Abstract
Objectives
To evaluate the discrimination, calibration and net benefit performance of the Prostate Cancer Prevention Trial Risk Calculator (PCPTRC) across five European Randomized study of Screening for Prostate Cancer (ERSPC), 1 United Kingdom, 1 Austrian and 3 US biopsy cohorts.
Methods
PCPTRC risks were calculated for 25,733 biopsies using prostate-specific antigen (PSA), digital rectal examination, family history and history of prior biopsy, and single imputation for missing covariates. Predictions were evaluated using the areas underneath the receiver operating characteristic curves (AUC), discrimination slopes, chi-square tests of goodness of fit, and net benefit decision curves.
Results
AUCs of the PCPTRC ranged from a low of 56% in the ERSPC Goeteborg Rounds 2-6 cohort to a high of 72% in the ERSPC Goeteborg Round 1 cohort, and were statistically significantly higher than that of PSA in 6 out of the 10 cohorts. The PCPTRC was well-calibrated in the SABOR, Tyrol and Durham cohorts. There was limited to no net benefit to using the PCPTRC for biopsy referral compared to biopsying all or no men in all five ERSPC cohorts and benefit within a limited range of risk thresholds in all other cohorts.
Conclusions
External validation of the PCPTRC across ten cohorts revealed varying degree of success highly dependent on the cohort, most likely due to different criteria for and work-up before biopsy. Future validation studies of new calculators for prostate cancer should acknowledge the potential impact of the specific cohort studied when reporting successful versus failed validation.
Keywords: receiver operating characteristic curve, risk, prostate cancer, calibration, net benefit
Introduction
The Prostate Cancer Prevention Trial (PCPT) Risk Calculator for Prostate Cancer (PCPTRC) was developed from 5,519 men from the placebo arm of the PCPT who underwent biopsy at or before the end of 7 years of annual prostate-specific antigen (PSA) and digital rectal exam (DRE) screening regardless of PSA or DRE indication [1]. Due to the required end-of-study biopsy, the PCPTRC is not subject to ascertainment bias, which results when only participants with clinical indications for biopsy are actually biopsied, while others are not and hence excluded from the study. However, there are several reasons why the PCPTRC may not be accurate when applied to other cohorts, including the use of a 6 core biopsy in place of more extended schemes and a majority of PSA values less than 3.0 ng/mL.
The purpose of the current study was to test the external validity of the PCPTRC for prostate cancer detection in ten European and US contemporary biopsy cohorts, some with more than 6-core biopsies performed and most with significant numbers of PSA values exceeding those in the PCPTRC cohort.
Methods
Data were included from ten European and US cohorts belonging to the Prostate Biopsy Collaborative Group (PBCG), where criteria for biopsy referral and sampling scheme have been previously summarized [2]. These included five screening cohorts from the European Randomized study of Screening for Prostate Cancer (ERSPC), three additional screening cohorts, San Antonio Biomarkers Of Risk of cancer study (SABOR), Texas, US, ProtecT, United Kingdom, and Tyrol, Austria, and two US clinical cohorts, Cleveland Clinic, Ohio, and the Durham VA, North Carolina. All cohorts except for ERSPC Goeteborg and Rotterdam Rounds 1 included some patients who had been previously screened. All biopsies after a positive biopsy for prostate cancer were excluded from the analysis.
Clinical characteristics of each cohort were summarized in terms of median and range (age and PSA) and by numbers (percent) in each category (DRE, family history, African origin, prior negative biopsy, prostate cancer, Gleason grade).
For each biopsy in the dataset, the PCPTRC risk of a positive biopsy was computed, requiring PSA, DRE, family history of prostate cancer (father, brother or son ever diagnosed with prostate cancer) and history of a prior negative prostate biopsy. Within-cohort multiple imputation was used when the percentage of missing data for a risk factor in a cohort was less than 100%. The number of iterations was set to 20 and PCPTRC risks were computed as the average over five imputations of the missing risk factor [3,4]. For cohorts with family history or DRE not recorded for any participants, single imputation of “no family history” and “negative DRE”, respectively, were used following recommendation by the online PCPTRC.
