Abstract
During the past decade, extensive progress has been made toward understanding the molecular basis for the regulation of apoptosis. In mammalian cells undergoing apoptosis, two distinct mechanisms or pathways are operated and are triggered by cell death stimuli from intra- or extra-cellular environments, namely the intrinsic or extrinsic pathways, resulting in mitochondrial membrane depolarization. Several lines of evidence from our laboratories and others have indicated that galectin-3 plays an important role in these pathways by binding to various ligands. Here the authors provide a brief discussion on the role of endogenous or extra-cellular galectin-3 in the regulation of apoptosis and how it could be used as a therapeutic target using natural plant products as its functional inhibitors.
Keywords: Extrinsic apoptotic pathways, Intrinsic apoptotic pathways, Galectin-3, Modified citrus pectin
Introduction
All multi-cellular organisms have endogenous mechanism for selectively killing their own cells. The physiological cell death that occurs in multi-cellular organisms is called “programmed cell death” or apoptosis and involves a complex network of biochemical pathways that normally ensure a homeostatic balance between cellular proliferation and turnover in nearly all tissues. Apoptosis is essential for the body, as its disregulation can lead to several diseases including cancer. During tumor progression cancer cells can develop ingenious mechanisms to escape the immune system most notably an increased resistance to apoptosis.
Two major pathways of apoptosis have been defined. The intrinsic pathway is mitochondria-mediated signaling pathway generated by signals arising within the cell, while the extrinsic or death receptor mediated pathway is triggered by the binding of death molecules to cell surface receptors. Both apoptotic signaling pathways involve mitochondrial membrane permeabilization, during which the mitochondrial outer membrane as well as inner membrane are permeabilized, resulting in the release of soluble proteins including cytotoxic proteins from the inter-membrane space (Green and Kroemer, 1998; Ferri and Kroemer, 2001). Such cytotoxic proteins include caspase independent death effectors (nucleases and proteases) as well as caspase activators namely cytochrome c (Patterson et al., 2000), which activates the apoptosome, a caspase activation complex including dATP, apoptosis protease activating factor (Apaf-1) and procaspase-9, which ultimately activates the effector caspase cascade (caspase 3 and 7), leading to cell death (Danial and Korsmeyer, 2004). Mitochondrial permeabilization, therefore, is recognized as a crucial checkpoint in programmed cell death of both normal and cancer cells. The death receptor pathway or the extrinsic pathway is activated upon interaction of death receptors at the cell surface with their cognate ligands on, for example, T cells, whereupon the adaptor protein FAS-associated death domain (FADD) and initiator caspase-8 or caspase-10 are recruited to the intracellular death domains of these receptors. Assembly of this death inducing signaling complex (DISC) leads to sequential activation of initiator and effector caspases and ultimately result in apoptotic death. In some cells the death receptor pathway relies on a mitochondrial amplification loop that is activated by caspase 8 mediated cleavage of the BH-3 only interacting domain death agonist Bid to a truncated form (tBid). Truncated Bid subsequently activates the mitochondrial pathway (Chowdhury et al., 2006). In addition there are other signaling pathways such as p53 dependent pathways, which get activated in response to stress. The mitochondrial sensitivity to apoptosis has been believed to be exquisitely regulated by the B-cell leukemia/lymphoma 2 (Bcl-2) family of pro- and anti-apoptotic proteins via their regulation of cytochrome c release (Breckenridge and Xue, 2004). The balance between the pro and anti apoptotic proteins determines whether the cell will enter apoptosis or not. Recently, another protein namely, galectin-3, which shares some structural properties with Bcl2 has been reported to play an important role in the apoptotic pathway.
