex, an assembly of seven proteins, has been determined at 2.0 angstrom resolution [39] and other ultrastructural approaches are being applied to determine targets on the complex for small molecule interventions.
Microtubules are polymerized at the microtubule organizing center near the nucleus by gamma tubulin resulting in microtubules oriented with their plus-ends at the cell periphery [41]. Microtubules are also formed from basal bodies at the cortex which produces flagella and cilia. Finally, microtubules are nucleated off the cortical membrane where the initiating factor(s) has not yet been identified. In this case, microtubules have their plus ends pointed towards the nucleus.
Location of the nucleation machinery determines the relationship of the orientation of filaments to the architecture of the cell. Both actin filaments and microtubules in cultured non-neuronal cells are usually plus end out. In axons of adult motor neurons and in sensory processes from neurons in the trigeminal ganglion, 95% of the microtubules are oriented with the plus end directed towards the synapse [42-45]. In contrast, microtubules in dendrites go in both directions [46]. The pattern of plus-end-out in long processes and bi-directional orientations in shorter, dendritic-like, processes holds true, at least for those few cultured neurons that have been studied: neurons from the hippocampus and from sympathetic ganglia [44,47-55].
ATOMIC STRUCTURE OF CYTOSKELETAL PROTEINS CAN BE USED TO IDENTIFY DRUG TARGET SITES
The crystal structures of the monomeric protein subunits of microtubules [56] and actin [57,58] have been solved, and are thus available for use to predict drug target sites. In addition, by comparisons of the atomic structure of the monomers with electron density mapping of filaments, the atomic structure of the filaments has been determined [59-63]. Recently, drug-induced changes in both filament systems have been studied in living cells using atomic force microscopy [64].
Actin is a highly conserved protein, with amino acid identities of 80-90% across species [65]. Thus, point mutations and amino acid substitutions in the actin sequence can be studied in the simple yeast by mutating the single actin gene, ACT1 [66]. Results from such yeast studies would be expected to be useful for predictions of drug targets across species, including human. A systematic dissection of the actin protein by such mutational screening in yeast has identified amino acid residues uniquely involved in specific actin-based activities that are not required for other tasks that actin performs within the cell [67]. These findings suggest that despite the fact that actin is ubiquitously expressed, drugs that alter specific aspects of actin dynamics can be found. This may also hold true for microtubules although similar studies for that filament system have not yet been done.
THE MOTORS
Kinesin, the first microtubule-based motor, is represented by 45 different isoforms in the human genome [68], and 21 in the worm, C. elegans [69]. The only known anterograde microtubule-based motors are a subset of the kinesins. Although the first kinesin to be identified was an anterograde motor, other family members were subsequently found that go in the retrograde direction on microtubules [70-72]. The intact conventional kinesin motor is a heterotetramer of two light chains and two heavy chains.
The crystal structure ofthe motor domain of kinesin has been solved [73-76]. In addition, the atomic structures of other members of the kinesin family have also been reported [77-79].
Transport along microtubules in the minus end direction (retrograde transport in the axon) could be mediated either by one of the retrograde kinesins or by dynein [72,80,81]. The atomic structure of dynein has not yet been reported, possibly due to its large size. Dynein is a very large multimer, with at least eight smaller subunits that are heterogenously expressed [82]. Its ability to bind to organelle cargo is mediated through an adaptor, dynactin, which is also a large multimer with many subunits [83].
Only one family of motor proteins, the myosins, has been identified as mediating transport along actin filaments. The crystal structure of the motor domain of the prototype of this family, myosin II or muscle myosin [84], has been compared to that of kinesin [74]. Also reminiscent of kinesins, myosins are heterodimers, some of which dimerize to form tetramers of two heavy chains and two light chains. There are at least 18 different classes of heavy chains, based on sequence comparison, and these classes are subdivided further into isoforms [85]. Function has not been assigned for many myosins and only the sequence is known. Most myosins move towards fast-growing (plus) ends of actin filaments[86]. Myosin VI, a non-muscle myosin, moves in the minus-end direction on actin [87] and is implicated in movements of membranous organelles in the cytoplasm [88]. Half of the axoplasmic nomnuscle myosin II is tightly associated with axoplasmic organelles [89], suggesting that this myosin plays a role in transport (see more below). Myosin V is also implicated in secretory vesicle transport in yeast, mouse, and squid [90].
THE UNANSWERED QUESTIONS CONCERNING THE CELL BIOLOGY OF TRANSPORT
There are key unanswered questions about the basic biology of the transport system in normal processes within cells. The most important of these is how motor and cargo recognize each other such that trafficking is directed appropriately. Recent evidence implicates Rab27 in recruitment of myosin V [91]. Kinectin, amyloid precursor protein, and drosophila Sunday Driver has also been suggested as a kinesin receptor on the endoplasmic reticulum [92-94]. Phospholipids may directly bind motors, although whether this interaction occurs physiologically inside cells is questionable [95]. Since the current understanding of the mechanism of motor recruitment is still rudimentary, it is not possible to use existing information to ferret out how pathogens interact with cellular motors. In fact, intracellular pathogens that move on filamentous tracks may prove good tools for the identification of normal cellular mechanisms of motor recruitment.
MODEL SYSTEMS
Herpesviruses, in particular the herpes simplex viruses (HSV) and pseudorabies virus, have long been known to traffic along nerves. In humans, HSV infects the epithelial cells of the mucous membrane and secondarily enters the nerve terminals. Once inside the nerve, the virus trafficks back to the nerve cell body in the trigeminal ganglion. In the neuronal nucleus, the virus can enter latency or replicate. Herpes simplex virus can also spread throughout the brain, possibly by transport in dendrites, which results in life-threatening encephalitis. Herpes simplex virus type 2, also known as genital herpes, can cross the placenta to infect the fetus or infect the infant during delivery. Birth defects and fatal encephalitis are frequent consequences.
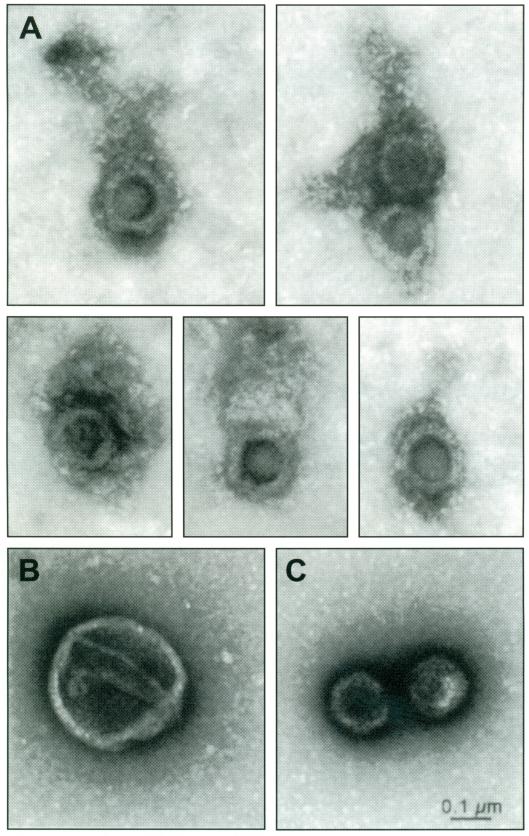
Herpesviruses are enveloped DNA viruses with four concentric compartments: an inner DNA-protein core, packaged in an icosahedral capsid, surrounded by an amorphous tegument and encased in a glycoprotein and lipid-containing membrane with a diameter of ~230 nm [96,97]. See Fig. (2). The human herpes simplex virus type 1 (HSV-1) is one of the largest viruses known, with a 150 kB linear double-stranded DNA genome encoding 89 genes. In addition to the structural proteins, HSV particles also contain proteins with enzymatic activity and proteins that act as transcription factors to turn up or down host and viral gene expression. During replication additional viral genes are expressed. Some of these proteins are packaged into the mature virion and others may have transient effects on the cell during virus replication, packaging and transport. After replication, the new viruses travel in the opposite direction back out the nerve to the mucosa where they emerge to cause a recurrent ulcer.
Fig. (2). Herpes simplex virus type I (HSV-1).
A. Gallery of detergent-stripped HSV. Note the halo of mesh-like material representing the tegument and attached to the orthogonal capsid. Such HSV viral particles stripped of envelope are transported in the retrograde direction when injected into the giant axon of the squid.
B. Enveloped virus. Intact virus is larger than stripped particles (~230 nm in diameter) and displays the typical studded smooth membrane surface, which is lost upon stripping with detergent and salt. These intact virions do not move in the retrograde direction when injected into the axon.
C. Naked nucleocapsids. Virus lacking both envelop and tegument appear as sharply delineated orthogonal particles without a halo. (From Bearer et al. 2000, Proc. Nat. A cad. Sci., by permission)
Note: Viral particles are significantly larger than most axoplasmic organelles shown in Fig. (1).
The life cycle of herpesviruses involves rounds of directed transport. Viral particles would be expected to display net movement in one direction or the other depending on where they are in the life cycle. A major question concerns what mechanism controls directionality of viral movements such that incoming viruses attain the nucleus and outward bound viruses reach the synapse. These are crucial events in the viral life cycle, and must be carefully regulated. Drug intervention of transport in either direction would be effective in eliminating the cycles of infections.
