Abstract
Recent advances with the type II clustered regularly interspaced short palindromic repeats (CRISPR) system promise an improved approach to genome editing. However, the applicability and efficiency of this system in model organisms, such as zebrafish, are little studied. Here, we report that RNA-guided Cas9 nuclease efficiently facilitates genome editing in both mammalian cells and zebrafish embryos in a simple and robust manner. Over 35% of site-specific somatic mutations were found when specific Cas/gRNA was used to target either etsrp, gata4 or gata5 in zebrafish embryos in vivo. The Cas9/gRNA efficiently induced biallelic conversion of etsrp or gata5 in the resulting somatic cells, recapitulating their respective vessel phenotypes in etsrpy11 mutant embryos or cardia bifida phenotypes in fautm236a mutant embryos. Finally, we successfully achieved site-specific insertion of mloxP sequence induced by Cas9/gRNA system in zebrafish embryos. These results demonstrate that the Cas9/gRNA system has the potential of becoming a simple, robust and efficient reverse genetic tool for zebrafish and other model organisms. Together with other genome-engineering technologies, the Cas9 system is promising for applications in biology, agriculture, environmental studies and medicine.
Keywords: CRISPR, Cas9, genome editing, knockin, conditional knockout
Introduction
Precise and efficient genome-engineering technologies, including the custom-made zinc-finger nucleases (ZFNs) and transcription activator-like effectors (TALENs), have been successfully applied in a variety of mammalian cells and model organisms, including fruit fly, zebrafish, frog, mouse, rat and pig1. A large body of investigations demonstrates that gene-specific alterations, including insertions or deletions (indels), can be generated via non-homologous end-joining (NHEJ) induced by ZFN- or TALEN-mediated double-stranded breaks (DSBs) in many mammalian cells and all model organisms tested. If a double- or single-stranded DNA donor is provided, precise nucleotide substitutions or insertions of up to ∼8.0 kb at or near the break site are achieved via homology-directed repair (HDR) in mammalian cells2,3,4,5,6 and in zebrafish embryos7, suggesting the room for improvement in target-specific gene knockin and knockout in vivo. Precise insertion of a reporter gene or mutant loxP (mloxP) site is especially important for investigating endogenous gene expression or generating conditional gene knockouts in model organisms such as zebrafish, in which gene targeting in the embryonic stem cells is unavailable7.
Recent advances in the study of prokaryotic clustered regularly interspaced short palindromic repeats (CRISPR) adaptive immune system provide an alternative genome editing approach. The CRISPR/CRISPR-associated (Cas) system protects bacteria and archaea from invading viruses and plasmids. Three major types of CRISPR have been categorized on the basis of locus organization and gene conservation8. Cas9 belongs to the best studied type II CRISPR/Cas system, and is thought to be the sole protein responsible for small interfering CRISPR RNA (crRNA)-guided silencing of foreign DNA9,10. A transactivating crRNA (tracrRNA) that is complementary to the repeat sequences in pre-crRNA is required for the silencing. Several studies have demonstrated that Cas9 guided by gRNA that comprises the fusion of crRNA to tracrRNA is sufficient to execute in vitro sequence-specific cleavage of target DNA11,12, and site-specific DNA cleavage in mammalian cells as reported most recently13,14. However, the applicability and efficiency of this system in model organisms, such as zebrafish, are little studied. Here, we report that the Cas9/gRNA system efficiently executes site-specific cleavage, leading to NHEJ-mediated indels and introduction of a small foreign DNA fragment into the host genome via HDR if a donor DNA is provided in mammalian cells or zebrafish embryos. Remarkably, the Cas9 nuclease-mediated DSBs efficiently cause biallelic conversion of etsrp or gata5 in the resulting somatic tissues, which results in similar phenotypes of their corresponding genetic mutants, etsrpy11 or fautm236a, in zebrafish15,16.
