Abstract
Proteomic characterization of protein complexes leverages the versatile platform of liquid chromatography-tandem mass spectrometry to elucidate molecular and cellular signaling processes underlying the dynamic regulation of macromolecular assemblies. Here, we describe a complementary proteomic approach optimized for immunoisolated protein complexes. As the relative complexity, abundance, and physiochemical properties of proteins can vary significantly between samples, we have provided (1) complementary sample preparation workflows, (2) detailed steps for HPLC and mass spectrometric method development, and (3) a bioinformatic workflow that provides confident peptide/protein identification paired with unbiased functional gene ontology analysis. This protocol can also be extended for characterization of larger complexity samples from whole cell or tissue Xenopus proteomes.
Keywords: Xenopus, Proteomics, Protein complex, Protein interactions, In-gel digestion, In-solution digestion, Mass spectrometry, HPLC, Bioinformatics
1. Introduction
Technological advances in sample preparation and instrumentation for application of mass spectrometry-based proteomics to biological samples has permitted an increasing depth of proteome coverage. For example, biochemical and analytical separation strategies, such as multidimensional chromatography (1, 2) and affinity enrichment (3, 4), have seen significant advances that include specific optimization for downstream mass spectrometry analysis. These methods are complemented by continued development of nanoscale liquid chromatography, affording improved separation efficiency (5), as well as mass spectrometry technology that provides improved mass accuracy, resolution, and ion transmission and fragmentation (6, 7). Collectively, these achievements have enabled proteomic analysis to answer diverse biological questions, ranging from large scale, discovery-based experiments (8) to targeted, hypothesis-driven studies (9–11).
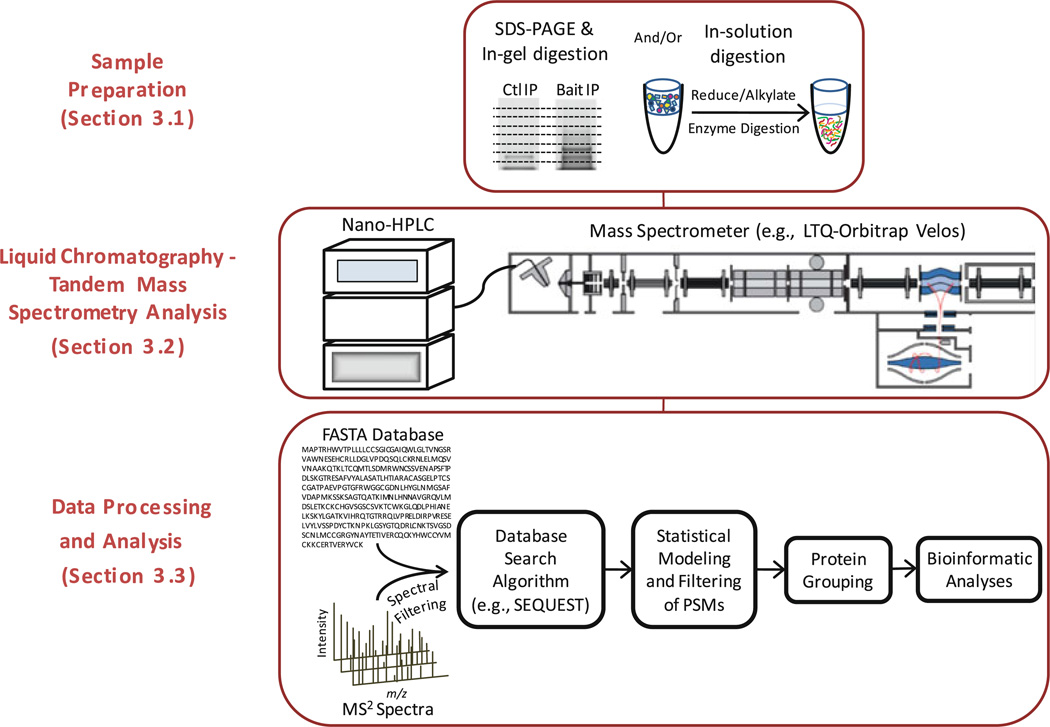
Leveraging several of these technologies, this chapter describes a mass spectrometry-based proteomics workflow (Fig. 1), tailored towards the analysis of protein complexes and assemblies obtained from affinity purification strategies ((12); see Chapter 21). Complementary sample preparation strategies are described (Subheading 3.1); a gel-based approach, which is readily used for optimization experiments and advantageous for complex samples (e.g., >1,000 proteins) of high dynamic range, and a solution-based approach that employs a single dimension of separation, providing greater reproducibility and optimal for lower complexity and sample amounts (13). Peptide analysis is performed by reverse phase nanoscale liquid chromatography coupled online to tandem mass spectrometry (Subheading 3.2). Data processing and analysis comprise the greatest overall proportion (based on time) of the workflow (Subheading 3.3). This multi-step process involves filtering of MS/MS spectra, matching of experimental spectra to theoretical spectra generated from in silico digestion of a protein sequence database. Next, database search software provides putative Peptide Spectrum Matches (PSMs). Then, post-search validation algorithms calculate p-values for all PSMs (i.e., the likelihood of an incorrect match by chance) as well as q-values, which allow filtering of PSMs to achieve a desired global false discovery rate (FDR). Peptides passing these criteria are then assembled into protein groups, requiring a minimum number of unique peptides (e.g., 2) to define a protein group. Finally, bioinformatic and data reduction strategies are presented to aid in transitioning from hypothesis-generating to hypothesis-driven studies. Overall, these strategies are applicable not only to affinity-purified samples, but to a broad range of cellular and organism proteomes.