Infection occurs at the synapse or close to it by a fusion between the viral envelope and the cell plasma membrane [98]. It is thought that most envelope components are left in the plane of the membrane, while the tegument and capsid are released into the cell cytoplasm. The movement of most of the tegument proteins is still poorly defined, although VP16 and VP22 have been studied in detail (see below).
After fusion of the viral envelope with the plasma membrane, HSV particles travel in the retrograde direction to the nucleus. After replication, emerging newly synthesized HSV travel in the anterograde direction. Thus, if the virus could be isolated at different times in its life cycle, the different mechanisms of transport might be discovered.
Recent advances in understanding transport of herpesviruses have come through three new approaches: 1) labeling of individual viral proteins with green fluorescent protein, 2) co-immunoprecipitation and re-association of viral proteins with known motors, and 3) reconstitution of viral transport in a powerful model, the giant axon of the squid.
RETROGRADE TRANSPORT OF HERPESVIRUSES
Fusion of the viral envelope with the plasma membrane liberates the nucleocapsid together with its halo of tegument proteins into the host cell cytoplasm [99]. Some of the tegument proteins may diffuse away from the nucleocapsid, while others remain closely associated.
Transport of herpesviruses is thought to be microtubule-based in both directions. Depolymerization of microtubules arrests viral movements both in cultured neurons [100] and in animal models [44,101]. However, actin depolymerization also affects viral transport [44]. Since we now know that the two filament systems are interdependent [29], it will be important to determine whether depolymerization of one affects the other. If this is the case, then the apparent effect of microtubule destabilizing agents on transport could actually be due to a concomitant disruption of actin filaments.
The most obvious candidate for a retrograde microtubule motor mediating transport of herpesviruses is dynein. Indeed, dynein has been found associated with intra-cytoplasmic viral particles by immuno-electronmicroscopy [99], and there are reports that over-expression of p50 glued subunit of the dynactin complex inhibits herpesvirus transport by disrupting the dynein-cargo association (Beate Sodeik, personal communication). However, some of the observed behaviors of viral transport are not consistent with a dynein motor, as we have described (see below). Retrograde kinesins also exist, and myosin-driven movements could supplement microtubule motor activity.
Direct imaging of retrograde transport in a mature axon in which microtubules are known to be polarized with the plus-end towards the synapse demonstrates that viral particles are transported in the retrograde direction at 2.2 μm/sec [97]. See Fig. (3). This transport is relentless with few stops and rare reversals. Unlike the saltatory movements mediated by dynein, the movements of viral particles were continuous and smooth. These direct observations of viral movements are made possible by using the giant axon of the squid, Loligo peallii. This axon has served as a powerful model for the discovery of the action potential of neuron [102,103], and for the identification of the first microtubule-based motor, kinesin [104].
Fig. (3). GFP-VP16 labeled herpes simplex virus type 1 particles.
Detergent-stripped human herpes simplex viral particles move in the retrograde direction when injected into the giant axon of the squid [97]. Shown are still frames from a video of GFP-labeled viral particles moving in the squid axon taken at 5 second intervals. Two viral particles (arrows) move across the screen relative to fixed patterns (horizontal arrow) in the background. The last panel shows the orientation of the axon, but is not drawn to scale. The two particles move on slightly different tracks but at a very similar velocity, ~2.2 μm/sec. (Supplementary video material available at http://biomed.brown.edu/Faculty/B/Bearer.html).
For HSV transport studies, the giant axon offers the advantage of being large enough to inject—it is 0.6 mm in diameter and up to 7 cm in length. Injection bypasses infection and allows a biochemical dissection of viral components uniquely involved in transport. HSV-1 labeled with a truncated amino terminus of the tegument protein, VP16, fused to GFP is bright enough to image single particles deep in the giant axon by confocal laser-scanning microscopy [97].
Herpes simplex virus type 1 is transported in the retrograde direction in the squid axon only after the envelope has been stripped with detergent prior to injection [97]. This stripping process exposes tegument which apparently is required for motor recruitment, since neither enveloped virus nor naked nucleocapsids move in the retrograde direction. This provides evidence that the viral tegument recruits the motor.
The tegument compartment of HSV-1 contains at least 21 viral proteins (Table 1). Since the function and distribution of several viral proteins have yet to be assigned, this list may not yet be complete. The structural proteins of the tegument bind to each other, to the capsid and to the envelope, mediating a dense matrix of proteinaceous material between the envelope and the capsid in the intact virus [105]. After stripping with detergent, the remaining tegument structure appears as a fine mesh attached to the nucleocapsids [97], See Fig. (2).
Table 1. Tegument proteins of Herpes Simplex Virus.
Listed are 21 proteins that have been assigned to the tegument compartment of HSV-1. Those marked with an asterisk are known to be located in the tegument compartment, between the capsid and the envelope. We have added additional viral proteins to this list which have been reported in the tegument compartment but are not defined as tegument proteins. These other proteins may be packaged into the tegument compartment as innocent bystanders, or they may play roles as yet undefined. Functionally relevant tegument proteins perform four main tasks: (1) structural integrity of the virion particle, both when devoid of envelope during retrograde transport and after replication during packaging; (2) recruitment of cellular motors for transport to the nucleus; (3) activation of initiating events in viral replication or latency, such as viral transcription or host cell gene silencing; and (4) activation of linkages between capsid to envelope glycoproteins during post-replicative packaging.
| Gene/protein | Size | Possible functions other than transport | Potential involvement in transport |
|---|---|---|---|
| US3 | 68kD | Phosphorylation of UL34 [172] | |
| US9* | 10kD | Unknown [173,174] | |
| US10* | 36kDa | Unknown [175] | |
| US11* | 21kD | Nuclear export [176]; Envelopment of capsid in cytoplasm [174]. Transcription: associates with ribosomal 60s subunit and binds RNA[177] | Transport of tegument: binds directly to kinesin heavy chain and the tegument protein, vp16 [114] |
| UL4 | 25kD | Unknown [174] | |
| UL11(gM) | 13-16kD | Capsid envelopment and exocytosis [178,179] | |
| UL13 (PK) | 57kD | Phosphorylation of tegument: substrates include viral ICP0, ICP22, UL49, UL41, UL47, gE, and host cell EF-1δ [174,179] | |
| UL14* | 32kDa | Unknown [180] | |
| UL17* | 78kD | Cleavage and packaging of viral DNA [181] | |
| UL21 | 62kD | Promoting polymerization of microtubules [182] | |
| UL36*(VP1/2; ICP1/2) | 273kD | Envelopment of nucleocapsids in cytoplasm [183] | |
| UL37* | 120kD | Shuttling between nucleus and cytoplasm [184] (minor component of tegument) | Retrograde transport: localizes at nucleus [184] |
| UL41*(VHS) | 58kD | Shutting down of host protein synthesis by degrading mRNA[179] | |
| UL46* (VP11/12) | 93/87kD | Modulating activity of UL48 [174] | |
| UL47* (VP13/14) | 82/81kD | Modulating activity of UL49 [174] | Retrograde transport: trafficks to nucleus in vivo [108,185,186] |
| UL48* (VP 16; ICP25; αTIF) | 65kD | Regulation of alpha viral genes and host genes [179] | Retrograde transport: trafficks to nucleus in vivo [185]; imaged by GFP-label in retrograde transport [97] |
| UL49* (VP22) | 38kD | Enhancement of cell to cell spread of virus [124] | Anterograde and retrograde transport: trafficks to nucleus in vivo [185], interacts with non-muscle myosin II [123] stabilizes microtubules and translocates to neighboring cells [115] |
| UL51 | 27-30kDa | Unknown [187] | |
| UL56 | 30kD | Virulence reduction [188] | |
| ICP0 (α0) | 110kD | Expression of immediate early genes [189]; Reactivation from latent infections in sensory ganglia[190,191] | |
| ICP4 (α4) | 175kD | Regulation of gene expression during viral replication [192] |
The squid giant axon offers a model system for reconstituting viral motility with single viral proteins, since we can bypass the infective process and deliver motile particles directly into the axoplasm by injection. In addition, the mature nature of the axon itself ensures that the transport system—tracks and motors—are most similar to those of the mature neurons that are natural hosts for the virus.
HSV proteins that might recruit retrograde motors have been studied by reciprocal pull down experiments. This approach identified UL34 as binding to dynein [106]. The amino terminus of the intermediate chain, IC-1a, of the dynein complex pulls down three proteins from extracts of herpes infected cells: VP5, UL31 and UL34. The interaction appears to be with the UL 34 subunit, since GST-UL34 reciprocally pulls down IC-1a after in vitro transcription-translation. GST-UL34 pulls down UL31 and VP5 from infected cell lysates. By immunofluorescence, UL34 overexpressed in cultured cells co-localizes with microtubules. UL31 depends on UL34 for its stability [106].