Discrimination was calculated via receiver operating characteristics curves (ROC). Areas underneath the ROC curve (AUC) were calculated for PCPTRC risks and compared to those for PSA (provided by Kattan et al., unpublished manuscript) for the same cohort using U-statistics [5]. Calibration plots were computed by comparing observed proportions of cancer to mean PCPTRC risks by PCPTRC deciles observed in the cohort [6]. The Hosmer-Lemeshow chi-square test was used to compare the same observed rates to predicted PCPTRC risks across the deciles. For this test a p<0.05 indicates a poor agreement between predicted PCPTRC risks and actual observed risk. Net benefit curves were computed by calculating for each possible PCPTRC risk of a positive biopsy (p) ranging from 0 to 100%, Sensitivity × %Cancer – False Positive Rate × %Non-cancer × [p/(1-p)], and were assessed for their gain over the corresponding net benefit curves of referring no patients to biopsy (horizontal line at 0) or simply referring all patients to biopsy, × %Cancer – %Non-cancer × [p/(1-p)] [7].
Results
Across the PBCG cohorts age was fairly consistent with a median in the early sixties (Table 1). Median PSA values ranged from 3.4 ng/mL in the SABOR cohort to 5.2 ng/mL in the Durham VA cohort, and rates of abnormal DRE, from a low of 10% in the Goeteborg Rounds 2-6 and Tyrol cohorts to a high of 31% in the Tarn cohort. Family history was only reported in half of the cohorts and those reported all fell at or below 11% except for SABOR at 29%. This again was an artifact of selection bias for the SABOR cohort since its protocol included a family history substudy that offered biopsy to men with PSA less than 4 ng/mL and a positive family history. African origin was not reported in the European cohorts but could be presumed to be negligible. The Durham VA cohort provided a contrast, with 45% of the cohort of African origin. This cohort also had the highest cancer rate of 47% exceeding the nine other cohorts with cancer rates ranging from 26 to 39%. Biopsy Gleason grade distributions indicated a majority of low grade cancers (Gleason 6 or less) in the ERSPC and SABOR screening cohorts, but only approximately half or less low grade cancers in the Tarn section of the ERSPC and the more clinical cohorts, Cleveland Clinic and Durham VA cohorts.
Table 1. Clinical characteristics of each cohort: age and PSA report median (range), the rest report number n (%). Biopsy gleason grade reports percent of cancers.
| Goeteborg Round 1 | Goeteborg Rounds 2-6 | Rotterdam Round 1 | Rotterdam Rounds 2-3 | Tarn | SABOR | Cleveland Clinic | ProtecT | Tyrol | Durham VA | |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of patients | 740 | 1241 | 2895 | 1494 | 298 | 392 | 2631 | 7324 | 4199 | 1856 |
| Number of biopsies | 740 | 1241 | 2895 | 1494 | 298 | 392 | 3286 | 7324 | 5644 | 2419 |
| Age | ||||||||||