Fig. 1.
Workflow summary for proteomic analysis of immunoisolated protein complexes; diagram of LTQ-Orbitrap Velos from Thermo Fisher Scientific.
2. Materials, Equipment, and Instrumentation
2.1. Sample Preparation for Mass Spectrometry
2.1.1. SDS-PAGE and In-Gel Protein Digestion
Electrophoresis and protein staining
Primary eluate from immunoaffinity purification (see Chapter 21, Subheading 3.3.2).
NuPAGE 4–12% Bis-Tris pre-cast SDS-PAGE gel, 10 well (Invitrogen).
Xcell SureLock Mini-Cell electrophoresis system (Invitrogen).
20× NuPAGE MOPS SDS Running Buffer (Invitrogen).
1× Running Buffer: Dilute 20× NuPAGE MOPS SDS Running Buffer to 1× in 700 mL of ultrapure water.
Precision Plus Protein Dual Color Molecular Weight Standards (BioRad).
4× NuPAGE LDS Sample Buffer (Invitrogen).
10× NuPAGE Sample Reducing Reagent (Invitrogen).
1× LDS Sampler Buffer: Mix 50 µL of 4× NuPAGE LDS Sampler buffer, 20 µL of 10× NuPAGE Sample Reducing Reagent, and 130 µL of ultrapure water.
SimplyBlue SafeStain (Invitrogen).
In-gel digestion and peptide extraction
Store stock solutions in glass containers that have been thoroughly rinsed with ultrapure water. Avoid using glassware that has been washed with detergents.
SimplyBlue SafeStain (Invitrogen).
Ultrapure dH2O (e.g., from a Milli-Q Purification System).
Rocking platform.
Sheet protector and Scanner (Officemax).
Solution basins (Fisher).
Axygen Maxymum Recovery microcentrifuge tubes, 1.5 mL (VWR).
Axygen 96-well plates with sealing mat (VWR).
Multichannel pipet (Fisher).
Non-autoclaved, low binding pipet tips, 200 µL (VWR).
Ceramic Plate, Forceps, Razor Blade, Windex.
Mickle Gel Slicer (Brinkmann).
Lens Paper (Fisher).
Water, HPLC grade (Fisher).
Acetonitrile (ACN), HPLC grade (Fisher).
Formic acid stock (FA), LC-MS grade, 99+ % (Pierce).
1% FA solution: Mix 1.0 mL of FA stock in 99.0 mL ultrapure water. Store at RT.
0.1 M ammonium bicarbonate (ABC): Dissolve 0.80 g of ABC solid in 0.1 L of HPLC grade water. Store at RT and use within 1 month, or sterile filter for long-term storage.
Destain solution: Mix 2 mL of ACN with 2 mL of 0.1 M ABC. Prepare fresh before use in a solution basin.
Rehydration solution: Mix 2.5 mL of ultrapure water with 2.5 mL of 0.1 M ABC. Prepare fresh before use in solution basin.
0.5 µg/µL Trypsin stock, sequencing grade (Promega). Store at −80°C, limit to <5 freeze/thaw cycles.
12.5 ng/µL Trypsin solution: Mix 8.5 µL of trypsin stock with 331.5 µL of rehydration solution. Prepare fresh immediately before use (step 16).
Vortex device with an adaptor for 96-well plates (Fisher).
Glass autosampler vials, MS Certified with 200 µL fused inserts and pre-slit caps (National Scientific).
50% ACN/0.5% FA solution. Mix 0.5 mL of ACN and 0.5 mL of FA solution (1%) in microfuge tube. Prepare fresh on Day 3.
Savant SpeedVac Concentrator (Thermo Scientific).
2.1.2. In-Solution Digestion (Alternative Workflow)
Unless otherwise stated, store solutions at RT in glass containers that have been thoroughly rinsed with ultrapure water.
Primary eluate from immunoaffinity purification (see Chapter 21, Subheading 3.3.2).
HPLC grade water (Fisher).
Dithiothreitol (DTT), solid (Invitrogen). Store at −20°C.
Tris-base, solid (Fisher).
Urea, solid (Sigma).
Trifluoroacetic acid (TFA), LC-MS grade (Fisher). Always use a glass syringe to pipet concentrated acid stocks.