Other HSV proteins implicated in motor recruitment are the tegument proteins VP22 and VP16. For each of these proteins, some molecules remain with the nucleocapsid after infection and some diffuse away from it to be independently delivered to the nucleus. VP22 localizes with microtubules and is transported inside cells in a microtubule-dependent manner (see below) [107]. VP16 is trafficked to the nucleus very early in infection [108] and therefore is likely to be delivered by active transport, possibly independent of other viral components.
There is some evidence to suggest that the presence in the axon of herpes structural proteins alters normal functioning [109]. Electrical potentials are not propagated properly. Viral competition for mitochondrial transport machinery would be expected to result in decreased motility of mitochondria. Energy production and increased local accumulation of toxic oxygen radicals, byproducts of mitochondrial metabolism, could result in neuronal death.
ANTEROGRADE TRANSPORT OF HERPES-VIRUSES
Newly replicated herpesviruses travel in the anterograde direction, out from the cell body towards the synapse. Rates of travel are similar to the velocities of axoplasmic organelles, with average velocities of ~2 μm/sec [110]. Microtubules are thought to be the tracks mediating this transport, although the interdependence of the microtubule and actin system suggests that both may serve as supports for transport [29].
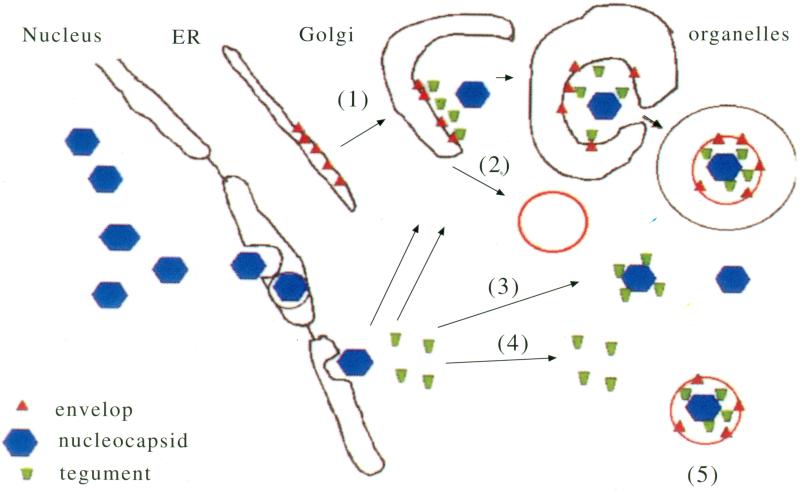
There is much doubt as to the composition of the viral particle that is transported, and at least three different hypotheses are currently in the literature [111-113]. See Fig. (4). One hypothesis proposes that each viral compartment is transported separately, with naked nucleocapsid, soluble tegument and viral glycoproteins of the envelope traveling independently to the synapse where the complete viral particle is assembled during budding. Another proposes that the virus travels intact, with the surface of the envelope exposed to the cytoplasm. How this would bud from the cell surface is a mystery. The third and most attractive hypothesis is that the virus buds into the late Golgi or a secretory vesicle in the perinuclear region and then travels inside this second membrane out the sensory process to the synapse [112]. In this third hypothesis, the virus would be secreted rather than budding from the cell surface. Electron-microscopic evidence exists for each of these proposals, and it remains possible that the virus uses all of these methods to arrive at the synapse.
Fig. (4). Hypothetical correlations between packaging and transport of herpes simplex virus.
Viral capsids (blue) are assembled in the nucleus and attain the cytoplasm by passing through the nuclear envelope. Tegument (green) is synthesized in the perinuclear region of the cytoplasm by normal cellular protein synthesis machinery. Viral glycoproteins (red) are synthesized in the rough endoplasmic reticulum (ER). Full assembly could occur as the glycoproteins pass through the Golgi network (1). In this case, the cytoplasmic tails of the viral envelop proteins are exposed to the cytoplasm, where they pick up newly synthesized tegument, which in turn associates with the capsid. Binding to the capsid pulls the Golgi membrane around the virus, enveloping it in a double membrane with the inner layer containing viral glycoproteins and the outer being derived from the Golgi. Alternatively, vesicles containing viral envelope components could bud from the Golgi independent of other viral structures (2). In this case, capsid with or without tegument (3), or tegument alone (4) would be trafficked outwards separately. Finally, intact enveloped virus without additional cellular membrane could be trafficked independent of cellular membrane components, although how this would be packaged is less easily predicted.
Given that the nature of the viral-cytoplasmic interface remains unclear, it has been difficult to identify viral proteins involved in anterograde motor recruitment. A recent study analyzing the interaction between kinesin and HSV-1 has demonstrated that the viral protein, US 11, interacts with the tail of the heavy chain of ubiquitous kinesin [114]. The carboxy-terminus of US11 overexpressed in bacteria was found to bind recombinant kinesin heavy chain. Two other viral proteins, VP16 and VP22, were also tested for kinesin binding and were only found to associate with kinesin in the presence of US11. Since US11 is a tegument protein, and not expected to be exposed on the surface of the viral envelope, these results provide strong support for the hypothesis that nucleocaspids are independently transported in the anterograde direction. Alternatively, this kinesin could be mediating delivery of the tegument to the relevant organelle for subsequent viral packaging during assembly in the perinuclear region.
VP22 is actively transpotied in both directions in neurons and cultured cells. VP22 transport can occur independently of other viral proteins. Whether VP22 “carries” the tegument-capsid with it in the infected cell is not known. VP22 has been implicated in other aspects of transport, including binding to microtubules and alteration of microtubule distribution [115], translocation to the nucleus during mitosis [116], and transference from an infected or over-expressing cell to a neighboring cell [117,118]. The domains in VP22 responsible for nuclear targeting and intercellular spread have been mapped as follows: intercellular transport- amino acids 81-195; binding and reorganization of cytoskeleton- amino acids 159-267; nuclear targeting, stabilization of microtubules- amino acids 81-121; and nuclear targeting and facilitation of intercellular transfer- amino acids 267-301 [118,119]. The chromosome binding and microtubule binding domains overlap. The VP22 nuclear targeting moiety is now available commercially in a plasmid for use as a fusion construct for the targeting of the fusion proteins to the nucleus (Invitrogen). The molecular mechanism by which VP22 mediates this targeting, which is likely to be involved in retrograde transport, is as yet unknown.
VP22 is known to be phosphorylated [120]. Phosphorylation correlates with viral packaging [121]. Substitution mutations of tyrosine 38 to phenylalanine abolish assembly-associated phosphorylation and decrease the amount of VP22 found in newly synthesized viral particles [121].
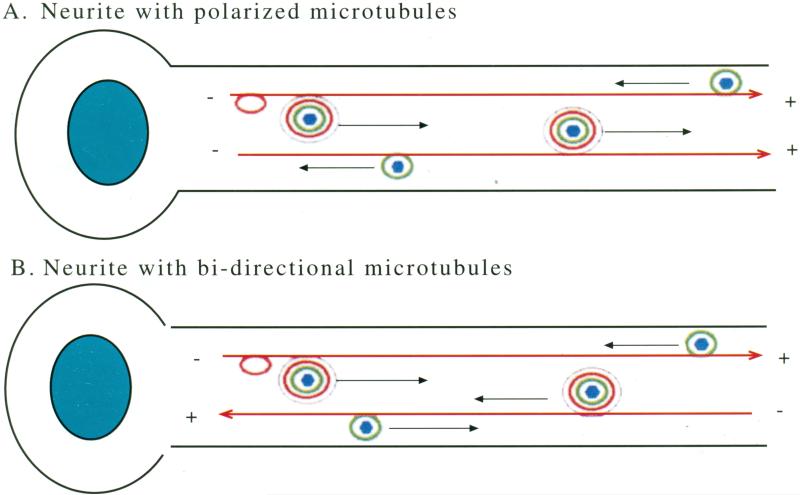
Analysis of anterograde transport of various viral proteins by immunofluorescence also supports separate transport pathways of the different viral compatiments. VP16 (tegument), gB or gC (envelope) and VP5 (capsid) travel at different rates from the cell body out the processes of cultured human fetal neurons [100]. Brefeldin A, an inhibitor of endoplasmic reticulum to Golgi processing, slowed envelope trafficking but not that of the other components. In pseudorabies virus, mis-sense mutations of US9, a viral membrane protein, but not of the glycoprotein, gE, results in the presence of capsid and tegument trafficking to the neurite in the absence of the glycoproteins gB, gC or gE [122]. Thus, US9 appears to be required for the anterograde trafficking of envelope glycoproteins but not of capsid and tegument. In these cultured neurons, it is possible that the microtubule system is not as polarized as it is in mature axons. Thus, the outbound movements detected in these cells could consist of both anterograde and retrograde motility on a bipolar microtubule system. See Fig. (5).
Fig. (5). Viral particles move in both directions on microtubules.
A. In the long neurites of polarized neurons, microtubules display uniform orientation with plus ends towards the synapse. In this case, after initial infection at the synapse, non-enveloped HSV particles move smoothly inwards towatds the cell body at the minus end of the microtubule tracks. Viral tegument (green) is likely to recruit retrograde motors that power this transport. After replication, newly synthesized virus (bue/green/red/black) would be expected to have net movement in the other direction, outwards on these polarized tracks. Viral glycoproteins incorporated into transport vesicle membranes (red) would also traffick outwards. Which component recruits the anterograde motors is not known.