| Median (range) | 61 (51,70) | 63 (53, 71) | 66 (55, 75) | 67 (59, 75) | 64 (55, 71) | 63 (50,75) | 64 (50,75) | 63 (50,72) | 63 (50,75) | 64(50,75) |
| PSA Median | 4.7 | 3.6 | 5.0 | 3.5 | 4.5 | 3.4 | 5.8 | 4.4 | 4.2 | 5.2 |
| Range | (0.5, 226.0) | (2.0, 88.8) | (0.0, 245.0) | (0.4, 99.5) | (1.6, 131.0) | (0.2,919.2) | (0.2,491.7) | (3.0, 847.0) | (0.1,3210.0) | (0.1,1355.6) |
| < 3 ng/mL | 33 (4%) | 205 (17%) | 147 (5%) | 417 (28%) | 26 (9%) | 166 (42%) | 337 (10%) | 0 (0%) | 1614 (29%) | 309 (13%) |
| ≥ 3 ng/mL | 707 (96%) | 1036(83%) | 2748 (95%) | 1077 (72%) | 272 (91%) | 226 (58%) | 2949(90%) | 7324 (100%) | 4030 (71%) | 2110 (87%) |
| DRE result | ||||||||||
| Normal | 614 (83%) | 1117 (90%) | 2137 (74%) | 1182 (79%) | 179 (60%) | 280 (71%) | 3083(94%) | 0 | 5076(90%) | 887(37%) |
| Abnormal | 126 (17%) | 124 (10%) | 758 (26%) | 312 (21%) | 92 (31%) | 112 (29%) | 203 (6%) | 0 | 568 (10%) | 265 (11%) |
| Unknown | 0 | 0 | 0 | 0 | 27 (9%) | 0 | 0 | 7324(100%) | 0 | 1267(52%) |
| Family history | ||||||||||
| No | 0 | 0 | 1708 (59%) | 875 (59%) | 0 | 280 (71%) | 1690(51%) | 5736(78%) | 0 | 0 |
| Yes | 0 | 0 | 328 (11%) | 160 (11%) | 0 | 112 (29%) | 373 (11%) | 454(6%) | 0 | 0 |
| Unknown | 740 (100%) | 1241 (100%) | 859 (30%) | 459 (31%) | 298 (100%) | 0 | 1223(37%) | 1134(15%) | 5644(100%) | 2419(100%) |
| African origin | ||||||||||
| No | 0 | 0 | 0 | 0 | 0 | 349 (89%) | 2818(86%) | 6933(95%) | 0 | 1218(50%) |
| Yes | 0 | 0 | 0 | 0 | 0 | 43 (11%) | 422 (13%) | 34 (0%) | 0 | 1079(45%) |
| Unknown | 740 (100%) | 1241 (100%) | 2895 (100%) | 1494 (100%) | 298 (100%) | 0 | 46 (1%) | 357 (5%) | 5644(100%) | 122 (5%) |
| Prior biopsy | ||||||||||
| Yes | 0 | 0 | 0 | 0 | 0 | 96(24%) | 1091(33%) | 0 | 1555(28%) | 568(23%) |
| No | 740(100%) | 1241(100%) | 2895(100%) | 1494(100%) | 298(100%) | 296(76%) | 2195(67%) | 7324(100%) | 4089(72%) | 1851(77%) |
| Cancer | 192 (26%) | 322 (26%) | 800 (28%) | 388 (26%) | 96 (32%) | 133 (34%) | 1292(39%) | 2570(35%) | 1562 (28%) | 1148(47%) |
| Biopsy Gleason grade | ||||||||||
| ≤ 6 | 152 (79%) | 269 (84%) | 508 (64%) | 297 (77%) | 42 (44%) | 95 (71%) | 669 (52%) | 1703(66%) | 911 (58%) | 606 (53%) |
| 7 | 33 (17%) | 45 (14%) | 234 (29%) | 78 (20%) | 37 (39%) | 28 (21%) | 478 (37%) | 729 (28%) | 319 (20%) | 387 (34%) |
| ≥ 8 | 7 (4%) | 8 (2%) | 52 (6%) | 13 (3%) | 14 (15%) | 7 (5%) | 145 (11%) | 138 (5%) | 137 (9%) | 141 (12%) |
| Unknown | 0 | 0 | 6 (1%) | 0 | 3 (3%) | 3(2%) | 0 | 0 | 195 (12%) | 14(1%) |
Table 2 gives the external validation report for the PCPTRC in terms of discrimination, calibration and clinical net benefit. AUCs of the PCPTRC ranged from a low of 56.2% in the Goeteborg Rounds 2-6 cohort to a high of 72.0% in the Goeteborg Round 1 cohort. While the AUC of the PCPTRC exceeded the AUC of PSA in all cohorts, it failed to be statistically significantly greater in 4 of the 10 cohorts: Rotterdam Rounds 2-3, Tarn, SABOR, and ProtecT, all screening rather than clinical cohorts.
Table 2. Discrimination, calibration and net benefit metrics.