Iodoacetamide (IAA), solid (Sigma). Store at 4°C.
1 M DTT: Dissolve 0.77 g DTT in 5 mL of HPLC grade water. Aliquot and store at −80°C.
0.2 M Tris–HCl, pH 8.0: Dissolve in 1.21 g Tris-base in 40 mL of HPLC grade water, pH to 8.0, and dilute to 50 mL. Filter sterilize.
0.05 M ABC: Dissolve 0.40 g of ABC solid in 0.1 L of HPLC grade water. Sterile filter, store at RT and use within 3 months.
1 M IAA (Sigma): Dissolve 0.185 g of IAA in 1 mL HPLC grade water. Dispense into 50 × 20 µL aliquots and store at −20°C.
50% ACN in HPLC grade water.
0.5 µg/µL Trypsin stock, sequencing grade (Promega). Store at −80°C, keep freeze/thaw cycles <5.
10% TFA. Mix 1 mL of 100% TFA with 9 mL of HPLC grade water. Store at 4°C.
Non-autoclaved, low binding pipet tips, 200 µL (VWR).
Amicon-500, 10 kDa ultra filtration devices (Millipore).
Refrigerated microcentrifuge capable of 14,000 × g (maintain at 20°C).
Eppendorf tube shaker (e.g., Tomy shaker).
Savant SpeedVac Concentrator (Thermo Scientific).
8 M Urea buffer (UB): Dissolve 0.48 g urea in 160 µL HPLC water and 0.5 mL 0.2 M Tris–HCl, pH, 8.0. Rock gently until dissolved. Yields 1 mL of solution, sufficient for one sample. Keep solution at RT. Do not heat urea-containing solutions above 37°C.
100 mM IAA solution: Prepare 200 µL of solution/sample by mixing 180 µL of UB with 20 µL of 1 M IAA.
5 ng/µL Trypsin solution: Prepare 100 µL/sample by mixing 10 µL of 0.5 µg/µL trypsin stock with 90 µL of 0.05 M ABC. Prepare fresh immediately before use (step 8).
Glass autosampler vials, MS Certified with 200 µL fused inserts and pre-slit caps (National Scientific).
2.2. Liquid Chromatography-Tandem Mass Spectrometry Analysis
2.2.1. NanoLC Configuration and Peptide Separation
Unless otherwise stated, all solutions should use LC-MS grade solvents.
Optima LC-MS grade solvents: water, ACN, and TFA (Fisher).
Formic acid (FA), LC-MS grade, 99+ % (Pierce).
Graduated glass cylinders, 100 and 500 mL.
Glass syringe with needle, 0.5 mL (Hamilton).
Nanoflow HPLC system, e.g., Dionex Ultimate, Waters nano Acuity, or Agilent 1200 series (see Note 1).
Mobile phase A (MPA): 0.1% FA/99.9% water. Dilute 0.5 mL FA with water to final volume of 0.5 L in a glass graduated cylinder. Transfer to amber bottle and store for up to 6 months (see Note 2).
Mobile phase B (MPB): 0.1% FA/97% ACN/2.9% water. Mix 0.5 mL FA and 14.5 mL water. Dilute with ACN to final volume of 0.5 L in a glass graduated cylinder. Transfer to amber bottle and store for up to 6 months (see Note 2).
Mobile phase C (MPC): 0.5% TFA/1% ACN/98.5% water. Mix 1.5 mL TFA and 3.0 mL ACN. Dilute with water to final volume of 0.3 L in a glass graduated cylinder. Prepare fresh and transfer to HPLC solvent bottle. Replace monthly.
Self-Pack IntegraFrit Columns, 360 µm OD × 100 µm ID (New Objective).
Micro Tool Kit (New Objective).
Trap column cartridge assembly (Upchurch).
Polyimide-coated fused silica capillary tubing (Upchurch).
Trap column, 360 µm × 100 µm ID × 2.5 cm. Construct by packing reverse phase resin (Magic C18 AQ, 3 µm, Michrom Bioresources) into Self-Pack IntegraFrit columns, cleaving to 2.5 cm length, and inserting into Trap column cartridge assembly (see Note 3).
Analytical column, 75 µm ID × 25 cm, e.g., Acclaim PepMap RSLC (Dionex) (see Note 3).
2.2.2. Mass Spectrometry Configuration and Peptide Analysis
LTQ-Orbitrap Velos hybrid mass spectrometer (Thermo fisher Scientific) (see Note 4).
Nanospray ESI source (Thermo fisher Scientific).
SilicaTip Emitter, Tubing (OD × ID) 360 µm × 50 µm; Tip (ID) 10 µm (New Objective).
2.3. Data Processing and Analysis
Multi-core/multi-CPU 64-bit PC workstation with at least 12 GB of RAM and 2 TB of storage (see Note 5).