Results
Cas9/gRNA ribonucleoprotein complex executes site-specific DNA cleavage in mammalian cells
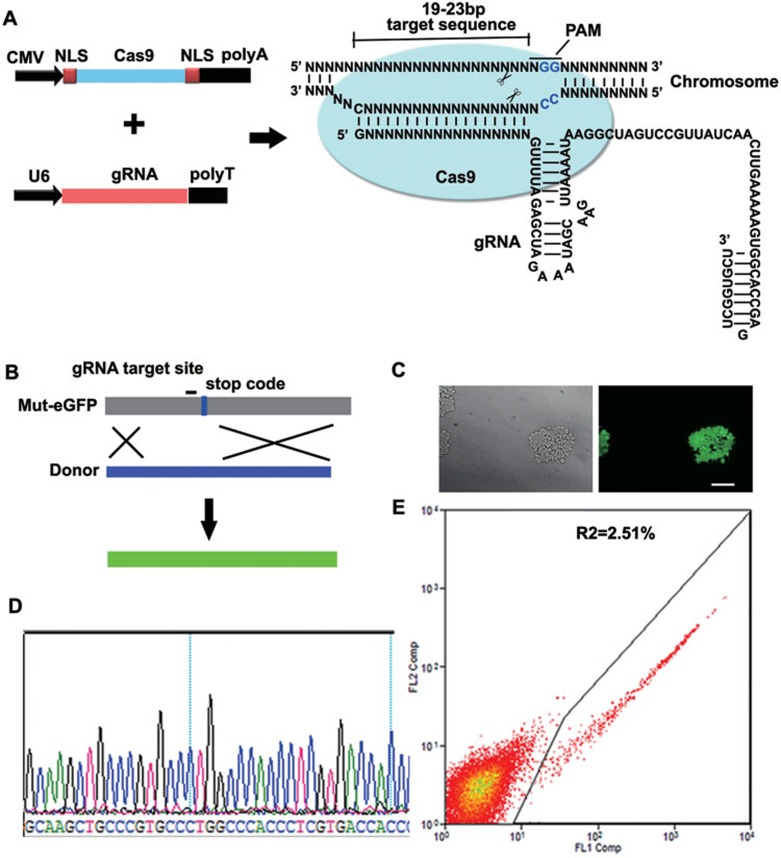
A previous study showed that a single gRNA efficiently guides Cas9 to catalyze in vitro site-specific cleavage of target DNA11. To test whether the Cas9/gRNA system can mediate site-specific cleavage in mammalian cells, we converted bacterial Cas9 cDNA into a humanized Cas9 cDNA by optimizing mammalian codons (Supplementary information, Table S1), and included nuclear localization signals (NLSs) at both ends of Cas9 to facilitate the entry of Cas9 protein into the nuclei of mammalian cells (Figure 1A). We also fused crRNA with a normally trans-encoded tracrRNA to form a gRNA that was driven by the human U6 promoter (Figure 1A and Supplementary information, Table S2). We designed a gRNA for targeting genome sequence (19-23 bp) immediately at the 5′ side of protospacer adjacent motif (PAM). Cas9 guided by gRNA unwinds and cleaves the double-stranded DNA of chromosomes in a sequence-specific manner. We evaluated the Cas9/gRNA-induced NHEJ in HEK293T cells, and found the efficiency to be 29% as determined by SURVEYOR (data not shown). To investigate Cas9/gRNA-induced HDR in HEK293T cells, we stably expressed a mutant eGFP construct that contains tens of mutant sequences and a stop codon in the middle (Figure 1B and Supplementary information, Table S6), thus generating a weak fluorescent signal in cells. We then introduced Cas9, gRNA and a donor DNA fragment into the cells, and identified the Cas9-mediated conversion of mutant eGFP into wild-type eGFP by detecting eGFP fluorescence (Figure 1C). This conversion was further confirmed by Sanger sequencing of positive cells (Figure 1D). Quantification of Cas9-mediated HDR was carried out by counting eGFP-positive cells by FACS after 48 h in culture, revealing 2.51% HDR efficiency (Figure 1E).
Figure 1.