| Cohort (n) | Discrimination AUC PCPTRC (%) (p-value for comparison to the AUC of PSA) | Calibration Risk range where PCPTRC primarily over-predicts Goodness of fit p-value | Net Benefit Range of PCPTRC risks of positive biopsy showing improved net benefit over the rules of biopsying everyone or no one (%) |
|---|---|---|---|
| ERSPC Goeteborg Round 1 (n=740) | 72.0 (<.0001) | Entire range p < .0001 | None |
| ERSPC Goeteborg Rounds 2-6 (n=1241) | 56.2 (<.0001) | Entire range p < .0001 | None |
| ERSPC Rotterdam Round 1 (n=2895) | 70.0 (<.0001) | Entire range p <.0001 | None |
| ERSPC Rotterdam Rounds 2-3 (n=1494) | 61.0 (.15) | Entire range p <.0001 | None |
| ERSPC Tarn (n=298) | 66.7 (.07) | No over-prediction p <.0001 | 27-35 |
| SABOR, US (n=392) | 65.4 (.20) | No over-prediction p = 0.24 | 15-45 |
| Cleveland Clinic, US (n=3286) | 58.8 (<.0001) | 50% and higher p < .0001 | 35-45 |
| ProtecT, UK (n=7324) | 63.9 (.14) | 50% and lower p < .0001 | 30-85 |
| Tyrol, Austria (n=5644) | 66.7 (<.0001) | Entire range p < .0001 | 18-41 |
| Durham VA, US (n=2419) | 71.5 (<.0001) | No over-prediction p = .0008 | 25-100 |
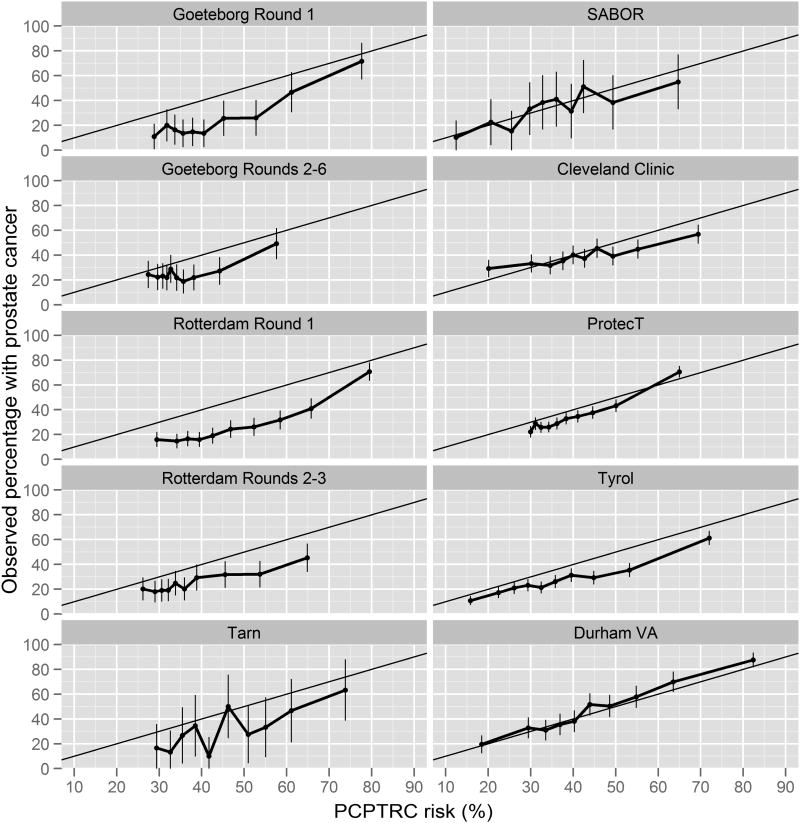
Calibration plots of Figure 1 indicate that the PCPTRC overestimated the risk of prostate cancer for men of low, medium and high risks for all of the ERSPC cohorts except for the Tarn sections, where 95% confidence intervals of the observed risks overlapped with predicted PCPTRC risks. The latter, however, could be attributed to the small sample size of the Tarn section (n=298), which results in wider confidence bands and greater chance for overlap. For similar reasons, the PCPTRC appeared calibrated for the SABOR cohort (n=392). The PCPTRC also over-predicted in risk ranges of practical relevance (below 50%) for the large Cleveland Clinic, Protect and Tyrol cohorts (Table 2). However, for the Durham cohort (n=2419), which had the highest cancer prevalence (47%), the PCPTRC was calibrated across all risks. The Hosmer-Lemeshow test rejected goodness-of-fit for all cohorts except for the SABOR cohort, but this test has the undesirable quality of being more likely to reject the null hypothesis of goodness-of-fit as the sample size increases so is not as objective a benchmark for calibration as the calibration plots.
Figure 1.
Calibration plots for the PCPTRC showing average PCPTRC risks for men grouped by their PCPTRC risk value (x-axis) compared to the actual percentage of these groups which were diagnosed with prostate cancer (y-axis). Perfect calibration would fall on the black diagonal line where predicted risks equal observed rates of prostate cancer.