Software for generating peaklists and scoring of PSMs, e.g., Proteome Discoverer 1.3 (Thermo fisher Scientific), Mascot 2.3 (Matrix Science), Scaffold 3.0 (Proteome Software), or COMPASS (14) (see Note 6).
Spreadsheet software (e.g., Microsoft Excel).
3. Methods
3.1. Sample Preparation for Mass Spectrometry
3.1.1. SDS-PAGE and In-Gel Protein Digestion
This protocol is performed over the course of 3 days. To minimize keratin and other environmental contaminants, it is recommended to wear a lab coat and hair protection, to avoid close contact and limit environmental exposure of the pre-case gel during sample loading and protein staining.
Electrophoresis and protein staining (Day 1)
If necessary, thaw primary eluate sample(s) at 70°C.
- Set up the Xcell SureLock Mini-Cell electrophoresis system:
- Remove the white strip and the comb from the NuPAGE 4–12% Bis-Tris pre-cast SDS-PAGE gel and rinse with ultrapure water. Place the gel in the apparatus, using a buffer dam for the opposing side, then lock the assembly in place.
- Fill the inner chamber with 200 mL of 1× Running Buffer.
- Fill the outer chamber with 500 mL of 1× Running Buffer.
Load samples into wells as follows: Lane 2, 10 µL Molecular Weight Standards. Lane 4, 90% of the eluate (~40 µL) (see Note 7).
Load 20 µL 1× LDS Sample Buffer into all empty wells.
Electrophorese for 5 min at 100 V, then 15–25 min at 200 V, or until the dye front has migrated approximately one third of the way down the gel (see Note 8).
Open gel cassette to expose gel and discard the wells. Working with wet gloves, transfer gel (by the thick ridge at the bottom) into a plastic tray containing ultrapure water. Remove bottom ridge.
Wash the gel 3 × 5 min with ultrapure H2O while rocking.
Dispense SimplyBlue SafeStain to cover the gel completely. Incubate for 1–3 h until the protein bands become clearly visible. Pour off the solution and replace it with ultrapure water. Destain 1–2 h at room temperature or overnight at 4°C with gentle rocking (see Note 9).
In-gel digestion (Day 2)
Working solution volumes are calculated for 16 samples (based on 90 µL of solution/well except where noted). For higher throughput, a 96-well plate with sealing mat is recommended (see Note 10).
All solutions can be added/removed from the wells using a multichannel pipettor. For each sample set, use a different set of multichannel tips, though one set of tips can be used for all steps performed on Day 2.
Wash the destained gel several times with ultrapure water to remove excess staining reagent.
Place the gel in a sheet protector. Keep the gel wet with a little ultrapure water. Scan the gel, save the image, and print it out as it will serve as a reference during processing of gel slices (step 7).
Wash the ceramic plate, forceps, blade of the gel slicer, and a razor blade first with Windex then with ultrapure water.
Place the gel on the plate. Cut out the entire vertical lane that contains the sample. Trim lane to remove lower portion just at the dye front and the top 2 mm near the well.
Place a small piece of lens paper on the cutting stage of the Mickle Gel Slicer, wet the lens paper with ultrapure water, and flatten it out. Place the excised gel lane on the lens paper.
Cut the gel lane with the Mickle Gel Slicer to 1 mm slices (approximately 20–30 slices). Push down any gel pieces that get stuck to the gel slicer blade with a pair of tweezers.
Lift the lens paper and flip the cut gel lane back onto the ceramic plate. Carefully remove the lens paper. Keep cut gel slices slightly wet for ease of sample handling.
Group together 2–3 gel pieces, starting from the bottom of the gel, and cut each 1 mm slice into three equal pieces (see Note 11).
Transfer gel pieces using forceps or the flat side of razor blade into one well. Do not exceed 3 × 1 mm slices/well.
Add 90 µL of destain solution to each well, seal plate, and agitate on a vortexer (medium setting) at room temperature (RT) for 10 min. Aspirate and discard. For all aspiration steps, make sure the gel pieces are not stuck to pipet tips when discarding waste solution as sample loss could occur.
Repeat step 10 with fresh destain solution. While destaining, prepare and aliquot rehydration solution and ACN (5 mL each) in separate solution basins.
Aspirate the destain solution, add 90 µL ACN to the gel pieces, seal plate with mat, and invert manually several times. Let it stand at RT until the gel pieces are white (<5 min).
Remove ACN and add 90 µL of rehydration solution, seal the plate, and manually invert several times. Incubate at RT for <5 min, or until the gel pieces have swelled and are translucent. Aspirate rehydration solution.
Repeat dehydration/rehydration (steps 12/13) once.
Add 90 µL of ACN to dehydrate as previously described (step 12). Aspirate ACN and let residual ACN evaporate.
While ACN is evaporating, prepare trypsin solution (12.5 ng/µL). Mix gently and add 20 µL of trypsin solution to the dried gel pieces. Let the gel pieces swell (<5 min).