Genome editing via the type II CRISPR system in mammalian cells. (A) Schematic diagrams showing the CRISPR system composed of Cas9 and gRNA. The expression of Cas9 protein flanked with a SV40 NLS at the N-terminus and a nucleoplasmin NLS at the C-terminus is driven by the CMV promoter, whereas the transcription of gRNA is driven by the human U6 promoter. gRNA is designed to target the genome sequence of 19-23 bp at the 5′ side of PAM (NGG). (B) A strategy of Cas9/gRNA-induced HDR used to convert a mutant eGFP (mut-eGFP) into wild-type eGFP. The mixture of Cas9/gRNA and a donor DNA fragment was delivered into a HEK293T cell line that stably expresses mutant eGFP. (C) Fluorescent image showing a successfully targeted clone of 293T cells that express the correct eGFP. Scale bar, 100 μm. (D) Sanger sequencing of the PCR amplicon confirmed the correct sequence of eGFP of the clone in C. (E) Isolating eGFP-positive cells by FACS demonstrated 2.51% HDR efficiency.
Cas9/gRNA induces indels in the etsrp locus in zebrafish
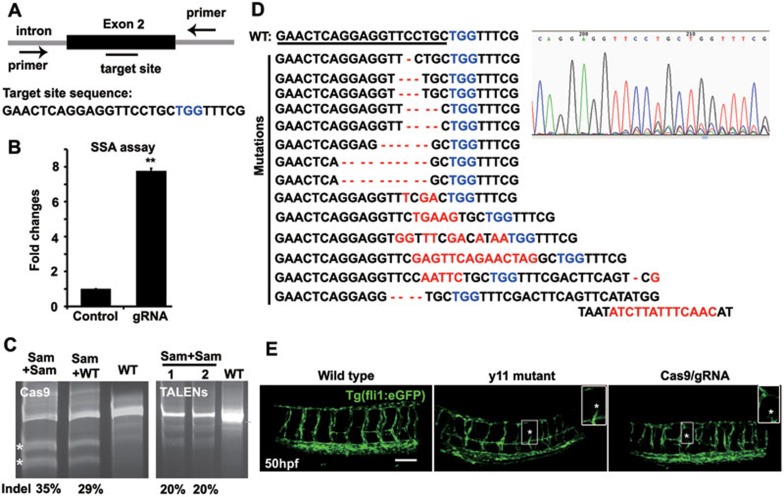
Encouraged by the efficiency of Cas9/gRNA in mammalian cells, we explored whether this nuclease complex can also catalyze site-specific DNA cleavage in zebrafish embryos in vivo. We first chose to target the etsrp locus, as it is a well-characterized gene in vascular endothelial development and its genetic mutant etsrpy11 is available15. We designed a gRNA to target etsrp (Figure 2A) and examined the efficiency of Cas9/gRNA in 293T cells by using the luciferase single-strand annealing (SSA) recombination assay. We found that the Cas9/gRNA efficiently generated DSBs and homologous recombination repair was executed in its targeted sequence, as demonstrated by an 8-fold increase in luciferase activity in the presence of both Cas9 and gRNA (Figure 2B). We microinjected Cas9 mRNA and gRNA into one-cell embryos of zebrafish. At 50 h post fertilization (hpf), we randomly selected 8 embryos, extracted their genomic DNAs for PCR amplification and confirmed the indels with two assays, SURVEYOR and Sanger sequencing. SURVEYOR assay revealed that the frequency of indels in 8 tested embryos was between 31.2% and 38.4%, comparable with that induced by TALENs (Figure 2C). Sanger sequencing of these 8 embryos (20-30 PCR amplicons were sequenced for each embryo) confirmed the site-specific indels (Figure 2D). The sequencing trace file revealed two or more peaks at the same location by Sanger sequencing of PCR fragments from a single embryo (Figure 2D), suggesting mixed DNA templates induced by Cas9/gRNA. A group of representative indels was presented in Figure 2D, including a 1-10 bp deletion or a 3-13 bp insertion. As expected, the Cas9/gRNA-mediated mutations occurred at or near the target site, further validating the sequence specificity of this targeting process. Furthermore, we found that over 38% of Cas9/gRNA-treated embryos (91 out of 237 embryos) had defects in the sprouting of intersegmental vessels, similar to those observed in etsrpy11 mutant embryos at 50 hpf (Figure 2E), suggesting efficient biallelic conversion of the etsrp locus in the resulting somatic cells. As observed in etsrpy11 mutant embryos, the intersegmental vessels were either absent or partially formed, with diverse sizes of vessel lumens in Cas9/gRNA-induced etsrp mutants. Further, SURVEYOR assay revealed that all of 30 randomly selected etsrpy11-like embryos carried indels (data not shown). Therefore, the Cas9/gRNA catalyzes site-specific DSBs in a highly efficient and sequence-specific manner, and even leads to biallelic conversion in somatic cells in zebrafish.