The last column of Table 2 shows the range of risk thresholds for which the PCPTRC had higher clinical net benefit than the alternative strategies of biopsying all or no men. A risk threshold is the minimum risk at which a patient and clinician would opt for biopsy and varies between individuals due to personal preference. One reasonable threshold is 20%, suggesting that it would be worth conducting no more than 5 biopsies to find one cancer; a reasonable range of thresholds might be 15% to 30%. There was limited (ERSPC Tarn, Cleveland Clinic, ProtecT) to no clinical benefit at all (other four ERSPC cohorts) to using the PCPTRC to determine a subgroup of men to be biopsied compared to biopsying all of those meeting cohort-specific criteria for biopsy. For the remaining three cohorts, SABOR, Tyrol, and Austria, there was clinical benefit observed at reasonable risk ranges: 15-45%, 18-41%, 25-100%, respectively (Table 2).
Discussion
Since its publication in 2006 and posting online for external validation, several single institution or study reports of successful or failed validation of the PCPTRC have appeared leading to confusion as to whether the tool can be recommended in practice [8-14]. By examining the spectrum of answers obtained in a wide variety of cohorts using three complementary validation metrics, this report illuminates the inherent variability of results of external validation by cohort and chosen metric. This variation is not unique to the PCPTRC but would rather extend to validation studies of all risk prediction tools and the rapidly increasing numbers of investigations of new markers for enhancing prostate cancer, including the urine/blood markers PCA3, AMACR, MMP-2, and GSTP1/RASSF1A methylation status [15, 16]. Indeed, these results are a convincing demonstration that properties such as “calibration [are] best seen not as a property of a prediction model, but of a joint property of a model and the particular cohort to which it is applied” [17].
The AUC appears to be the most ubiquitous criterion implemented for validation in urologic research, but even in the absence of a calculator, the AUC for PSA itself evaluated across the ten cohorts of this study varied from no utility at all (AUC=50.9%, Goeteborg Rounds 2-6) to fairly decent performance (AUC=67.0%, Goeteborg Round 1) (data provided by Kattan). The AUC suffers an additional disadvantage by being influenced by selection of patients for inclusion based on PSA; including only patients with PSA exceeding 3 ng/mL will downwardly influence the AUC. The PCPTRC amounts to a weighted average of PSA along with the dichotomous (yes versus no) risk factors of DRE, family history and prior biopsy, and as such its AUC typically tracks that of PSA in the same cohort. Accordingly the AUC of the PCPTRC was also lowest in Goeteborg Rounds 2-6 (AUC=56.2%) and highest in Goeteborg Round 1 (AUC=72.0%). In these two cohorts along with four others, the AUC of the PCPTRC was statistically significantly higher than that of PSA. As noted by Kattan [18], because of unmeasurable cohort differences, head-to-head comparisons of markers or calculators within cohorts and not across cohorts is key for unbiased comparisons.
Calibration plots confirmed an earlier PBCG observation that for most cohorts, the PCPTRC tends to give prostate cancer risk predictions that are too high, overestimating actual risks both in the PSA < 4 ng/mL range, the range on which the PCPTRC was largely developed, and grossly overestimating outside this range [2]. The calibration plots revealed that the PCPTRC was better calibrated for cohorts with larger prevalences of cancer, in particular the Durham VA clinical cohort. A limitation of all results is that single imputation had to be performed for missing risk factors in several cohorts, and this would affect calibration. For example family history was not recorded in five of the cohorts, so for these cohorts the optimal value “no family history” was used for all participants. Unfortunately, calibration performed with the actual values of family history for men in these cohorts would be worse, since even with the favorable risk outcome imputed into the calculator, the PCPTRC still overestimated risks. Additionally, because the lowest PCPTRC risks observed in many of the cohorts fell near 30%, the current study provides no assessment of calibration of PCPTRC for lower risks that might be of greatest interest for decision-making concerning a biopsy.