Galectin-3 is a member of a family of carbohydrate-binding proteins consisting so far of 14 reported members, characterized by a conserved amino acid sequence defined by structural similarities in their carbohydrate-binding domain and affinity for β-galactoside containing glycoconjugates (Barondes et al., 1994). Galectin-3 is the unique member of this family having a chimeric structure consisting of a amino terminal domain of 12 amino acids containing a serine phosphorylation site that regulates its cellular targeting, a α- collagen like sequence of about 110 amino acids that serves as a substrate for matrix metalloproteases, and a carboxy terminal domain of about 130 amino acids that contains a single carbohydrate recognition domain (Barondes et al., 1994; Ochieng et al., 1994; Gong et al., 1999). Galectin-3 is predominantly localized in the cytoplasm; may translocate to the perinuclear membrane, nucleus and/or get secreted from the cytoplasm (Moutsatsos et al., 1987; Hughes, 1999). By its ability to bind to N-lactosamine residues present on the intracellular, extracellular and cell surface associated glycoconjugates, galectin-3 is involved with a number of cellular functions like cell growth, cell adhesion, cell differentiation, tumor progression, angiogenesis and metastasis (Liu et al., 2002; Takenaka et al., 2004; Dumic et al., 2006). Galectin-3 is phosphorylated by casein kinase I in vitro at serine 6 amino acid (Huflejt et al., 1993) and it exists in this phosphorylated form inside the cell (Cowles et al., 1990).
There are numerous data on roles of galectin-3 in regulation of cellular homeostasis. This protein was shown to modulate cell growth, to control the cell cycle, and to be involved in the regulation of apoptosis. Galectin-3 has been shown to translocate either from the cytosol or from the nucleus to the mitochondria following exposure to apoptotic stimuli (Yu et al., 2002) and to block changes in the mitochondrial membrane potential, thereby preventing apoptosis (Matarrese et al., 2000). It is indicated that galectin-3 might exert its anti-apoptotic activity by interacting with other apoptosis regulators that function in the mitochondria. When the serine 6 residue is mutated to alanine or glutamic acid, galectin-3 can not be phosphorylated or transported to the nuclei and its anti-apoptotic activity is decreased (Yoshii et al., 2002) indicating that phosphorylation at serine 6 is important for its anti-apoptotic activity.
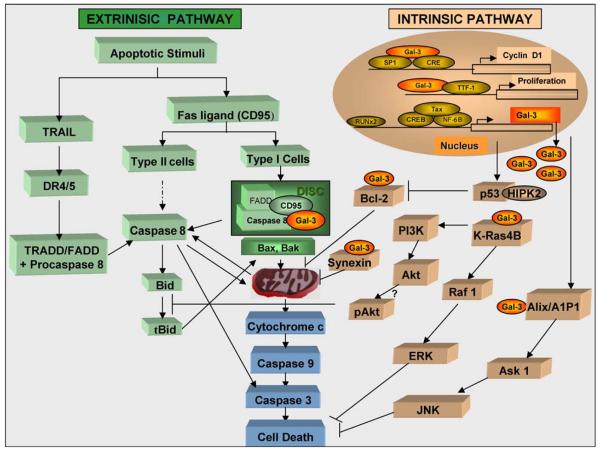
Here we provide an overview of the data relating to its functions in the intrinsic and extrinsic pathways of apoptosis (Fig. 1). Some of the data were obtained using exogenously added galectin-3 stimulating the effects of the extra-cellular galectin-3, while others were demonstrated by over-expressing the protein, or inhibiting its expression by anti-sense transfections or using specific inhibitors.
Fig. 1.
A model for the regulation of extrinsic and intrinsic apoptotic pathways by galectin-3
Discussion
Role of galectin-3 in intrinsic apoptotic pathways
Many drugs, toxins or radiation induce a signal inside the cell that provokes or initiates the apoptotic response. The exact pathway of galectin-3 in apoptosis has not been fully elucidated, but it binds to various ligands in response to apoptotic insults in different cell lines. One of the master regulators of apoptosis is p53, which is a product of a tumor suppressor gene (Vousden, 2000). A stress signal (either external or internal to the cells) is transmitted to the p53 protein by post-translational modification. This results in activation of the p53 protein as a transcriptional factor, which is instrumental in the activation of the mitochondrial pathway of apoptosis. Several proteins have been identified that are necessary for p53 to mediate the apoptotic response (Appella and Anderson, 2000). One such protein is homeodomain-interacting protein kinase 2 (HIPK2), which is a Ser/Thr kinase that binds to and activates p53 by phosphorylating it at Ser46 (D'Orazi et al., 2002; Hofmann et al., 2002), and is relevant to p53 mediated apoptosis. It was further shown that cooperation between p53 and HIPK2 leads to repression of galectin-3, when p53 dependent apoptosis is induced (Cecchinelli et al., 2006). So galectin-3, like Bcl2, is one of the target genes for p53 and it is a mediator of p53 dependent apoptosis.