B. In neurites with bi-directional microtubules, individual viral particles can move in either direction relative to the cell body because the direction of movement for any particular motor is dependent on the orientation of the microtubule track. Thus, no matter which motor a viral particle attracts, it would be capable of moving in either direction within the neurite. Such bi-directional capability could be useful for viral spread throughout the CNS.
The packaging of virus that occurs in the perinuclear region may also involve myosin II, an actin-based motor. Myosins may also be involved in transport along the axon. We have recently shown that nonmuscle mysosin II is strongly associated with axoplasmic organelles [89]. A large proportion of this neuronal myosin (47% of that found in the neuronal process) is associated with membrane bound organelles. Myosin II co-purifies with VP22, one of the viral tegument proteins, from infected BHK cells [123], and VP22 affinity columns yield four proteins, including myosin II. Confirmation that this biochemical affinity has a physiological purpose is the fact that myosin II re-localizes from actin bundles to perinuclear granules in cells infected with virus. Inhibition of myosin II activity with butanedione monoxime results in increased numbers of viral particles in the perinuclear region and decreased viral particle release. These data suggest that actin-based motility is necessary for processing of viral particles into a form that can be transported out the neuronal process and may also be involved in axonal transport in both directions.
Despite these many activities of VP22, there is some evidence to suggest that VP22 is not required for viral synthesis and packaging. HSV packages normally in the absence of VP22 [124]. Furthermore, deletion of VP22 in pseudorabies does not effect virulence or neuronal spread in rodents [125].
Since transport appears to be dependent on packaging, it would be useful to know more about packaging. For example, what is the relative abundance of each viral protein in the perinuclear region and where do associations between viral proteins occur? If all viral components are equally abundant in the perinuclear region of infected cells, it is reasonable to propose that those that bind would assemble there. This is because biochemical interactions are dependent on relative concentrations as well as on binding affinities. During assembly, interactions between tegument and capsid proteins and the cytoplasmic domains of the envelope glycoproteins may reveal which components recruit anterograde motors. For example, UL20 interacts with gK [126], UL13 interacts with the gE-gi duo [127]; and UL49 (VP22) interacts with gM [128]. UL13 may serve as a linker between gE/gi and UL41 and UL49 (VP22). How the other glycoproteins attach to tegument is uncertain, although UL48 is thought to mediate associations with gB, gH/gL and gD. Other tegument proteins bind both tegument and capsid. Binding between the tegument proteins UL20, UL13, UL49 and UL41 is likely to mediate a bridge between the capsid and the envelope [112].
Outward bound virus may be capable of retrograde movements. Recent observations in cultured chick dorsal root neurons using GFP-labeled pseudodrabies virus show that newly synthesized virus traveling in the anterograde direction also reverse direction briefly to move in the retrograde direction [110]. The velocity of these reversals is slightly slower (1.9 μm/sec) than the retrograde transport of envelope-stripped human HSV-1 in the mature squid axon (2.2 μm.sec) [97]. There are several intersting explanations for these reversals of direction during outward transport, including:
The microtubule tracks may not be uniformly polarized with the plus ends outwards towards the synapse. See Fig. (5). In this case, the virus-motor interaction would be stable, and reversals would simply be a matter of brief track switching. Indeed, the polarity of microtubules in cultured sensory neurons from the dorsal root ganglion is assumed to be plus end out, but we were unable to find experimental documentation for this assumption. Furthermore, even in mature axons where the majority of microtubules are plus-end distal, 5-7% have reverse polarity [40-45].
Individual viral particles either are capable of exchanging an anterograde motor for a retrograde motor, or recruit both types of motors during transit outwards. This interpretation goes counter to the simple hypothesis that in-coming virus presents a different motor binding site to the cytoplasm than outward bound virus. Control of directional transport could be more complex than the single motor recruitment hypothesis predicts.
Individual viral particles are switching cytoskeletal tracks. This is likely because microtubule and microfilament systems are closely associated in the axon [29]. In squid axoplasm, single organelles are observed to move along both actin and microtubule tracks [19-21], and in cultured chick neurons mitochondria can move on either MTs or microfilaments [22]. Like endogenous organelles, the virus may be capable of recruiting both actin and microtubule-based motors. The general observation in neurons that long-range transport occurs on MTs while short-range transport is carried out on actin filaments may also be applicable to these viruses. Here the virus may be traveling in the anterograde direction on microtubules and occasionally shuttling onto the closely associated actin microfilaments. Depending on the myosin and the polarity of the microfilament, the virus may reverse direction during its brief detour on the actin filament. This also explains the difference in velocity observed between the long range movement and the brief reversals.
Other host factors in addition to motors may be involved in anterograde transport of the virus. Since perinuclear host factors would not be expected at the distal viral entry point, co-option of such factors during viral packaging could provide the means for selective recruitment of anterograde motors by newly emerging virus for its outbound journey.
Since viral glycoproteins are apparently synthesized and processed along with cellular glycoproteins in the Golgi network, other host proteins and membrane lipids may be selectively or randomly packaged into the viral envelope. Indeed, selective recruitment of host factors mediating anterograde transport of nascent intact virus may be a mechanism for viral transport, particularly if the viral envelope is exposed to the cytoplasm. Alternatively, virus enclosed within a secretory vesicle may need to “zipper” itself into the vesicle through interactions between the outer surface of the viral envelope and the lumenal surface of the vesicle. See Fig (4). How host membrane components might be selectively recruited into the virus is not known, although detection of host membrane constituents in viral particles has been useful in determining the location of viral envelope synthesis [129].
ADENOVIRUS AND VACCINIA VIRUS AND MICROTUBULE-BASED TRANSPORT
Adenovirus appears to use motor-powered transport mechanisms to move about within cells. The virus was long ago recognized as binding to microtubules [130]. Infection with adenovirus results in a reorganization of the cytoskeleton, both actin and microtubules [131]. Adenovirus moves in both directions on microtubules inside cells [132]. Dynein-dependent movements occur upon lysis from the endosomal compartment [133]. Activation by the virus of cellular MAPK phosphorylations apparently boost the cytoplasm-to-nuclear transport. However, access to the perinuclear region for replication in cultured cells does not appear to require microtubules [134]. When it does not need to travel long distances to arrive at the nucleus for replication, microtubule-based transport may be dispensable.
Vaccinia virus also motors to and from the perinuclear region on microtubules in a process thought to be powered by dynein and kinesin [135-137]. Movement from the perinuclear region to the cortex is saltatory and occurs at an average velocity of>2 μm/sec [137]. The VV A27L gene product is required for transport of intracellular mature virions, which are not enveloped [138]. The A36R viral protein is required for microtubule-based transport of intracellular enveloped virions (IEVs) out to the cell periphery [135]. Mutations in tyrosine 112 and 132, which are thought to be phosphorylated, impairs formation of actin tails but does not inhibit microtubule movements [137]. Immunofluorescence with anti-kinesin light chain antibodies reveals co-localization of conventional kinesin with IEV during transport [135]. Like adenovirus, infection with vaccinia virus also results in disruption of the host cell microtubule network [139].
Use of cellular transport may be a widespread phenomenon among intracellular microbes, especially those that infect the brain. For example, histochemical data of West Nile virus encephalitis shows viral particles within neuronal processes at great distances from the replication site in the cell body [140].
ACTIN-MEDIATED PROPULSION OF INTRACELLULAR PATHOGENS
Actin polymerization also provides the force to propel microorganisms through cellular cytoplasm. This is an exciting new area of research and many reviews have appeared on this topic recently [141,142]. We will just touch upon a few of the critical points here. Seminal observations on the intracellular motility of the gram negative bacterium, Listeria monocytogenes [143] demonstrated that actin filaments were present in the phase- and electron-dense material found at the rear of motile intracellular bacteria. The bacterial ActA protein is required for this intracellular movement [144]. It was subsequently found that ActA, a protein expressed on the surface of the bacterium, recruits the cellular Arp2/3 complex as well as VASP and possibly other poly-proline binding proteins [145]. This complex of proteins is joined by cofilin, coronin, and capping protein [146], as well as other as yet undefined cellular factors which act as a machine that polymerizes and depolymerizes actin filaments. Addition of actin monomers at their barbed ends which abuts the bacterial surface pushes the bacterium forward [147,148].
Many reviews have been written on the interactions between the ActA protein of Listeria and the actin polymerization machinery of the host cell. It now appears that other intracellular bacteria, including Shigella flexneri, Samonella typhimuriam, and Rickettsia co-opt the same machinery, albeit by grabbing on to different subunits of the Arp2/3 polymerization machine [149]. Each microorganism has a unique surface protein that recruits this cellular machinery. Key to all actin-based motility is activation of the Arp2/3 complex. Indeed, the discovery of Arp2 from platelets [36] and the isolation of the Arp2/3 complex as the cellular factor required for Listeria-based nucleation of actin filaments [38], represents a major breakthrough in identifying the key regulator of actin polymerization not only for pathogens but also for normal cellular processes. The first evidence that Arp2/3 complex is critical for cellular behavior is the finding that Arp2/3 is required for human platelet shape change, an actin-dependent process which is the initial step in platelet thrombus formation [35]. All of the proteins co-opted by pathogens to stimulate actin polymerization are present in human platelets and play key roles in platelet physiology [150].