Add ~30 µL of rehydration solution to the gel pieces (use enough volume to ensure all gel pieces are fully covered). Seal the plate and incubate overnight at 37°C.
Peptide extraction (Day 3)
In the morning, add 30 µL 1% FA to each sample and incubate for 4 h at RT on bench. CRITICAL STEP: Transfer the extracted peptides to pre-labeled autosampler vials. Make sure gel pieces are not transferred into autosampler vials. Store vials at 4°C.
Add 30 µL of 50% ACN/0.5% FA solution to wash the gel pieces. Incubate for 2 h at RT on bench.
Transfer and combine the respective washes with their corresponding FA extracts from step 1.
Speedvac the samples to ~10–12 µL, avoiding concentration to dryness (see Note 12). If necessary, adjust volumes with 1% FA. Proceed to Subheading 3.2. Store at 4°C in autosampler vials if the sample will be analyzed for mass spectrometry analysis within the next 2 days, otherwise store at −20 or −80°C for long-term storage.
3.1.2. In-Solution Digestion (Alternative Workflow)
All procedures are performed at either RT (20°C) or higher (where indicated) to avoid precipitation of detergent or urea from solutions. For this purpose, preset the temperature of the microcentrifuge to 20°C. This protocol is performed over the course of 2 days.
In-solution digestion (Day 1)
If necessary, thaw eluate samples previously prepared by immunoaffinity purification (see Chapter 21, Subheading 3.3.2) by heating the sample at 70°C for 10 min.
Mix each eluate sample (max. 150 µg total protein in <100 µL) with 400 µL of UB in an ultra filtration device. Centrifuge at 14,000 × g for 30 min.
Discard flow-through. Add 400 µL of UB to the top of the filter unit and centrifuge at 14,000 × g for 20 min.
Discard the flow-through. Add 200 µL of IAA solution and mix on an eppendorf tube shaker, medium setting, for 1 min.
Incubate for 15 min at RT protected from light. Centrifuge the filter units at 14,000 × g for 15 min. Discard flow-through.
Add 200 µL of 0.05 M ABC to the top of the filter and centrifuge at 14,000 × g for 15 min.
Repeat step 6 an additional two times discarding flow-through after each centrifugation. Ensure that the retained volume is ~30 µL. During the centrifugation steps rinse a fresh collection tube with 200 µL 50% ACN and discard ACN.
Transfer filter units to the pre-rinsed collection tubes and add 100 µL of 5 ng/µL trypsin (see Note 13). Mix on a tube shaker at medium speed for 1 min.
Seal caps with parafilm and incubate in a water bath at 37°C for at least 4 h to overnight.
In-solution digestion (Day 2)
Centrifuge the filter units at 14,000 × g for 15 min.
Add 50 µL of HPLC water to top of filter unit and centrifuge at 14,000 × g for 10 min.
Repeat step 2.
Retain flow-through and discard filter unit. Transfer flow-through to an autosampler vial containing 22 µL of 10% TFA.
Concentrate samples by vacuum centrifugation until total volume is between 10 and 30 µL. Final volume will be dependent upon the expected peptide yield (see Note 14).
3.2. Liquid Chromatography-Tandem Mass Spectrometry Analysis
3.2.1. NanoLC Configuration and Peptide Separation
Recommended solvent configuration for dual-pump LC system: Connect nanopump (NP) A and B solvent lines to 100 mL of MPA and MPB, respectively. Replace MPA and MPB monthly with solvent from stock solution. Connect loading pump (LP) solvent line to 300 mL of MPC. The LP flow path carries the sample to the trap column, while the NP delivers solvents to either (1) the analytical column alone or (2) both the trap and analytical column. These two conditions can be achieved using a six-port, two-position valve. Before sample injection, the trap column is equilibrated in 100% MPC for 1 min at 5 µL/min. The analytical column mobile phase should be held at 96% MPA/4% MPB by the NP at a flow rate of 250 nL/min.
Using a 20 µL sample loop and 25 µL syringe, injection volumes between 5 and 10 µL should be attainable. Optimally, the total amount of peptides analyzed on-column should be ≤ 4 µg (≈ 2 µg provides optimal chromatography). On-column sample load should be achieved by keeping injection volumes constant, while adjusting the total sample volume.
Desalting: Upon injection, the sample is washed onto the trap column and desalted in 100% MPC for 5 min at 5 µL/min. All solvent is directed to waste. The analytical column should remain under the same conditions as in step 2.
Elution: Using a two-position valve, the trap column is placed in line with the analytical column. Desalted peptides bound to the trap column are eluted and resolved over the analytical column at 250 nL/min using an increasing percent MPB delivered by the NP. For samples from in-gel digestion, a 90 min, discontinuous linear elution gradient is performed as follows: 4–20% MPB over 50 min, followed by 20–40% MPB over 40 min. For insolution digestion samples, a 180 min continuous linear elution gradient is performed as follows: 4–40% MPB over 180 min.