Ras proteins (H-Ras, K-Ras, N-Ras) are members of large super family of small GTPases, which function as core signaling molecules and are activated by convergent signaling pathways after extra-cellular stimuli. Activated Ras, in turn, regulates a diversity of downstream cytoplasmic signaling cascades and can induce different biological consequences such as cell proliferation, senescence, survival, or death, depending on the cellular context (Mitin et al., 2005). Galectin-3 acts as a selective binding partner of activated K-Ras. Co-transfectants of K-Ras/galectin-3 in HEK-293 cells, but not of H-Ras/galectin-3 exhibited enhanced and prolonged EGF-stimulated increases in Ras-GTP, Raf-1 activity and phosphotidylinositol 3 kinase (PI3K) activity. However, ERK activity was attenuated and PI3K activity was augmented in K-Ras/galectin-3 co-transfectants (Elad-Sfadia et al., 2004).
In a more recent study, it was demonstrated that over-expression of galectin-3 in BT-549 human breast cancer cells coincided with a significant increase in K-Ras-GTP coupled with loss in N-Ras-GTP, whereas the non-oncogenic galectin-3 mutant proteins (S6E and G182A) failed to induce the Ras isoform switch. (Shalom-Feuerstein et al., 2005). These findings suggest that the level of galectin-3 defines outputs of oncogenic K-Ras protein.
NF-kB (nuclear factor kB) belongs to a family of heterodimeric transcription factors that play a role in inflammatory and stress responses as well as in tumor cell resistance to apoptosis (Delhalle et al., 2004). Expression of galectin-3 in human T lymphotrophic virus-1 (HTLV-1) infected T-cells was increased by the galectin-3 promoter through interaction with NF-kB (Hsu et al., 1996). Inhibition of NF-kB by a specific proteosomal inhibitor attenuated the induction of galectin-3 in glioblastoma cells (Dumic et al., 2000). The regulation of galectin-3 expression through the NF-kB transcription factor was shown to be mediated by nucling, a novel apoptosis associated protein, which interferes with NF-kB via the nuclear translocation process of NF-kB/p65, thus inhibiting galectin-3 expression at both the protein and mRNA levels (Liu et al., 2004).
ALG-2 linked protein × (Alix) or ALG-2 interacting protein-1 (AIP-I) is a galectin-3 binding cytoplasmic protein in Jurkat cells (Liu et al., 2002), which interacts with the cell death related calcium binding protein ALG-2 (Missotten et al., 1999; Vito et al., 1999), which has recently been reported to inhibit paraptosis, a form of programmed cell death (Sperandio et al., 2004). Alix/AIP-1 contains a proline glycine alanine and tyrosine-rich sequence in the N terminal region which is highly homologous to the tandem repeat sequence in the N-terminal part of galectin-3.
Bcl2 translocation to the mitochondrial membrane leads to anti apoptotic activity resulting from blocking cytochrome c release (Burlacu, 2003). It was reported that galectin-3 can also inhibit cytochrome c release followed by activation of the caspase cascade when it prevents nitric oxide induced apoptosis in human breast carcinoma BT-549 cells (Moon et al., 2001). Moreover, galectin-3 can bind to the Bcl-2 protein in vitro and might mimic the ability of the Bcl2 family members by forming heterodimers (Xu et al., 1995). Thus, galectin-3 might be a mitochondrial associated apoptotic regulator through interaction with Bcl-2 in the cytoplasm. It was also reported that synexin, a Ca+2 and a phospholipid binding protein, is required for galectin-3 prevention of mitochondrial damage followed by cytochrome c release after treatment of BT-549 cells with cisplatin (Yu et al., 2002). Furthermore, galectin-3 failed to translocate to the perinuclear membranes and exert antiapoptic effects, when the expression of synexin was down-regulated (Yu et al., 2002).