WASp/Scar family of proteins is critical to Arp2/3 activation [151]. WASp, Wiskott-Aldrich syndrome protein, itself may be activated by cdc42, a member of the rho family of GTPases [152]. While the ActA protein of Listeria apparently mimics the effect of N-WASp (neural WASp) on Arp2/3, the Shigella IcsA surface protein recruits and activates N-WASp which secondarily activates Arp2/3 [153]. Arp2/3 is likely to be activated by other signals as well, since human platelets that require Arp2/3 activity for actin polymerization in response to thrombin have very little N-WASp [35]. WASp is abundant in platelets but this is not sufficient to stimulate actin polymerization by Shigella in platelet extracts [153].
Other proteins in addition to Arp2/3 that regulate actin tail formation include vasodilator-stimulated phosphoprotein (VASP). VASP was initially isolated from platelets as the major substrate of GMP/AMP kinases [154]. It binds the poly-proline repeats in the Listeria ActA protein [155-158]. There are four poly-proline repeats in the central domain of ActA. Decreasing the number of these domains decreases the rate of bacterial motility and produces other abnormalities in the actin tails, including discontinuous tails, longer than normal pseudopodial projections, and formation of the tail from the side of the oblong bacterium instead at the end [159]. Deletion ofthe Arp2/3-binding domain of ActA does not remove all actin-dependent movements, while deletion of both poly-proline repeats and Arp2/3 binding domain does. Deletion of the Arp2/3 binding domain also results in a decrease in actin polymerization as shown by the pyrene assay of actin polymerization. This effect can be eliminated by addition of excess purified VASP. These studies indicate that VASP contributes to the motility of intracellular bacteria by binding to the poly-proline repeats and possibly recruiting Arp2/3.
Other VASP family members include MENA and EVL. In the presence of VASP, the rate of fibroblast migration is decreased while the rate of Listeria intracellular motility is increased [160]. Therefore, VASP family members negatively regulate fibroblast migration which contrasts with their positive role in Listeria motility. We have proposed that this effect is due to the proven ability of VASP to bundle and stabilize actin filaments which prevents them from being severed by gelsolin [161]. By blocking the severing of actin, excess VASP creates abnormally stable filaments that cannot be remodeled for some motile events but can serve as stable supports for other types of movement.
Ena/VASP proteins have three domains, the atomic structure of one of these, the Ena/VASP homology 1 (EVH1) domains has been determined [162,163] The other two domains, a poly-proline rich mid-section and the carboxy terminus, have distinct functions. While the poly-proline domain in VASP recruits VASP to the ActA poly-proline repeats, other domains in VASP may be responsible for its effect on actin polymerization at the cell cortex [160]. These other domains could be involved in recruitment of Arp2/3. The actin-binding domain of VASP is located in the carboxy-terminus [164], and VASP apparently forms tetramers which explains its bundling propetiies. In vitro, purified VASP stimulates actin polymerization on its own [161]. Thus, at least for Listeria, two different and complementary pathways exist to promote intracellular motility: one pathway would involve direct recruitment Arp2/3 and the other the recruitment of VASP which might secondarily activate Arp2/3.
Virus also appear to recruit the Arp2/3 complex to nucleate actin filaments at the cell surface. Vaccinia virus was the first virus observed to use this type of motility [138,165], but other members of this family of viruses, including the smallpox virus, may share this behavior. Vaccinia virus apparently recruits the Arp2/3 complex via an upstream tyrosine phosphorylation event which occurs at the membrane[166]. Tyrosine phosphorylation of the vaccinia A36R protein, possibly via host cell src-like kinases, recruits the adaptors Nck, WASP-interacting protein (WIP) and N-WASP which activate the Arp2/3 complex [166,167].
Originally thought to move through the host cell cytoplasm by forming actin tails, it is now known that vaccinia only induces actin tails after exocytosis from the cell [135,168]. Remaining associated with the cell surface, the virus induces actin-based filopodial projections in the cytoplasm on the inner side of the membrane behind it. The filopodia propel viral particles into the extracellular space, towards other cells, thereby propagating the infection. This process improves cell to cell spread, as demonstrated by the reduced plaque size of vaccinia mutants deficient in inducing actin tails [138,165,169-171].
CONCLUSIONS
Intracellular transport plays a significant role in microbial pathogenesis. Molecules from both the host and the pathogen present possible sites of pharmacologic intervention. In the host cell, the filamentous tracks, composed of actin filaments and microtubules, and the motors, including myosins, kinesins and dynein, may be drug targets. While these structural and motor proteins play major roles in cellular physiology, unique sites are likely to be found that interact with pathogen factors but are not required for cellular function. Specific molecules used by the pathogen to recruit the cellular motility machinery are just now being identified. These include the ActA protein of Listeria, and the IcsA protein from Shigella. For intracellular viruses such as HSV and vaccinia, specific molecules involved in the process of transport have yet to be found. The tegument proteins of herpesviruses are likely to be responsible for recruitment of retrograde motors.
Understanding the mechanisms of intracellular transport utilized by pathogens will be useful far beyond the identification of targets for antimicrobial therapies. The zip code that herpesviruses use to travel to the brain could be used to target drugs or genes to the central nervous system. Since crucial aspects of intracellular transport remain to be uncovered, using these pathogens as tools will elucidate important questions in cell biology.
ACKNOWLEDGEMENTS
Due to the breadth of the two fields, cytoskeleton and intracellular pathogens, covered in this review, it was not possible to cite every contribution. We apologize to those whose work was not cited this time around. We appreciate discussion about Herpes virus with Xandra Breakfield and Jennifer LaVail, and the support of our long-time collaborator, Thomas S Reese. This work was funded in part by Public Health Service Award NIGMS R01 47368 from the National Institutes of Health (E.L.B.) and the Molecular and Cellular Biology and Biochemistry Graduate Program at Brown University (P.S.).
ABBREVIATIONS (IN ORDER OF OCCURRENCE)
- HSV
herpes simplex virus
- NGF
nerve growth factor
- CNS
central nervous system
- KI
potassium iodide
- RNA
ribonucleic acid
- mRNA
messenger ribonucleic acid
- IFT
intraflagellar transport particles
- NTP
nucleotide triphosphate
- GFP
green fluorescent protein
- DNA
deoxyribonucleic acid
- HSV-1
herpes simplex virus, type I
- GST
glutathione-S-transferase
- BHK cells
baby hamster kidney cells
- VV
vaccinia virus
- IEV
intracellular enveloped virions
- VASP
vasodilator-stimulated phosphoprotein
- WASp
Wiskott-Aldrich syndrome protein
- N-WASp
Neural Wiskott-Aldrich syndrome protein
- AMP
adenine monophosphate
- GMP
guanine monophosphate
- MENA
mouse homolog of drosophila enabled
- EVL
enabled- and VASP-like protein
- EVH1
Ena/VASP homology domain 1
- WIP
WASp-interacting protein
Biography

Footnotes
During preparation of this manuscript, another actin nucleator in addition to Arp 2/3 has been identified: fonnin. Formins have conserved roles in cell polarity, cytokinesis, and formation of actin- and microtubule-based cytoskeletal structures [193, 194, 195]. Formins are required for limb development in mouse and drosophila [195, 196]. In vitro, yeast formin polymerizes microfilaments independently of Arp2/3 [197, 198]. Interactions between fonnin and intracellular pathogens have yet to be identified.