Washing and Re-equilibration: Columns are washed for 5 min at 60% MPB, followed by 10 min at 97% MPB, and then re-equilibrated at 4% MPB for 15 min. The LC system is now ready to perform the next analysis. The total run time is calculated by adding the gradient time + 35 min (see Note 15).
Quality Controls: A standard, known sample should be injected at regular intervals to monitor LC and MS performance. Tryptic digests of a known protein, such as enolase, or a complex peptide mixture from whole cell lysate digestion can be used to monitor chromatographic reproducibility and overall depth of analysis. Also, between analyses of different biological samples, a “blank” or “wash” run should be conducted to minimize carryover of abundant analytes. Optionally, a high organic (60% ACN/0.5% TFA) solution can be injected to aid in removal of abundant analytes from the trap/analytical column.
3.2.2. Mass Spectrometry Configuration and Peptide Analysis
For global proteomic analysis of complex biological samples, data-dependent acquisition using collision-induced dissociation (CID) on a hybrid mass spectrometer, such as the LTQ-Orbitrap Velos, is the preferred method. The following steps summarize recommended instrument parameters for a hybrid LTQ-Orbitrap Velos mass spectrometer. While acquisition methods are often instrument-specific, the overall goal of the method should be to optimize the duty cycle or cycle time of the method, often de fined as the time between repeated full scan MS acquisition events or the time to accomplish a single round of peptide fragmentation events. For most nanoLC chromatographic separations, an optimal cycle time should average ≤4 s.
MS1 (Precursor Ion) Acquisition: Precursor ions are detected in the Orbitrap analyzer at a resolution of 60,000 at m/z = 400. It is recommended that the “Preview mode for FTMS master scan” is disabled, so that data-dependent selection of precursors is performed using the MS1 analytical scan collected at maximum resolution (see Note 16).
MS2 (Fragment Ion) Acquisition: In data-dependent experiments, precursor ions detected in the MS1 spectra are selected for fragmentation primarily as a function of intensity. A data-dependent CID Top 20 method is created, which selects the top 20 most intense precursor ions for acquisition of MS2 (tandem or MS/MS) CID spectra. To optimize MS2 acquisition, a maximum injection time of 100 ms, AGC target value of 5,000, and one microscan is recommended. To increase the dynamic range in abundance over which fragmentation spectra can be acquired, the dynamic exclusion function should be enabled (see Note 17). Using these settings, a single MS1 scan at 60,000 resolution and 20 tandem (MS2) scans, corresponding to a single acquisition cycle, can be completed in about 4 s.
Alternating MS1–MS2 (Top 20) cycles are repeated continuously throughout the LC gradient.
3.3. Data Processing and Analysis
To date, there are several well-developed software suites that process and analyze tandem mass spectra from large-scale, bottom-up proteomic experiments and provide a list of high confidence protein identifications. An overview of this workflow is presented, and for each stage of analysis a brief description is provided below. Although the spectral filters and algorithms may differ between software, the conceptual workflow is similar. For clarity, the individual steps described below use Proteome Discoverer and Scaffold as representative software.
Peptide spectrum matching by database searching
Database Creation: The UniProt knowledgebase of protein sequences in FASTA format are recommended as two levels of redundancy are available: the Swiss-Prot database, which is manually curated and reviewed, and the TrEMBL database which is automatically annotated and not reviewed (http://www.uniprot.org/downloads). For samples derived from well-annotated organisms, such as Homo sapiens, the Swiss-Prot sequences alone are adequate. For the other organisms, such as Xenopus, both the Swiss-Prot and TrEMBL sequences must be utilized. It is recommended that common contaminants, such as keratins, cell culture serum components, immunoglobulins, and digestion enzymes, be included in the database. For convenience, a custom database containing protein sequences from H. sapiens, Mus musculus, Xenopus laevis, and Xenopus tropicalis appended with common contaminants has been made available at http://www.xenbase.org.
Spectral Filtering: MS2 spectra should be filtered before submitting to a database search engine. While some software has built-in spectral filters (e.g., to remove noise), others allow specification of individual spectrum filtering. At minimum, spectra should be filtered based on minimum number of peaks ( N = 30) and intensity ( N = 200).
Sequence-to-Spectrum Matching: Putative PSMs are generated by submission of filtered spectra to the database search algorithm (e.g., SEQUEST or Mascot), which assigns scores based on the similarity between the experimental and theoretical tandem mass spectra. The search algorithm should be configured to specify the following parameters: precursor tolerance, 10 ppm; fragment tolerance, 0.5 Da; static modification, cysteine carbamidomethylation. Methionine oxidation and serine, threonine, and tyrosine phosphorylation should be specified as dynamic modifications.