Figure 2.
Cas9/gRNA induces indels in the etsrp locus in zebrafish. (A) Cartoon showing the position of the target site and its sequence in the etsrp locus in zebrafish. (B) The SSA recombination assay showing luciferase activity with and without cleavage of the target site by the Cas9/gRNA system in 293T cells. The luciferase activity increased by 8-fold in the group with gRNA (Cas9 + gRNA) compared with the control group (Cas9 + empty vector). Error bars indicate SD, n = 3. **P < 0.01. (C) Representative SURVEYOR assay showing 35% efficiency of Cas9-mediated cleavage in a single embryo and 20% by TALENs-mediated cleavage in 2 embryos at 50 hpf. The indels of 8 embryos were measured with a frequency of 31.2% - 38.4%. * indicates the cleavage bands. Sam, sample treated by Cas9/gRNA or TALENs; WT, wild type. (D) Representative Sanger sequencing results of the PCR amplicons from 8 individual embryos at 50 hpf showing indels (red) induced by Cas9/gRNA in the targeted etsrp locus. PCR fragments were amplified and cloned into the pEASY vector from an individual embryo for Sanger sequencing (20-30 clones for each embryo). TGG (blue) is the PAM sequence. Deletion is represented by a dashed line and insertion is highlighted in red. (E) Fluorescent images showing abnormal intersegmental vessels that were similar in the etsrpy11 mutant and Cas9/gRNA-induced mutant, compared with the wild-type transgenic embryo at 50 hpf. * indicates defects in intersegmental vessel sprouting that are enlarged in the insets. Scale bar, 100 μm.
Cas9/gRNA induces indels in the gata4 and gata5 loci in zebrafish
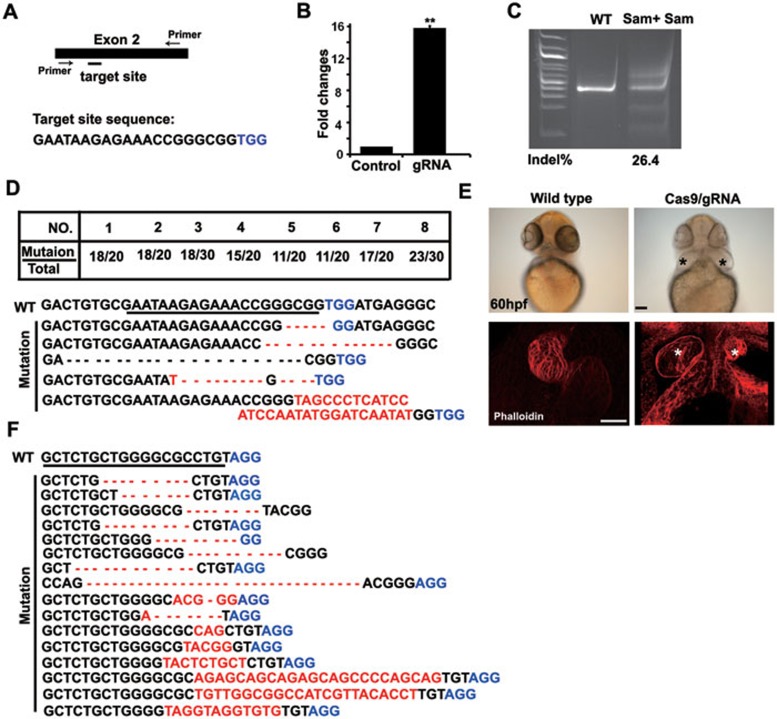
To test whether Cas9/gRNA system can be broadly used for generating sequence-specific DSBs in zebrafish, we chose to target the gata5 and gata4 loci, two well-known essential genes in endoderm and heart development16. A previous study reported that the fautm236a mutant has a G-to-A substitution at the third intron splice site of gata5, leading to two abnormal splicing variants that encode truncated proteins. In fautm236a mutants, the lateral cardiac mesoderm is normally formed, but the subsequent myocardial migration and fusion into the heart tube in the midline is affected, leading to cardia bifida in 0% to 45% of the mutants16. We designed a gRNA for targeting gata5 (Figure 3A) and confirmed the sequence-specific cleavage by SSA recombination assay (Figure 3B). We then microinjected Cas9 mRNA and gRNA into one-cell embryos of zebrafish. At 50 hpf, we found ∼26.4% indels estimated by SURVEYOR (Figure 