Galectin-3 is not a member of the Bcl2 family, but it shares significant structural properties with Bcl-2. Both proteins are rich in proline, glycine, and alanine amino acid residues in their N terminal and contain the Asp-Trp-Gly-Arg (NWGR) domain in the C terminal domain (Yang et al., 1996; Akahani et al., 1997). This motif designated as the anti-death motif is found in the BH-1 domain of Bcl2 and shown to be critical for the anti-apoptotic function of this protein (Hanada et al., 1995). This sequence is highly conserved among galectin-3 during evolution and is also essential for the carbohydrate-binding activity of this lectin (Yang et al., 1998). It was reported that an amino acid substitution of Gly to Ala in the NWGR motif of galectin-3 abrogated its apoptosis resistance properties (Akahani et al., 1997). This suggests that NWGR motif in galectin-3 is also equally significant for its anti-apoptotic activity as it is for Bcl2.
Role of galectin-3 in extrinsic apoptotic pathways
The induction of the extrinsic pathway is initiated by the binding of death receptors (Fas (apo-1 or CD95), tumor necrosis factor receptor-1 (TNFR-1/p55/CD120) and interferon and TRAIL (TNF related apoptosis inducing ligand or Apo2-L) receptors to their ligands in the plasma membrane of the cell (Ashkenazi and Dixit, 1998) (Schulze-Osthoff et al., 1998). The death receptors are trans-membrane proteins with cystein rich extra-cellular domains that interact with ligands. Based on receptor type there are two major signaling sub-types: the fas mediated signaling path and TRAIL mediated signaling path. Fas are glycosylated type-1 transmembrane receptors that either activate mitochondria dependent or mitochondria independent signaling pathway in response to ligand (Peter and Krammer, 1998; Nagata, 2000; Peter and Krammer, 2003). The selection of the pathway depends upon the amount of active caspase 8 produced at the death inducing signaling complex (DISC) (Scaffidi et al., 1998). The mitochondria independent pathway is activated by the DISC prior to loss of mitochondrial trans-membrane potential, which is characterized by a very efficient recruitment of both the Fas-associating protein with the death domain (FADD) and caspase 8 to CD95, leading to DISC formation. This step is dependent on the availability of F-actin. The DISC for mation in the mitochondria-dependent pathway forms very inefficiently without the involvement of F-actin (Algeciras-Schimnich et al., 2002) and is triggered by activated caspase 8 through the cleavage of the c terminal fragment of the BH3 only member of the Bcl-2 family from protein Bid to truncated Bid (tBid), which translocates to the outer membrane of the mitochondria, allowing the loss of mitochondrial trans-membrane potential and inducing cytochrome c release. Thus, Bid mediates the cross talk from the extrinsic to the intrinsic form of cell death (Li et al., 1998; Luo et al., 1998).
It was shown recently, that a major difference between the mitochondria independent (type I) and dependent (type II) pathways is the presence of endogenous galectin-3. Type I T-cell lymphoma cell lines SKW6.4 or H9 cells express high endogenous levels of galectin-3, whereas type II cells like Jurkat or CEM cells do not. In these cells CD95 apoptotic signaling pathway in both type I and type II cells is determined partially by endogenous galectin-3. In addition, over-expression of galectin-3 in type II CEM cells results in binding to the CD95 receptor and change the pattern of signaling pathway similar to the type I (Fukumori et al., 2004). Surprisingly, when galectin-3 was added to these cells exogenously, there was an induction of apoptosis, which varied from type I to type II cells. The levels of endogenous galectin-3 also affected the way a particular T cell lymphoma cell line reacted to the extra-cellular galectin-3. It was found that galectin-3 null T cells like Jurkat, CEM and MOLT-4 cells were significantly more sensitive to exogenous galectin-3 compared to SKW6.4 and H9 cells, which have very high expression of endogenous galectin-3. These differences might be caused from a balance between the anti-apoptotic activity of intracellular galectin-3 and pro-apoptotic activity of extra-cellular galectin-3. Extra-cellular galectin-3 can bind to the cell surface glycoproteins CD29 and CD7 in a CD29/CD7 complex, which triggers the activation of intracellular apoptotic signaling toward the mitochondria leading to cytochrome c release followed by activation of caspase 3 and apoptosis. Addition of neutralizing antibody against CD7 and/or CD29 markedly inhibited apoptosis induced by extra-cellular galectin-3 (Fukumori et al., 2003).