REFERENCES
- 1.Weiss P, Hiscoe HP. J. Exp. Zool. 1948;107:315–395. doi: 10.1002/jez.1401070302. [DOI] [PubMed] [Google Scholar]
- 2.Tytell M, Black MM, Gamer JA, Lasek RJ. Science. 1981;214:179–181. doi: 10.1126/science.6169148. [DOI] [PubMed] [Google Scholar]
- 3.Schnapp BJ, Reese TS. J. Cell. Biol. 1982;94:667–669. doi: 10.1083/jcb.94.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papasozomenos SC, Autilio-Gambetti L, Gambetti P. Brain Res. 1983;278:232–235. doi: 10.1016/0006-8993(83)90243-3. [DOI] [PubMed] [Google Scholar]
- 5.Lalli G, Schiavo G. J. Cell. Biol. 2002;156:233–239. doi: 10.1083/jcb.200106142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parton RG, Simons K, Dotti CG. J. Cell. Biol. 1992;119:123–137. doi: 10.1083/jcb.119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacInnis BL, Campenot RB. Science. 2002;295:1536–1539. doi: 10.1126/science.1064913. [DOI] [PubMed] [Google Scholar]
- 8.Shah JV, Cleveland DW. Curr. Opin. Cell Biol. 2002;14:58–62. doi: 10.1016/s0955-0674(01)00294-0. [DOI] [PubMed] [Google Scholar]
- 9.Pfister KK. Mol. Neurobiol. 1999;20:81–91. doi: 10.1007/BF02742435. [DOI] [PubMed] [Google Scholar]
- 10.Brown A. Nat. Rev. Mol. Cell Biol. 2000;1:153–156. doi: 10.1038/35040102. [DOI] [PubMed] [Google Scholar]
- 11.Nixon RA. Curr. Opin. Cell Biol. 1998;10:87–92. doi: 10.1016/s0955-0674(98)80090-2. [DOI] [PubMed] [Google Scholar]
- 12.Galbraith JA, Gallant PE. J. Neurocytol. 2000;29:889–911. doi: 10.1023/a:1010903710160. [DOI] [PubMed] [Google Scholar]
- 13.Scott JR. Br. Med Bull. 1993;49:778–791. doi: 10.1093/oxfordjournals.bmb.a072646. [DOI] [PubMed] [Google Scholar]
- 14.Boldogh IR, Yang HC, Pon LA. Traffic. 2001;2:368–374. doi: 10.1034/j.1600-0854.2001.002006368.x. [DOI] [PubMed] [Google Scholar]
- 15.Sheetz MP, Block SM, Spudich JA. Methods Enzymol. 1986;134:531–544. doi: 10.1016/0076-6879(86)34118-1. [DOI] [PubMed] [Google Scholar]
- 16.Sheetz MP, Spudich JA. Nature. 1983;303:31–35. doi: 10.1038/303031a0. [DOI] [PubMed] [Google Scholar]
- 17.Brady ST. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 18.Schnapp BJ, Vale RD, Sheetz MP, Reese TS. Cell. 1985;40:455–462. doi: 10.1016/0092-8674(85)90160-6. [DOI] [PubMed] [Google Scholar]
- 19.Kuznetsov SA, Langford GM, Weiss DG. Nature. 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- 20.Bearer EL, DeGiorgis JA, Bodner RA, Kao AW, Reese TS. Proc. Natl. Acad. Sci. USA. 1993;90:11252–11256. doi: 10.1073/pnas.90.23.11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bearer EL, DeGiorgis JA, Medeiros NA, Reese TS. Cell Motil. Cytoskeleton. 1996;33:106–114. doi: 10.1002/cm.970330202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris RL, Hollenbeck PJ. J. Cell. Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheetz MP, Vale R, Schnapp B, Schroer T, Reese T. Ann. NY Acad. Sci. 1987;493:409–416. doi: 10.1111/j.1749-6632.1987.tb27227.x. [DOI] [PubMed] [Google Scholar]
- 24.Kozminski KG, Forscher P, Rosenbaum JL. Cell Motil. Cytoskeleton. 1998;39:347–348. [PubMed] [Google Scholar]
- 25.Signor D, Wedaman KP, Orozco JT, Dwyer ND, Bargmann CI, Rose LS, Scholey JM. J. Cell. Biol. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. J. Cell. Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallee RB, Bloom GS. Annu. Rev. Neurosci. 1991;14:59–92. doi: 10.1146/annurev.ne.14.030191.000423. [DOI] [PubMed] [Google Scholar]
- 28.Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. J. Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bearer EL, Reese TS. J. Neurocytol. 1999;28:85–98. doi: 10.1023/a:1007025421849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menzel D, Schliwa M. Eur. J. Cell. Biol. 1986;40:286–295. [PubMed] [Google Scholar]
- 31.Watters JW, Dewar K, Lehoczky J, Boyartchuk V, Dietrich WF. Curr. Biol. 2001;11:1503–1511. doi: 10.1016/s0960-9822(01)00476-6. [DOI] [PubMed] [Google Scholar]
- 32.Westphal M, Jungbluth A, Heidecker M, Muhlbauer B, Heizer C, Schwartz JM, Marriott G, Gerisch G. Curr. Biol. 1997;7:176–183. doi: 10.1016/s0960-9822(97)70088-5. [DOI] [PubMed] [Google Scholar]
- 33.Fischer M, Kaech S, Knutti D, Matus A. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- 34.Carminati JL, Stearns T. J. Cell. Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Kim ES, Bearer EL. Blood. 2002;99:4466–4474. doi: 10.1182/blood.v99.12.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bearer EL. Cell Motil. Cytoskeleton. 1995;30:50–66. doi: 10.1002/cm.970300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. J. Cell. Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch MD, Iwamatsu A, Mitchison TJ. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 39.Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 40.Volkmann N, Amann KJ, Stoilova-McPhie S, Egile C, Winter DC, Hazelwood L, Heuser JE, Li R, Pollard TD, Hanein D. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- 41.Oakley BR. Curr. Top. Dev. Biol. 2000;49:27–54. doi: 10.1016/s0070-2153(99)49003-9. [DOI] [PubMed] [Google Scholar]
- 42.Heidemann SR, Landers JM, Hamborg MA. J. Cell. Biol. 1981;91:661–665. doi: 10.1083/jcb.91.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baas PW, Ahmad FJ. J. Cell. Biol. 1993;120:1427–1437. doi: 10.1083/jcb.120.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Topp KS, Meade LB, La Vail JH. J. Neurosci. 1994;14:318–325. doi: 10.1523/JNEUROSCI.14-01-00318.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viancour TA, Forman DS. J. Neurocytol. 1987;16:69–75. doi: 10.1007/BF02456698. [DOI] [PubMed] [Google Scholar]
- 46.Mandell JW, Banker GA. Neurobiol. Aging. 1995;16:229–237. doi: 10.1016/0197-4580(94)00164-v. discussion 238. [DOI] [PubMed] [Google Scholar]
- 47.Caceres A, Banker GA, Binder L. J. Neurosci. 1986;6:714–722. doi: 10.1523/JNEUROSCI.06-03-00714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baas PW, Slaughter T, Brown A, Black MM. J. Neurosci. Res. 1991;30:134–153. doi: 10.1002/jnr.490300115. [DOI] [PubMed] [Google Scholar]
- 49.Ginzburg I. Trends Biochem. Sci. 1991;16:257–261. doi: 10.1016/0968-0004(91)90099-h. [DOI] [PubMed] [Google Scholar]
- 50.Dotti CG, Banker G. J. Cell Sci. Suppl. 1991;15:75–84. doi: 10.1242/jcs.1991.supplement_15.11. [DOI] [PubMed] [Google Scholar]
- 51.Baas PW, Black MM, Banker GA. J. Cell. Biol. 1989;109:3085–3094. doi: 10.1083/jcb.109.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baas PW, Deitch JS, Black MM, Banker GA. Proc. Natl. Acad Sci. USA. 1988;85:8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burton PR. Brain Res. 1988;473:107–115. doi: 10.1016/0006-8993(88)90321-6. [DOI] [PubMed] [Google Scholar]
- 54.Sharp DJ, Yu W, Baas PW. J. Cell. Biol. 1995;130:93–103. doi: 10.1083/jcb.130.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharp DJ, Yu W, Ferhat L, Kuriyama R, Rueger DC, Baas PW. J. Cell. Biol. 1997;138:833–843. doi: 10.1083/jcb.138.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nogales E, Wolf SG, Downing KH. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 57.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 58.Schroder RR, Manstein DJ, Jahn W, Holden H, Rayment I, Holmes KC, Spudich JA. Nature. 1993;364:171–174. doi: 10.1038/364171a0. [DOI] [PubMed] [Google Scholar]
- 59.Nogales E. Annu. Rev. Biophys. Biomol. Struct. 2001;30:397–420. doi: 10.1146/annurev.biophys.30.1.397. [DOI] [PubMed] [Google Scholar]
- 60.Milligan RA, Whittaker M, Safer D. Nature. 1990;348:217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- 61.Holmes KC, Popp D, Gebhard W, Kabsch W. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 62.Meurer-Grob P, Kasparian J, Wade RH. Biochemistry. 2001;40:8000–8008. doi: 10.1021/bi010343p. [DOI] [PubMed] [Google Scholar]
- 63.Orlova A, Galkin VE, VanLoock MS, Kim E, Shvetsov A, Reisler E, Egelman EH. J. Mol. Biol. 2001;312:95–106. doi: 10.1006/jmbi.2001.4945. [DOI] [PubMed] [Google Scholar]
- 64.Rotsch C, Radmacher M. Biophys. J. 2000;78:520–535. doi: 10.1016/S0006-3495(00)76614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hightower RC, Meagher RB. Genetics. 1986;114:315–332. doi: 10.1093/genetics/114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wertman KF, Drubin DG, Botstein D. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belmont LD, Drubin DG. J. Cell. Biol. 1998;142:1289–1299. doi: 10.1083/jcb.142.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miki H, Setou M, Kaneshiro K, Hirokawa N. Proc. Natl. Acad. Sci. USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siddiqui SS. Traffic. 2002;3:20–28. doi: 10.1034/j.1600-0854.2002.30104.x. [DOI] [PubMed] [Google Scholar]