Clinical net benefit is a more recently proposed validation metric that seeks to quantify the net benefit to a patient for using a particular decision rule to opt for a prostate biopsy, specifically, by choosing a threshold risk and deciding to undergo biopsy only if risk predicted by the decision rule exceeds this value. For each possible threshold, the net benefit of using the PCPTRC along with this threshold for referral to biopsy is assessed relative to just the rule of referring everyone in the cohort for biopsy. The five ERSPC cohorts had per protocol referral men for biopsy for PSA exceeding 3 ng/mL (4 ng/mL in some sections at some years) and there was no observed benefit to using the PCPTRC for these men with primarily high risks to begin with. In contrast, net benefit of using the PCPTRC at thresholds 15 to 45% was observed in the SABOR cohort, a cohort with lower PSA values, and most similar in nature to the PCPT cohort as described above. Among the remaining cohorts there was only limited net benefit at limited ranges of PCTPRC thresholds.
In sum this study has shown that the PCPTRC may not be universally applicable, that in the population of men with elevated PSA (above 3.0 ng/mL) who would most seriously consider prostate biopsy, the PCTPRC may overestimate the risk of finding prostate cancer. This result could be due to that the PCPTRC was fit on a different population of men, primarily healthy men with PSA less than 3.0 ng/mL. The accuracy of the PCPTRC on such a healthy population of men is not ruled out by the current validation study since no cohorts of this type were included. We are investigating a modification to the PCPTRC tailored to men more commonly referred to biopsy.
Findings of this study have implications for other risk prediction tools beyond the PCPTRC. It is typical in urologic research to declare definitive success or failure of a tool based on a single validation measure evaluated on data from a single institution. However, if validation is a function of both the model and the cohort being studied, there are two consequences. First, those proposing models must explore the properties of the model in different cohorts, and investigate the aspects of a cohort that affect model performance. Second, clinicians should be cautious in using a model unless it has been shown to provide added value, such as benefit, in a very similar population to the one in which it is being used clinically.
Acknowledgments
Statistics supported in part by funds from David H. Koch provided through the Prostate Cancer Foundation, the Sidney Kimmel Center for Prostate and Urologic Cancers SPORE grant from the U.S. National Cancer Institute [P50-CA92629]and a Cancer Center Support Grant for the Cancer Therapy and Research Center at the University of Texas Health Science Center at San Antonio [P30-CA054174]. Grants to support the work of the ERSPC include: European Union Grants SOC 95 35109, SOC 96 201869 05F022, SOC 97 201329, SOC 98 32241, the 6th Framework Program of the EU: PMark:LSHC-CT-2004-503011;The Dutch Cancer Society (KWF 94-869, 98-1657, 2002-277, 2006-3518); The Netherlands Organisation for Health Research and Development (ZonMW-002822820, 22000106, 50-50110-98-311); The Prostate Cancer Research Foundation of Rotterdam (SWOP); Beckman-Coulter-Hybritech Inc; Abbott Pharmaceuticals, Sweden; Af Jochnick's foundation; Catarina and Sven Hagstroms family foundation; Gunvor and Ivan Svensson's foundation; Johanniterorden, King Gustav V Jubilée Clinic Cancer Research Foundation; Sahlgrenska University Hospital; Schering Plough, Sweden, Swedish Cancer Society (Contract numbers 09 0107, 080315 and 083455); Wallac Oy, Turkku, Finland. The Tyrol study is supported by the International Agency for Research on Cancer, Lyon and the Tyrolean Prostate Cancer Early Detection Group. The SABOR project is supported by the San Antonio Center of Biomarkers of Risk for Prostate Cancer CA086402. The ProtecT study is funded by the UK NIHR Health Technology Assessment Programme (projects 96/20/06, 96/20/99).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Thompson IM, Ankerst DP, Chi C, Goodman PJ, Tangen CM, Lucia MS, Feng Z, Parnes HL, Coltman CA., Jr Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529–534. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 2.Vickers AJ, Cronin AM, Roobol MJ, Hugosson J, Jones JS, Kattan MW, Klein E, Hamdy F, Neal D, Donovan J, Parekh DJ, Ankerst D, Bartsch G, Klocker H, Horninger W, Benchikh A, Salama G, Villers A, Freedland SJ, Moreira DM, Schroeder FH, Lilja H. The relationship between prostate-specific antigen and prostate cancer risk: the Prostate Biopsy Collaborative Group. Clinical Cancer Research. 2010;16:4374–4381. doi: 10.1158/1078-0432.CCR-10-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Buuren S. Multiple imputation of discrete and continuous data by fully conditional specification. Statistical Methods in Medical Research. 2007;16:219–242. doi: 10.1177/0962280206074463. [DOI] [PubMed] [Google Scholar]