A similar mitochondrial independent event appears to be triggered when TRAIL binds to a different family of death inducing receptors. TRAIL is a trans-membrane protein that functions by binding to two closely related death receptors, DR4 and DR5 (Chaudhary et al., 1997; Griffith and Lynch, 1998) which leads to cleavage and activation of caspase-8, resulting in Bid cleavage, a Bcl-2 inhibitory protein, triggering mitochondrial depolarization (Suliman et al., 2001). Akt, a serine/threonine kinase was assumed to be involved in resistance to TRAIL-mediated apoptosis observed in certain types of tumors. Over-expression of galectin-3 in human breast carcinoma cell line BT-549 promotes TRAIL-induced cytotoxicity in a way to suppress Akt activity by dephosphorylation (Lee et al., 2003). On the contrary, over-expression of galectin-3 in J82 human bladder carcinoma cells rendered them resistant to TRAIL induced apoptosis, whereas phosphatidylinositol3 kinase PI3K inhibitors (wortmannin and LY 294002) blocked the protecting effect of galectin-3. High levels of constitutively active Akt were seen in the galectin-3 over-expressing J82 cells suggesting that galectin-3 involves Akt as a modulator molecule in protecting bladder carcinoma cells from TRAIL induced apoptosis (Oka et al., 2005). It is reasonable to assume that observed inconsistencies are probably due to use of different cell lines. Our recent observations (unpublished) indicate that galectin-3 regulates Akt activity via phosphate and tension homologue deleted on chromosome 10 (PTEN).
Therapeutic implications
Despite the fact that some of the signaling pathways in which galectin-3 is involved, have been identified, the precise mechanism of regulation of apoptosis by galectin-3 remains elusive. However, its interactions with a variety of ligands indicate its importance in various biological functions. Many of these interactions can be inhibited by specific carbohydrate inhibitors. In fact, peptides specific to galectin-3 carbohydrate recognition domain significantly inhibit the adhesion of a human breast carcinoma cell line to endothelial cells in vitro (Zou et al., 2005). In addition, the galectin-3 C-terminal domain fragment significantly suppresses tumor growth and inhibits metastasis in a mouse model of human breast cancer (John et al., 2003). Modified citrus pectin (MCP), which is modified from a natural plant polysaccharide, citrus pectin and is a competitive inhibitor of galectin-3, has been reported to inhibit galectin-3 mediated cellular functions (Inohara and Raz, 1994; Nangia-Makker et al., 2002). Injection of MCP treated mouse melanoma cells B16 F1 into mice (Platt and Raz, 1992) or the oral administration of MCP to male Copenhagen rats injected with prostate cancer cell line MAT-LyLu (Pienta et al., 1995) reduced spontaneous lung colonization. When MCP was fed to the mice injected with breast cancer cells MDA-MB-435 or colon carcinoma cells LSLiM6, there was a reduced tumor growth as well as metastasis (Nangia-Makker et al., 2002). In none of these studies apoptosis of the tumor cells was analyzed. It is assumed that MCP inhibits the formation of tumor cell emboli in blood circulation, as a result the tumor cells die and are not able to successfully metastasize. Whether this cell death is because of apoptosis, or immune surveillance, is not easy to detect. Another possibility for cell death in the blood circulation is their failure to adhere to the extra cellular matrix, and under this condition, they may die as a result of anoikis. We have shown that over-expression of galectin-3 protects cells from anoikis (Kim et al., 1999). Moreover, it was reported that rats fed on a 15% citrus pectin enriched diet showed a higher apoptotic index in their colon (Avivi-Green et al., 2000). In another study, treatment of human colonic adenocarcinoma cell line HT29 with dietary pectin and its degradation products (pectic ologisaccharides) resulted in reduced proliferation and increased caspase 3 activity and DNA laddering (Olano-Martin et al., 2003).
It was found that galectin-3 expression resulted in enhanced adhesion to laminin, fibronectin and vitronectin (Ochieng et al., 1998; Matarrese et al., 2000). Since increased cell adhesion is known to protect cells from apoptosis, the resistance to apoptosis resulting from galectin-3 over-expression could be due to increased cell adhesion. MCP interferes with the adhesion of galectin-3 or galectin-3 expressing cells to extra-cellular matrix proteins (Inohara and Raz, 1994; Nangia-Makker et al., 2002), thus leading the cells to apoptosis. Although these results do not provide a complete picture, they do emphasize the emergence of galectin-3 as a promising molecular target for inhibition of apoptosis. Specific low molecular weight inhibitors of galectin-3, or a sub-fraction of MCP with low molecular weight which could easily enter the cell and block its carbohydrate binding or other functions, could give a better understanding of its functions and clinical implications.