3C). Eight Cas9-induced mutants, randomly selected with two small hearts, showed site-specific indels with frequencies from 50% to 90%, as demonstrated by Sanger sequencing (Figure 3D). Furthermore, we found that 24% (13/54) of the Cas9/gRNA-treated embryos (Figure 3E) had the cardia bifida phenotype at 60 hpf as in fautm236a mutant embryos (data not shown), suggesting efficient biallelic conversion of gata5 in the resulting somatic cells.
Figure 3.
Cas9/gRNA induces indels in the gata5 and gata4 loci in zebrafish. (A) Cartoon showing the position of the gRNA-targeting site and its sequence in the gata5 locus in zebrafish. (B) SSA recombination assay showing luciferase activity with and without cleavage of the target site by the Cas9/gRNA system in 293T cells. The luciferase activity increased by 16-fold in the group with gRNA (Cas9 + gRNA) compared with the control group (Cas9 + empty vector). Error bars indicate SD, n = 3. **P < 0.01. (C) Representative SURVEYOR assay showing the efficiency of Cas9-mediated cleavage, 26.4% in a single embryo at 50 hpf. (D) Representative Sanger sequencing results of the PCR amplicons of 8 individual embryos at 50 hpf, showing indels induced by Cas9/gRNA in the targeted gata5 locus. Twenty to thirty clones were sequenced for each embryo. Table summarizes the frequencies of site-specific indels, ranging from 50% (11/20) to 90% (18/20), in 8 individual embryos tested by Sanger sequencing. TGG (blue) is the PAM sequence. Deletion is represented by a dashed line and insertion is highlighted in red. (E) Two small hearts were formed in a Cas9/gRNA-induced mutant (right), which phenocopied that in the fautm236a genetic mutant, whereas a single heart formed in control embryos at 60 hpf. The heart was stained with phalloidin. * indicates two small hearts. Scale bar, 100 μm. (F) Representative Sanger sequencing results of the PCR amplicons of 8 individual embryos at 50 hpf, showing indels induced by Cas9/gRNA in the targeted gata4 locus as in D. AGG (blue) is the PAM sequence. AGG in 2 out of 16 sequencing results was deleted.
Microinjection of Cas9 mRNA and gRNA (targeting gata4) into one-cell embryos led to site-specific indels, which were confirmed by Sanger sequencing performed at 50 hpf (Figure 3F). A group of representative indels from eight embryos were presented, including a 7-32 bp deletion or a 1-24 bp insertion. Therefore, the Cas9/gRNA system can serve as an alternative and efficient approach for generating genetic mutants, complementing the applications of ZFNs and TALENs in zebrafish.