- 70.Kamal A, Goldstein LS. Curr. Opin. Cell Biol. 2000;12:503–508. doi: 10.1016/s0955-0674(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 71.Goldstein LS, Philp AV. Annu. Rev. Cell Dev. Biol. 1999;15:141–183. doi: 10.1146/annurev.cellbio.15.1.141. [DOI] [PubMed] [Google Scholar]
- 72.Hanlon DW, Yang Z, Goldstein LS. Neuron. 1997;18:439–451. doi: 10.1016/s0896-6273(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 73.Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 74.Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozielski F, Sack S, Marx A, Thormahlen M, Schonbrunn E, Biou V, Thompson A, Mandelkow EM, Mandelkow E. Cell. 1997;91:985–994. doi: 10.1016/s0092-8674(00)80489-4. [DOI] [PubMed] [Google Scholar]
- 76.Gulick AM, Song H, Endow SA, Rayment I. Biochemistry. 1998;37:1769–1776. doi: 10.1021/bi972504o. [DOI] [PubMed] [Google Scholar]
- 77.Hirose K, Lowe J, Alonso M, Cross RA, Amos LA. Mol. Biol. Cell. 1999;10:2063–2074. doi: 10.1091/mbc.10.6.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kikkawa M, Okada Y, Hirokawa N. Cell. 2000;100:241–252. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- 79.Stone DB, Hjelm RP, Jr., Mendelson RA. Biochemistry. 1999;38:4938–4947. doi: 10.1021/bi982374z. [DOI] [PubMed] [Google Scholar]
- 80.Paschal BM, Vallee RB. Nature. 1987;330:181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- 81.Schnapp BJ, Reese TS. Proc. Natl. Acad. Sci. USA. 1989;86:1548–1552. doi: 10.1073/pnas.86.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chuang JZ, Milner TA, Sung CH. J. Neurosci. 2001;21:5501–5512. doi: 10.1523/JNEUROSCI.21-15-05501.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. J. Cell. Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 85.Sellers JR. Biochim. Biophys. Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 86.Higuchi H, Endow SA. Curr. Opin. Cell Biol. 2002;14:50–57. doi: 10.1016/s0955-0674(01)00293-9. [DOI] [PubMed] [Google Scholar]
- 87.Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 88.Lantz VA, Miller KG. J. Cell. Biol. 1998;140:897–910. doi: 10.1083/jcb.140.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.DeGiorgis JA, Reese TS, Bearer EL. Mol. Biol. Cell. 2002;13:1046–1057. doi: 10.1091/mbc.01-06-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reck-Peterson SL, Provance DW, Jr., Mooseker MS, Mercer JA. Biochim. Biophys. Acta. 2000;1496:36–51. doi: 10.1016/s0167-4889(00)00007-0. [DOI] [PubMed] [Google Scholar]
- 91.Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenldns NA, Hammer JA., III Nat. Cell. Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- 92.Toyoshima I, Yu H, Steuer ER, Sheetz MP. J. Cell. Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar J, Yu H, Sheetz MP. Science. 1995;267:1834–1837. doi: 10.1126/science.7892610. [DOI] [PubMed] [Google Scholar]
- 94.Goldstein LS. Proc. Natl. Acad Sci. USA. 2001;98:6999–7003. doi: 10.1073/pnas.111145298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fields BN, Knipe DM, Howley PM. Fields virology. 3rd ed. Vol. 2. Lippincott-Raven Publishers; Philadelphia: 1996. [Google Scholar]
- 97.Bearer EL, Breakefield XO, Schuback D, Reese TS, LaVail JH. Proc. Natl. Acad. Sci. USA. 2000;97:8146–8150. doi: 10.1073/pnas.97.14.8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnson DC, Wittels M, Spear PG. J. Virol. 1984;52:238–247. doi: 10.1128/jvi.52.1.238-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sodeik B, Ebersold MW, Helenius A. J. Cell. Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miranda-Saksena M, Armati P, Boadle RA, Holland DJ, Cunningham AL. J. Virol. 2000;74:1827–1839. doi: 10.1128/jvi.74.4.1827-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Topp KS, Bisla K, Saks ND, Lavail JH. Neuroscience. 1996;71:1133–1144. doi: 10.1016/0306-4522(95)00497-1. [DOI] [PubMed] [Google Scholar]
- 102.Huxley AF. J. Physiol. 2002;538:2. doi: 10.1113/jphysiol.2001.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hodgkin AL, Huxley AF. J. Physiol. 1952;116:449–472. doi: 10.1113/jphysiol.1952.sp004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vale RD, Reese TS, Sheetz MP. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. J. Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ye GJ, Vaughan KT, Vallee RB, Roizman B. J. Virol. 2000;74:1355–1363. doi: 10.1128/jvi.74.3.1355-1363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Elliott G, O'Hare P. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 108.Morrison EE, Stevenson AJ, Wang YF, Meredith DM. J. Gen. Virol. 1998;79(Pt 10):2517–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- 109.White BH, Cummins TR, Wolf DH, Waxman SG, Russell DS, Kaczmarek LK. J. Neurophysiol. 2002;87:2149–2157. doi: 10.1152/jn.00498.2001. [DOI] [PubMed] [Google Scholar]
- 110.Smith GA, Gross SP, Enquist LW. Proc. Natl. Acad. Sci. USA. 2001;98:3466–3470. doi: 10.1073/pnas.061029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tomishima MJ, Smith GA, Enquist LW. Traffic. 2001;2:429–436. doi: 10.1034/j.1600-0854.2001.020701.x. [DOI] [PubMed] [Google Scholar]
- 112.Mettenleiter TC. J. Virol. 2002;76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Holland DJ, Miranda-Saksena M, Boadle RA, Armati P, Cunningham AL. J. Virol. 1999;73:8503–8511. doi: 10.1128/jvi.73.10.8503-8511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Diefenbach RJ, Miranda-Saksena M, Diefenbach E, Holland DJ, Boadle RA, Armati PJ, Cunningham AL. J. Virol. 2002;76:3282–3291. doi: 10.1128/JVI.76.7.3282-3291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Elliott G, O'Hare P. J. Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kotsakis A, Pomeranz LE, Blouin A, Blaho JA. J. Virol. 2001;75:8697–8711. doi: 10.1128/JVI.75.18.8697-8711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brewis N, Phelan A, Webb J, Drew J, Elliott G, O'Hare P. J. Virol. 2000;74:1051–1056. doi: 10.1128/jvi.74.2.1051-1056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aints A, Guven H, Gahrton G, Smith CI, Dilber MS. Gene Ther. 2001;8:1051–1056. doi: 10.1038/sj.gt.3301493. [DOI] [PubMed] [Google Scholar]
- 119.Martin A, O'Hare P, McLauchlan J, Elliott G. J. Virol. 2002;76:4961–4970. doi: 10.1128/JVI.76.10.4961-4970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elliott G, O'Reilly D, O'Hare P. Virology. 1996;226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 121.Ren X, Harms JS, Splitter GA. J. Virol. 2001;75:9010–9017. doi: 10.1128/JVI.75.19.9010-9017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tomishima MJ, Enquist LW. J. Cell. Biol. 2001;154:741–752. doi: 10.1083/jcb.200011146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van Leeuwen H, Elliott G, O'Hare P. J. Virol. 2002;76:3471–3481. doi: 10.1128/JVI.76.7.3471-3481.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pomeranz LE, Blaho JA. J. Virol. 2000;74:10041–10054. doi: 10.1128/jvi.74.21.10041-10054.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.del Rio T, Werner HC, Enquist LW. J. Virol. 2002;76:774–782. doi: 10.1128/JVI.76.2.774-782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dietz P, Klupp BG, Fuchs W, Kollner B, Weiland E, Mettenleiter TC. J. Virol. 2000;74:5083–5090. doi: 10.1128/jvi.74.11.5083-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ng TI, Ogle WO, Roizman B. Virology. 1998;241:37–48. doi: 10.1006/viro.1997.8963. [DOI] [PubMed] [Google Scholar]
- 128.Wu SX, Zhu XP, Letchworth GJ. J. Virol. 1998;72:3029–3036. doi: 10.1128/jvi.72.4.3029-3036.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van Genderen IL, Brandimarti R, Torrisi MR, Campadelli G, van Meer G. Virology. 1994;200:831–836. doi: 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- 130.Miles BD, Luftig RB, Weatherbee JA, Weil1ing RR, Weber J. Virology. 1980;105:265–269. doi: 10.1016/0042-6822(80)90177-4. [DOI] [PubMed] [Google Scholar]
- 131.Staufenbiel M, Epple P, Deppert W. J. Virol. 1986;60:1186–1191. doi: 10.1128/jvi.60.3.1186-1191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Suomalainen M, Nakano MY, Keller S, Boucke K, Stidwill RP, Greber UF. J. Cell. Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E, Crystal RG. Hum. Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 134.Glotzer JB, Michou AI, Baker A, Saltik M, Cotten M. J. Virol. 2001;75:2421–2434. doi: 10.1128/JVI.75.5.2421-2434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rietdorf J, Ploubidou A, Reckmann I, Holmstrom A, Frischknecht F, Zettl M, Zimmermann T, Way M. Nat. Cell Blot. 2001;3:992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- 136.Ploubidou A, Moreau V, Ashman K, Reckmann I, Gonzalez C, Way M. EMBO J. 2000;19:3932–3944. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moss B, Ward BM. Nat. Cell Biol. 2001;3:E245–246. doi: 10.1038/ncb1101-e245. [DOI] [PubMed] [Google Scholar]