Acknowledgements
This article was written while the author (A.R.) was supported by NIH grant R37CA46120-19.
Bibliography
- Akahani S, Nangia-Makker P, Inohara H, Kim HR, Raz A. Cancer Res. 1997;57(23):5272–5276. [PubMed] [Google Scholar]
- Algeciras-Schimnich A, Shen L, Barnhart BC, Murmann AE, Burkhardt JK, Peter ME. Mol Cell Biol. 2002;22(1):207–220. doi: 10.1128/MCB.22.1.207-220.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appella E, Anderson CW. Pathol Biol (Paris) 2000;48(3):227–245. [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM. Science. 1998;281(5381):1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- Avivi-Green C, Madar Z, Schwartz B. Int J Mol Med. 2000;6(6):689–698. doi: 10.3892/ijmm.6.6.689. [DOI] [PubMed] [Google Scholar]
- Barondes SH, Cooper DN, Gitt MA, Leffler H. J Biol Chem. 1994;269(33):20807–20810. [PubMed] [Google Scholar]
- Breckenridge DG, Xue D. Curr Opin Cell Biol. 2004;16(6):647–652. doi: 10.1016/j.ceb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Burlacu A. J Cell Mol Med. 2003;7(3):249–257. doi: 10.1111/j.1582-4934.2003.tb00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchinelli B, Lavra L, Rinaldo C, Iacovelli S, Gurtner A, Gasbarri A, Ulivieri A, Del Prete F, Trovato M, Piaggio G, Bartolazzi A, Soddu S, Sciacchitano S. Mol Cell Biol. 2006;26(12):4746–4757. doi: 10.1128/MCB.00959-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J, Hood L. Immunity. 1997;7(6):821–830. doi: 10.1016/s1074-7613(00)80400-8. [DOI] [PubMed] [Google Scholar]
- Chowdhury I, Tharakan B, Bhat GK. Cell Mol Biol Lett. 2006;11(4):506–525. doi: 10.2478/s11658-006-0041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles EA, Agrwal N, Anderson RL, Wang JL. J Biol Chem. 1990;265(29):17706–17712. [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell. 2004;116(2):205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Delhalle S, Blasius R, Dicato M, Diederich M. Ann N Y Acad Sci. 2004;1030:1–13. doi: 10.1196/annals.1329.002. [DOI] [PubMed] [Google Scholar]
- D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, Piaggio G, Fanciulli M, Appella E, Soddu S. Nat Cell Biol. 2002;4(1):11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- Dumic J, Dabelic S, Flogel M. Biochim Biophys Acta. 2006;1760(4):616–635. doi: 10.1016/j.bbagen.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Dumic J, Lauc G, Flogel M. Cell Physiol Biochem. 2000;10(3):149–158. doi: 10.1159/000016345. [DOI] [PubMed] [Google Scholar]
- Elad-Sfadia G, Haklai R, Balan E, Kloog Y. J Biol Chem. 2004;279(33):34922–34930. doi: 10.1074/jbc.M312697200. [DOI] [PubMed] [Google Scholar]
- Ferri KF, Kroemer G. Bioessays. 2001;23(2):111–115. doi: 10.1002/1521-1878(200102)23:2<111::AID-BIES1016>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Fukumori T, Takenaka Y, Oka N, Yoshii T, Hogan V, Inohara H, Kanayama HO, Kim HR, Raz A. Cancer Res. 2004;64(10):3376–3379. doi: 10.1158/0008-5472.CAN-04-0336. [DOI] [PubMed] [Google Scholar]
- Fukumori T, Takenaka Y, Yoshii T, Kim HR, Hogan V, Inohara H, Kagawa S, Raz A. Cancer Res. 2003;63(23):8302–8311. [PubMed] [Google Scholar]