Cas9/gRNA induces insertion of mloxP at the break site of etsrp in zebrafish
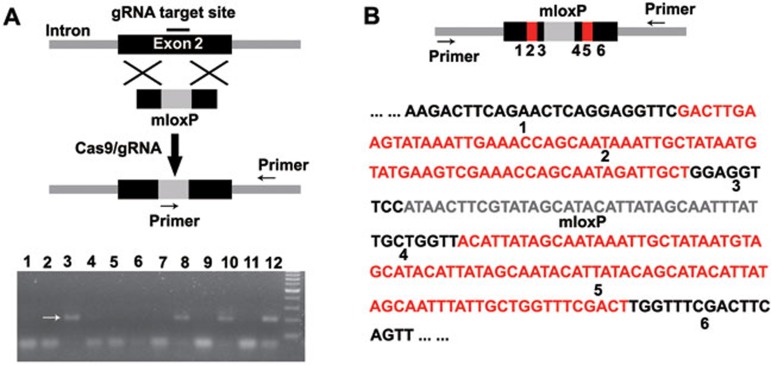
Recent reports demonstrate that single-stranded DNA (ssDNA) can be an effective donor for HDR-based genome editing by ZFN- or TALEN-induced DSBs in mammalian cells and zebrafish embryos5,6,7. Encouraged by the efficiency of Cas9/gRNA-induced DSBs in zebrafish embryos, we therefore determined whether a single-stranded oligo containing a mloxP site as described7 can act as a donor to introduce an mloxP site into the etsrp locus (Figure 4A). In order to prevent cleavage of the donor DNA, neither arm of the donor DNA had gRNA-targeting sites. Microinjection of Cas9 mRNA, gRNA (targeting etsrp) and the single-stranded donor DNA (targeting etsrp) into one-cell embryos of zebrafish led to the insertion of mloxP into the genome, which was detected by genotyping performed at 50 hpf with the forward primer matching the mloxP sequence and the reverse primer matching the downstream sequence of the donor sequence (Figure 4A). Four out of 12 randomly selected embryos had correct mloxP insertion. To further confirm the insertion of mloxP into the etsrp locus, we performed genotyping of the 4 embryos with insertion of the mloxP site by PCR and Sanger sequencing, with both primers targeting the 5′ and 3′ regions outside of the homologous arms in the donor DNA (Figure 4B). Sanger sequencing revealed that one embryo contains the site-specific integration of mloxP sequence, and left and right arm sequences from the donor DNA. However, we also observed unexpected sequences adjacent to the mloxP site (Figure 4B), suggesting the existence of unknown repair mechanisms in addition to HDR. The other three embryos were also confirmed to contain site-specific integration of mloxP sequence by Sanger sequencing, though with partial deletions of the mloxP (data not shown). We believe that obtaining clone(s) with the exactly correct site-specific integration of mloxP sequence is likely feasible upon screening (using PCR and sequencing analyses) of a sufficient number of embryos. Together, these data demonstrate the insertion of mloxP into the break site induced by Cas9/gRNA at the estrp locus.
Figure 4.
Targeted insertion of mloxP mediated by Cas9/gRNA in zebrafish. (A) Cartoon showing the principle of inserting mloxP into a target site in the etsrp locus. The primers for primary PCR genotyping of zebrafish embryos with the expected mloxP insertion being marked. Gel image showing 4 out of 12 randomly selected Cas9/gRNA-treated embryos at 50 hpf that had the expected mloxP insertion (arrows). (B) Sanger sequencing result of the PCR amplicon from one representative Cas9/gRNA-injected zebrafish embryo at 50 hpf, with the correct insertion of mloxP in the etsrp locus as domonstrated in A. PCR fragments were amplified from the tested embryo and cloned into the pEASY vector for sequencing (20-30 clones per embryo), and one clone was confirmed to have the correct insertion of mloxP in the etsrp locus. 1, 3, 4 or 6 represents the original sequence in the left or right arm, whereas 2 or 5 highlights unexpected sequences (red).
Discussion
The major findings of this work are (1) the Cas9/gRNA nuclease complex efficiently generates site-specific cleavage (∼35%) that is accompanied by a variety of indels in the etsrp, gata4 and gata5 loci in zebrafish; (2) Cas9/gRNA induces biallelic conversion of etsrp and gata5 in the resulting somatic cells in ∼30% of embryos; and (3) Cas9/gRNA generates DSBs to induce HDR at or near the break site in mammalian cells and zebrafish embryos. While we were preparing this manuscript, Joung and colleagues reported the generation of somatic mutations in 9 loci by the Cas9/gRNA nuclease complex in zebrafish17. However, our work presents two additional important findings: the biallelic knockouts by Cas9/gRNA and site-specific insertions of mloxP for the potential generation of conditional knockouts. Compared with ZFN or TALEN, the Cas9/gRNA system is an easier and economical way to design and manufacture genome editing tools, and has comparable efficiency in generating site-specific indels13,14 (Figure 2C). However, the off-target effects of the Cas9/gRNA system remains to be addressed prior to large-scale implementation in genome editing.