Statistical Modeling of PSMs: Most search engine raw scores do not provide a direct statistical assessment of confidence, i.e., the probability of an incorrect match. To accurately estimate error rates for large-scale proteomic datasets, it is essential to utilize a post-processing probabilistic algorithm, such as Percolator (15) or PeptideProphet (16) to re-rank PSM scores (see Note 18). SEQUEST search results (*.msf files from Proteome Discoverer) are analyzed by Scaffold as follows: (a) Search result files from each sample are loaded into distinct biological samples. For ingel digestion, the MuDPIT option in Scaffold is used to combine search result files corresponding to multiple gel fractions from the biological sample. (b) Two additional features of Scaffold that should be routinely enabled are (i) an X! Tandem re-search with the subset database option and (ii) high mass accuracy scoring. The former conducts a second search against a subset database containing only those proteins identified in the initial SEQUEST search. (c) Additional dynamic modifications, such as deamidation of asparagine and glutamine, should be included for X! Tandem re-searches. Optionally, other biological or chemical modifications of interest not included in the primary SEQUEST search can be added, such as acetylation or ubiquitination of lysine. (d) After input spectra are scored by X! Tandem, both SEQUEST and X! Tandem PSMs are analyzed by PeptideProphet to calculate estimated local FDRs (probability of an incorrect match of a single PSM or protein). The global FDR (error rate within a group of PSMs or proteins) is estimated from the statistical modeling of PSM score distributions (see Note 19).
Protein level filtering and bioinformatics analysis
Scaffold protein identifications should be filtered, removing keratins and contaminant proteins (Accession prefix “CON_”).
Protein identifications are exported from Scaffold to a xls (tab-separated) format by the command: “Export,” “To Excel,” “Current View….” The desired export values (unique peptides, assigned spectra, etc.) can be controlled under “Display Options.” It is recommended that separate protein identification spreadsheets containing unique peptides and unweighted spectrum count are exported.
Assessment of Bait-Specific Protein Interactions: Using a spreadsheet program, open Excel file containing unweighted spectrum count values. Calculate fold enrichment in spectrum counts: Divide spectral counts for each protein in the bait IP by the corresponding spectral counts in the control IP (e.g., GFP). Proteins with ≥5 spectral counts in one condition and either unique to bait IP or ≥2.0–2.5-fold change in spectral counts are the best candidates for specific protein interactions (see Note 20).
- Bioinformatic Analyses: Identification of targets that are of biological interest can be pursued by two approaches
- Protein-Specific: Accession numbers for proteins of interest can be queried through protein knowledge bases such as UniProt (http://www.uniprot.org), Genecard (http://www.genecards.org/), or the Comprehensive Resource of Mammalian protein complexes (CORUM) (http://mips.helmholtz-muenchen.de/genre/proj/corum). This approach should be used with the caveat that it may lead to biased interpretation of the data.
- Global: Accession numbers for enriched proteins are submitted to functional enrichment software, which often converts accessions to gene-centric identifiers (e.g., gene symbols) for gene ontology and functional annotation mapping. The functional relationship between candidate interactions can be examined using STRING (http://www.string-db.org/). To examine over- or under-representation of functional annotations in a set of candidate interactions vs. a reference genome, the FatiGO tool within the Babelomics software suite can be used (http://babelomics.bioinfo.cipf.es/). Alternatively, a highly versatile open-source software platform called Cytoscape (http://www.cytoscape.org) can be used with functional enrichment plugins, such as BiNGO (http://www.psb.ugent.be/cbd/papers/BiNGO/Home.html) to perform both functional enrichment analyses and data visualization within a single software environment.
Footnotes
While many HPLC configurations can be used, the system should have the capacity for nanoliter per minute flow rates (ideally ≤500 nL/min). Also, the system should have dedicated nano and capillary pump capabilities that allow for shorter overall run times.
Mobile phases should be replenished from the stock solution monthly.
Many chemistries for column packing material are available and should be selected based on relative retention of desired analytes. For example, Magic C18 AQ resin was selected for the trap column for its increased retention of hydrophilic peptides, such as shorter length phosphopeptides. The Acclaim PepMap material was selected for the analytical column as it is available in sub-2 µm particle size. When selecting a column, instrument pressure limits should be noted as smaller particle sizes and longer columns increase operating pressures.
While several mass spectrometer configurations are adequate for these experiments, a hybrid instrument such as the LTQ-Orbitrap Velos offers (1) resolution up to 100,000, (2) <2 ppm mass accuracy (external calibration) in the FT Orbitrap detector, and (3) attomol sensitivity in the linear ion trap detector.
While a computing cluster is preferred, the high initial and maintenance costs often make this option impractical. As a second option, a multi-CPU or multi-core workstation can be purchased for about $5,000. If multi-threaded applications will be used (e.g., Mascot Server), then a dual CPU system should be considered. Otherwise, maximizing the overall processing speed of a single multi-core CPU should be the goal. For a more details on hardware specifications see http://www.matrixscience.com/help/pc_specs.html.