- Gong HC, Honjo Y, Nangia-Makker P, Hogan V, Mazurak N, Bresalier RS, Raz A. Cancer Res. 1999;59(24):6239–6245. [PubMed] [Google Scholar]
- Green D, Kroemer G. Trends Cell Biol. 1998;8(7):267–271. doi: 10.1016/s0962-8924(98)01273-2. [DOI] [PubMed] [Google Scholar]
- Griffith TS, Lynch DH. Curr Opin Immunol. 1998;10(5):559–563. doi: 10.1016/s0952-7915(98)80224-0. [DOI] [PubMed] [Google Scholar]
- Hanada M, Aime-Sempe C, Sato T, Reed JC. J Biol Chem. 1995;270(20):11962–11969. doi: 10.1074/jbc.270.20.11962. [DOI] [PubMed] [Google Scholar]
- Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Nat Cell Biol. 2002;4(1):1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- Hsu DK, Hammes SR, Kuwabara I, Greene WC, Liu FT. Am J Pathol. 1996;148(5):1661–1670. [PMC free article] [PubMed] [Google Scholar]
- Huflejt ME, Turck CW, Lindstedt R, Barondes SH, Leffler H. J Biol Chem. 1993;268(35):26712–26718. [PubMed] [Google Scholar]
- Hughes RC. Biochim Biophys Acta. 1999;1473(1):172–185. doi: 10.1016/s0304-4165(99)00177-4. [DOI] [PubMed] [Google Scholar]
- Inohara H, Raz A. Glycoconj J. 1994;11(6):527–532. doi: 10.1007/BF00731303. [DOI] [PubMed] [Google Scholar]
- John CM, Leffler H, Kahl-Knutsson B, Svensson I, Jarvis GA. Clin Cancer Res. 2003;9(6):2374–2383. [PubMed] [Google Scholar]
- Kim HR, Lin HM, Biliran H, Raz A. Cancer Res. 1999;59(16):4148–4154. [PubMed] [Google Scholar]
- Lee YJ, Song YK, Song JJ, Siervo-Sassi RR, Kim HR, Li L, Spitz DR, Lokshin A, Kim JH. Exp Cell Res. 2003;288(1):21–34. doi: 10.1016/s0014-4827(03)00211-8. [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. Cell. 1998;94(4):491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- Liu FT, Patterson RJ, Wang JL. Biochim Biophys Acta. 2002;1572(2–3):263–273. doi: 10.1016/s0304-4165(02)00313-6. [DOI] [PubMed] [Google Scholar]
- Liu L, Sakai T, Sano N, Fukui K. Biochem J. 2004;380(Pt 1):31–41. doi: 10.1042/BJ20031300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Cell. 1998;94(4):481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- Matarrese P, Fusco O, Tinari N, Natoli C, Liu FT, Semeraro ML, Malorni W, Iacobelli S. Int J Cancer. 2000;85(4):545–554. [PubMed] [Google Scholar]
- Missotten M, Nichols A, Rieger K, Sadoul R. Cell Death Differ. 1999;6(2):124–129. doi: 10.1038/sj.cdd.4400456. [DOI] [PubMed] [Google Scholar]
- Mitin N, Rossman KL, Der CJ. Curr Biol. 2005;15(14):R563–574. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Moon BK, Lee YJ, Battle P, Jessup JM, Raz A, Kim HR. Am J Pathol. 2001;159(3):1055–1060. doi: 10.1016/S0002-9440(10)61780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsatsos IK, Wade M, Schindler M, Wang JL. Proc Natl Acad Sci USA. 1987;84(18):6452–6456. doi: 10.1073/pnas.84.18.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S. Exp Cell Res. 2000;256(1):12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- Nangia-Makker P, Hogan V, Honjo Y, Baccarini S, Tait L, Bresalier R, Raz A. J Natl Cancer Inst. 2002;94(24):1854–1862. doi: 10.1093/jnci/94.24.1854. [DOI] [PubMed] [Google Scholar]
- Ochieng J, Fridman R, Nangia-Makker P, Kleiner DE, Liotta LA, Stetler-Stevenson WG, Raz A. Biochemistry. 1994;33(47):14109–14114. doi: 10.1021/bi00251a020. [DOI] [PubMed] [Google Scholar]