- 4.Janssen KJM, Donders ART, Harrell FE, Jr, Vergouwe Y, Chen Q, Grobbee DE, Moons KGM. Missing covariate data in medical research: To impute is better than to ignore. Journal of Clinical Epidemiology. 2010;63:721–727. doi: 10.1016/j.jclinepi.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 5.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 6.Steyerberg EW. Clinical Prediction Models. Springer; New York, New York: 2010. pp. 270–271. [Google Scholar]
- 7.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Medical Decision Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavadas V, Osório L, Sabell F, Teves F, Branco F, Silva-Ramos M. Prostate Cancer Prevention Trial and European Randomized Study of Screening for Prostate Cancer Risk Calculators: A performance comparison in a contemporary screened cohort. European Urology. 2010;58:551–558. doi: 10.1016/j.eururo.2010.06.023. [DOI] [PubMed] [Google Scholar]
- 9.Eyre SJ, Ankerst DP, Wei JT, Nair PV, Regan MM, Bueti G, Tang J, Rubin MA, Kearney M, Thompson IM, Sanda MG. Validation in a multiple urology practice setting of the Prostate Cancer Prevention Trial calculator for predicting prostate cancer detection. Journal of Urology. 2009;182:2653–2658. doi: 10.1016/j.juro.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez DJ, Han M, Humphreys EB, Mangold LA, Taneja SS, Childs SJ, Bartsch G, Partin AW. Predicting the outcome of prostate biopsy: comparison of a novel logistic regression-based model, the prostate cancer risk calculator, and prostate-specific antigen level alone. British Journal of Urology International. 2009;103:609–614. doi: 10.1111/j.1464-410X.2008.08127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen CT, Yu C, Moussa A, Kattan MW, Jones JS. Performance of Prostate Cancer Prevention Trial Risk Calculator in a contemporary cohort screened for prostate cancer and diagnosed by extended prostate biopsy. Journal of Urology. 2010;183:529–533. doi: 10.1016/j.juro.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira M, Marques V, Carvalho AP, Santos A. Head-to-head comparison of two online nomograms for prostate biopsy outcome prediction. British Journal of Urology International. 2011;107:1780–1783. doi: 10.1111/j.1464-410X.2010.09727.x. [DOI] [PubMed] [Google Scholar]
- 13.Parekh DJ, Ankerst DP, Higgins BA, Hernandez J, Canby-Hagino E, Brand T, Troyer DA, Leach RJ, Thompson IM. External validation of the Prostate Cancer Prevention Trial Risk Calculator in a screened population. Urology. 2006;68:1153–1155. doi: 10.1016/j.urology.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 14.van den Bergh RC, Roobol MJ, Wolters T, van Leeuwen PJ, Schroder FH. The Prostate Cancer Prevention Trial and European Randomized Study of Screening for Prostate Cancer risk calculators indicating a positive prostate biopsy: a comparison. British Journal of Urology International. 2008;102:1068–1073. doi: 10.1111/j.1464-410X.2008.07940.x. [DOI] [PubMed] [Google Scholar]
- 15.Ankerst DP, Groskopf J, Day JR, Blase A, Rittenhouse H, Pollock BH, Tangen C, Parekh D, Leach RJ, Thompson I. Predicting prostate cancer risk through incorporation of prostate cancer gene 3. Journal of Urology. 2008;180:1303–1308. doi: 10.1016/j.juro.2008.06.038. [DOI] [PubMed] [Google Scholar]
- 16.Prior C, Guillen-Grima F, Robles JE, Rosell D, Fernandez-Montero JM, Agirre X, Catena R, Calvo A. Use of a combination of biomarkers in serum and urine to improve detection of prostate cancer. World Journal of Urology. 2010;28:681–686. doi: 10.1007/s00345-010-0583-x. [DOI] [PubMed] [Google Scholar]
- 17.Vickers AJ, Cronin AM. Everything you always wanted to know about evaluating prediction models (but were too afraid to ask) Urology. 2010;76:1298–1301. doi: 10.1016/j.urology.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kattan MW. Factors affecting the accuracy of prediction models limit the comparison of rival prediction models when applied to separate data sets. European Urology. 2011;59:566–567. doi: 10.1016/j.eururo.2010.11.039. [DOI] [PubMed] [Google Scholar]