Currently there are many excellent options for open-source or commercial software suites that include database search engines. The most critical aspect when selecting a software suite or database search tool is the capability to accurately estimate probability values (e.g., q-values or posterior error probabilities) of individual PSMs. All software cited above satisfies this requirement.
If loading additional eluates, skip two lanes between the samples. Reserve the remaining eluate (10%) for western blot analysis.
For more complex samples or greater protein load, resolving proteins for entire gel length may be beneficial.
If necessary, the stained gel can be stored in ultrapure water at 4°C, wrapped tightly, up to 2–3 weeks until proceeding to ingel digestion, though some sample loss may occur.
Axygen microfuge tubes can be used as an alternative to a 96-well plate.
Do not mince gel slices as this increases likelihood of pipeting gel pieces into autosampler vials during extraction.
If samples are concentrated to dryness, add 30 µL of 50% ACN/0.5% FA solution, vortex briefly, and SpeedVac to reduce the volume as above.
Assuming an enzyme:protein ratio of 1:50–1:100, then the working solution of trypsin is sufficient for digestion of 25–50 µg of protein. If more protein is digested then increase concentration accordingly.
If peptide yield is known or can be estimated based on input material, concentrate sample to a final volume containing a peptide concentration of 0.5 µg/µL. Peptide concentration can be estimated by measuring A280 on a Nanodrop and dividing by 1.1 (extinction coefficient estimated based on the tryptophan frequency in mammalian proteins from a 0.1% protein solution).
Gradient and total run times provide a starting point for method development and should be optimized for specific LC hardware configurations, chromatographic material, and sample complexity.
For some instrument configurations, disabling the “preview scan” function may cause cycle times to routinely exceed 4 s. If this occurs, then decreasing resolution to 30,000 is recommended.
Recommended dynamic exclusion settings for a 90 min gradient are as follows: a repeat count of 1, repeat duration of 30, exclusion list size of 500, exclusion duration 70, and exclusion mass width 10 ppm. The exclusion duration should be adjusted based on chromatographic peak widths.
Percolator has been implemented in Mascot 2.3 and Proteome Discoverer 1.3, while PeptideProphet is available using either Scaffold (http://www.proteomesoftware.com) or the Trans-Proteomic Pipeline open-source software (http://tools.proteomecenter.org/wiki/index.php?title=Software:TPP).
PeptideProphet and ProteinProphet probability filters should be selected to reduce FDR to less than 1% at both the peptide and protein level. While confidence thresholds can vary between datasets, a general starting point is 99.0% protein probability, 95% peptide probability, and a minimum of 2 peptides/protein. For a more detailed discussion of error rate estimation see ref. (17).
Spectral counting analysis is a computationally simple method to evaluate which proteins may have large relative abundance differences between control and experimental samples. However, in the absence of biological replicates or independent validation assays, the statistical power of these analyses is limited and the results should be interpreted conservatively.
References
- 1.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Chang-Wong T, Tang HY, Speicher DW. Comparison of extensive protein fractionation and repetitive LC-MS/MS analyses on depth of analysis for complex proteomes. J Proteome Res. 2010;9(2):1032–1040. doi: 10.1021/pr900927y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 4.Fang X, Zhang WW. Affinity separation and enrichment methods in proteomic analysis. J Proteomics. 2008;71(3):284–303. doi: 10.1016/j.jprot.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Kocher T, Swart R, Mechtler K. Ultrahigh-pressure RPLC hyphenated to an LTQ-Orbitrap Velos reveals a linear relation between peak capacity and number of identified peptides. Anal Chem. 2010;83(7):2699–2704. doi: 10.1021/ac103243t. [DOI] [PubMed] [Google Scholar]
- 6.Hossain M, Kaleta DT, Robinson EW, Liu T, Zhao R, Page JS, et al. Enhanced sensitivity for selected reaction monitoring mass spectrometry-based targeted proteomics using a dual stage electrodynamic ion funnel interface. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M000062-MCP201. M000062–MCP000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, et al. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8(12):2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 9.Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics. 2010;9(5):851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of Scerevisiae by targeted proteomics. Cell. 2009;138(4):795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selimi F, Cristea IM, Heller E, Chait BT, Heintz N. Proteomic studies of a single CNS synapse type: the parallel fiber/Purkinje cell synapse. PLoS Biol. 2009;7(4):e83. doi: 10.1371/journal.pbio.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greco TM, Yu F, Guise AJ, Cristea IM. Nuclear import of histone deacetylase 5 by requisite nuclear localization signal phosphorylation. Mol Cell Proteomics. 2011;10(2) doi: 10.1074/mcp.M110.004317. M110.004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer T, Greco TM, Enquist LW, Cristea IM. Proteomic characterization of pseudorabies virus extracellular virions. J Virol. 2011;85(13):6427–6441. doi: 10.1128/JVI.02253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ. COMPASS: a suite of pre- and post-search proteomics software tools for OMSSA. Proteomics. 2011;11(6):1064–1074. doi: 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 16.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 17.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics. 2010;73(11):2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]