- 138.Sanderson CM, Way M, Smith GL. J. Virol. 1998;72:1235–1243. doi: 10.1128/jvi.72.2.1235-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ploubidou A, Way M. Curr. Opin. Cell Biol. 2001;13:97–105. doi: 10.1016/S0955-0674(00)00180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shieh WJ, Guarner J, Layton M, Fine A, Miller J, Nash D, Campbell GL, Roehrig JT, Gubler DJ, Zaki SR. Emerg. Infect. Dis. 2000;6:370–372. doi: 10.3201/eid0604.000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ireton K, Cossart P. Annu. Rev. Genet. 1997;31:113–138. doi: 10.1146/annurev.genet.31.1.113. [DOI] [PubMed] [Google Scholar]
- 142.Cossart P. Cell Microbial. 2000;2:195–205. doi: 10.1046/j.1462-5822.2000.00053.x. [DOI] [PubMed] [Google Scholar]
- 143.Tilney LG, Portnoy DA. J. Cell. Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- 145.Portnoy DA, Schreiber RD, Connelly P, Tilney LG. J. Exp. Med. 1989;170:2141–2146. doi: 10.1084/jem.170.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.David V, Gouin E, Trays MV, Grogan A, Segal AW, Ampe C, Cossart P. J. Cell Sci. 1998;111(Pt 19):2877–2884. doi: 10.1242/jcs.111.19.2877. [DOI] [PubMed] [Google Scholar]
- 147.Dabiri GA, Sanger JM, Portnoy DA, Southwick FS. Proc. Natl. Acad. Sci. USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Theriot JA, Mitchison TJ, Tilney LG, Portnoy DA. Nature. 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- 149.Gouin E, Gantelet H, Egile C, Lasa I, Ohayon H, Villiers V, Gounon P, Sansonetti PJ, Cossart P. J. Cell Sci. 1999;112(Pt 11):1697–1708. doi: 10.1242/jcs.112.11.1697. [DOI] [PubMed] [Google Scholar]
- 150.Bearer EL, Prakash JM, Li Z. Int. Rev. Cytol. 2002;217:137–182. doi: 10.1016/s0074-7696(02)17014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Machesky LM, Insall RH. Curr. Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 152.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kitschner MW. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 153.Egile C, Loisel TP, Lautent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. J. Cell. Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Halbrugge M, Walter U. Eur. J. Biochem. 1989;185:41–50. doi: 10.1111/j.1432-1033.1989.tb15079.x. [DOI] [PubMed] [Google Scholar]
- 155.Sanger JM, Sanger JW, Southwick FS. Infect. Immun. 1992;60:3609–3619. doi: 10.1128/iai.60.9.3609-3619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chakraborty T, Ebel F, Domann E, Niebuhr K, Gerstel B, Pistor S, Temm-Grove CJ, Jockusch BM, Reinhard M, Walter U, et al. EMBO J. 1995;14:1314–1321. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lasa I, David V, Gouin E, Marchand JB, Cossart P. Mol. Microbiol. 1995;18:425–436. doi: 10.1111/j.1365-2958.1995.mmi_18030425.x. [DOI] [PubMed] [Google Scholar]
- 158.Smith GA, Theriot JA, Portnoy DA. J. Cell. Biol. 1996;135:647–660. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lauer P, Theriot JA, Skoble J, Welch MD, Portnoy DA. Mol. Microbiol. 2001;42:1163–1177. doi: 10.1046/j.1365-2958.2001.02677.x. [DOI] [PubMed] [Google Scholar]
- 160.Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- 161.Bearer EL, Prakash JM, Manchester RD, Allen PG. Cell Motil. Cytoskeleton. 2000;47:351–364. doi: 10.1002/1097-0169(200012)47:4<351::AID-CM8>3.0.CO;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Prehoda KE, Lee DJ, Lim WA. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 163.Fedorov AA, Fedorov E, Gertler F, Almo SC. Nat. Struct. Biol. 1999;6:661–665. doi: 10.1038/10717. [DOI] [PubMed] [Google Scholar]
- 164.Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cudmore S, Cossart P, Griffiths G, Way M. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 166.Frischlmecht F, Cudmore S, Moreau V, Reckmann I, Rottger S, Way M. Curr. Biol. 1999;9:89–92. doi: 10.1016/s0960-9822(99)80020-7. [DOI] [PubMed] [Google Scholar]
- 167.Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M. Nat. Cell Biol. 2000;2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- 168.Hollinshead M, Rodger G, Van Eijl H, Law M, Hollinshead R, Vaux DJ, Smith GL. J. Cell. Biol. 2001;154:389–402. doi: 10.1083/jcb.200104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sanderson CM, Hollinshead M, Smith GL. J. Gen. Virol. 2000;81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- 170.Zhang WH, Wilcock D, Smith GL. J. Virol. 2000;74:11654–11662. doi: 10.1128/jvi.74.24.11654-11662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wolffe EJ, Weisberg AS, Moss B. Virology. 1998;244:20–26. doi: 10.1006/viro.1998.9103. [DOI] [PubMed] [Google Scholar]
- 172.Purves FC, Spector D, Roizman B. J. Virol. 1992;66:4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jun PY, Strelow LI, Herman RC, Marsden HS, Eide T, Haarr L, Leib DA. J. Gen. Virol. 1998;79(Pt 7):1603–1611. doi: 10.1099/0022-1317-79-7-1603. [DOI] [PubMed] [Google Scholar]
- 174.Roizman B. Proc. Natl. Acad. Sci. USA. 1996;93:11307–11312. doi: 10.1073/pnas.93.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yamada H, Daikoku T, Yamashita Y, Jiang YM, Tsurumi T, Nishiyama Y. J. Gen. Virol. 1997;78(Pt 11):2923–2931. doi: 10.1099/0022-1317-78-11-2923. [DOI] [PubMed] [Google Scholar]
- 176.Duc Dodon M, Mikaelian I, Sergeant A, Gazzolo L. Virology. 2000;270:43–53. doi: 10.1006/viro.2000.0275. [DOI] [PubMed] [Google Scholar]
- 177.Roller RJ, Roizman B. J. Virol. 1992;66:3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Baines JD, Roizman B. J. Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fields BN, Knipe DM, Howley PM, Griffin DE. Fields virology. 4th ed. Vol. 2. Lippincott Williams & Wilkins; Philadelphia: 2001. [Google Scholar]
- 180.Cunningham C, Davison AJ, MacLean AR, Taus NS, Baines JD. J. Virol. 2000;74:33–41. doi: 10.1128/jvi.74.1.33-41.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Salmon B, Cunningham C, Davison AJ, Harris WJ, Baines JD. J. Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Takakuwa H, Goshima F, Koshizuka T, Murata T, Daikoku T, Nishiyama Y. Genes Cells. 2001;6:955–966. doi: 10.1046/j.1365-2443.2001.00475.x. [DOI] [PubMed] [Google Scholar]
- 183.Desai PJ. J. Virol. 2000;74:11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schmitz JB, Albright AG, Kinchington PR, Jenkins FJ. Virology. 1995;206:1055–1065. doi: 10.1006/viro.1995.1028. [DOI] [PubMed] [Google Scholar]
- 185.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. Mol. Celt Biol. 2000;20:4922–4931. doi: 10.1128/mcb.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Donnelly M, Elliott G. J. Virol. 2001;75:2566–2574. doi: 10.1128/JVI.75.6.2566-2574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. J. Gen. Virol. 1998;79(Pt 12):3027–3031. doi: 10.1099/0022-1317-79-12-3027. [DOI] [PubMed] [Google Scholar]
- 188.Kehm R, Rosen-Wolff A, Darai G. Virus Res. 1996;40:17–31. doi: 10.1016/0168-1702(96)80248-6. [DOI] [PubMed] [Google Scholar]
- 189.Mossman KL, Smiley JR. J. Virol. 1999;73:9726–9733. doi: 10.1128/jvi.73.12.9726-9733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhu XX, Chen JX, Young CS, Silverstein S. J. Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Halford WP, Schaffer PA. J. Virol. 2001;75:3240–3249. doi: 10.1128/JVI.75.7.3240-3249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xiao W, Pizer LI, Wilcox KW. J. Virol. 1997;71:1757–1765. doi: 10.1128/jvi.71.3.1757-1765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Feierbach B, Chang F. Curr. Biol. 2001;11:656–665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- 194.Chang F, Drubin D, Nurse P. J. Cell. Biol. 1997;137:69–82. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Castrillon DH, Wasserman SA. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- 196.Woychik RP, Maas RL, Zeller R, Vogt TF, Leder P. Nature. 1990;346:850–853. doi: 10.1038/346850a0. [DOI] [PubMed] [Google Scholar]
- 197.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. Nat. Cell Biol. 2002 doi: 10.1038/ncb834. (in press) [DOI] [PubMed] [Google Scholar]
- 198.Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]