- Ochieng J, Leite-Browning ML, Warfield P. Biochem Biophys Res Commun. 1998;246(3):788–791. doi: 10.1006/bbrc.1998.8708. [DOI] [PubMed] [Google Scholar]
- Oka N, Nakahara S, Takenaka Y, Fukumori T, Hogan V, Kanayama HO, Yanagawa T, Raz A. Cancer Res. 2005;65(17):7546–7553. doi: 10.1158/0008-5472.CAN-05-1197. [DOI] [PubMed] [Google Scholar]
- Olano-Martin E, Rimbach GH, Gibson GR, Rastall RA. Anti-cancer Res. 2003;23(1A):341–346. [PubMed] [Google Scholar]
- Patterson SD, Spahr CS, Daugas E, Susin SA, Irinopoulou T, Koehler C, Kroemer G. Cell Death Differ. 2000;7(2):137–144. doi: 10.1038/sj.cdd.4400640. [DOI] [PubMed] [Google Scholar]
- Peter ME, Krammer PH. Curr Opin Immunol. 1998;10(5):545–551. doi: 10.1016/s0952-7915(98)80222-7. [DOI] [PubMed] [Google Scholar]
- Peter ME, Krammer PH. Cell Death Differ. 2003;10(1):26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, Naik H, Akhtar A, Yamazaki K, Replogle TS, Lehr J, Donat TL, Tait L, Hogan V, Raz A. J Natl Cancer Inst. 1995;87(5):348–353. doi: 10.1093/jnci/87.5.348. [DOI] [PubMed] [Google Scholar]
- Platt D, Raz A. J Natl Cancer Inst. 1992;84(6):438–442. doi: 10.1093/jnci/84.6.438. [DOI] [PubMed] [Google Scholar]
- Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, Debatin KM, Krammer PH, Peter ME. Embo J. 1998;17(6):1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Osthoff K, Ferrari D, Los M, Wesselborg S, Peter ME. Eur J Biochem. 1998;254(3):439–459. doi: 10.1046/j.1432-1327.1998.2540439.x. [DOI] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Cooks T, Raz A, Kloog Y. Cancer Res. 2005;65(16):7292–7300. doi: 10.1158/0008-5472.CAN-05-0775. [DOI] [PubMed] [Google Scholar]
- Sperandio S, Poksay K, de Belle I, Lafuente MJ, Liu B, Nasir J, Bredesen DE. Cell Death Differ. 2004;11(10):1066–1075. doi: 10.1038/sj.cdd.4401465. [DOI] [PubMed] [Google Scholar]
- Suliman A, Lam A, Datta R, Srivastava RK. Oncogene. 2001;20(17):2122–2133. doi: 10.1038/sj.onc.1204282. [DOI] [PubMed] [Google Scholar]
- Takenaka Y, Fukumori T, Raz A. Glycoconj J. 2004;19(7–9):543–549. doi: 10.1023/B:GLYC.0000014084.01324.15. [DOI] [PubMed] [Google Scholar]
- Vito P, Pellegrini L, Guiet C, D'Adamio L. J Biol Chem. 1999;274(3):1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- Vousden KH. Cell. 2000;103(5):691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Xu XC, el-Naggar AK, Lotan R. Am J Pathol. 1995;147(3):815–822. [PMC free article] [PubMed] [Google Scholar]
- Yang RY, Hill PN, Hsu DK, Liu FT. Biochemistry. 1998;37(12):4086–4092. doi: 10.1021/bi971409c. [DOI] [PubMed] [Google Scholar]
- Yang RY, Hsu DK, Liu FT. Proc Natl Acad Sci USA. 1996;93(13):6737–6742. doi: 10.1073/pnas.93.13.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Fukumori T, Honjo Y, Inohara H, Kim HR, Raz A. J Biol Chem. 2002;277(9):6852–6857. doi: 10.1074/jbc.M107668200. [DOI] [PubMed] [Google Scholar]
- Yu F, Finley RL, Jr, Raz A, Kim HR. J Biol Chem. 2002;277(18):15819–15827. doi: 10.1074/jbc.M200154200. [DOI] [PubMed] [Google Scholar]
- Zou J, Glinsky VV, Landon LA, Matthews L, Deutscher SL. Carcinogenesis. 2005;26(2):309–318. doi: 10.1093/carcin/bgh329. [DOI] [PubMed] [Google Scholar]