Our work demonstrates the potential applicability of the Cas9/gRNA system in generating zebrafish mutants. This Cas9/gRNA system induced indels in F0 mutants comparable to, if not better than, that by TALENs (Figure 2C) and ZFNs in zebrafish18,19,20,21, therefore we expect that it should be feasible to generate heritable F1 mutants with the Cas9/gRNA system. The zebrafish embryos with Cas9/gRNA-induced indels (F0) are being raised to adulthood (currently ∼1 month old) in our laboratory for the next step of experiments to test germline transmission. The germline transmission efficiency of Cas9-induced mutations needs immediate attention and investigation.
Given the simplicity of designing gRNAs14, we anticipate that the Cas9/gRNA system will add another simple and efficient genetic tool for reverse genetic studies in zebrafish and other model organisms. Zebrafish is a powerful model organism for studying molecular and genetic mechanisms in the fields of developmental and regenerative biology. However, conditional knockout technology in zebrafish is not available. The precise insertion of the mloxP site induced by Cas9/gRNA system that we found here, together with that by TALENs7, shed light on and show great promise in developing conditional knockout zebrafish in the near future. In addition, exploiting the family of Cas9 enzymes and its differing PAM requirements13 will expand and maximize the usage of the CRISPR system for genome editing.
Materials and Methods
Zebrafish strains
Etsrpy11-mutant zebrafish were obtained from Dr Brant Weinstein at the National Institutes of Health15. fautm236a-mutant zebrafish were from the Zebrafish International Resource Center16. The transgenic line Tg(fli1:EGFP) in which EGFP is driven by the zebrafish fli1 promoter22,23 was crossed with heterozygous etsrpy11 zebrafish. Tg(fli1:EGFP)/+; etsrpy11 transgenic zebrafish were identified by genotyping. Wild-type TL, transgenic and mutant zebrafish were raised and handled in accordance with the institutional guidelines on animal usage and maintenance of Peking University.
Cas9/gRNA design
The Cas9 gene sequence was codon-optimized and ordered from Biomed (Beijing). The full-length products were cloned into pcDNA3.1+ (Invitrogen). The human U6 promoter and gRNA scaffold were from Biomed, and cloned into pUC19 vector. The sequences of both Cas9 and gRNA are listed in Supplementary information, Tables S1, S2 and S3.
Cell culture, primer, DNA transfection and FACS analysis
HEK293T cell line was cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum under humidified conditions in 95% air and 5% CO2 at 37 °C. HEK293T cells were seeded into 48-well plates (Thermo). After 24 h, cells were transfected using Lipofectamine 2000 (Invitrogen) with 400 ng plasmids, following the manufacturer's recommended protocol. 48 h later, cells were digested and analyzed by FACS (BD). The sequences of primers and etsrp mloxP donor are listed in Supplementary information, Table S4.
In vitro synthesis of capped Cas9 mRNA and injection of zebrafish embryos
The full length of humanized Cas9 cDNA was cloned into the PXT7 vector and linearized, and capped mRNA was synthesized using Ambion mMESSAGE mMACHINE mRNA transcription synthesis kits. Cas9 mRNA (300 ng/μl) and gRNA (20 ng/μl) were co-injected into one-cell-stage wild-type or Tg(fli1:EGFP) transgenic embryos. Single-stranded donor oligonucleotides were also co-injected for examining HDR. Injected embryos were incubated at 28.5 °C for examination of phenotypes. Embryos were fixed for phalloidin staining at 60 hpf, or were collected for making genomic DNA for genotyping at 50 hpf. Photographs were taken using a Leica DM5000B fluorescence microscope and/or a Zeiss 700 confocal microscope.
Luciferase SSA recombination assay
The luciferase SSA recombination assay was performed according to the methods previously described19. In brief, the luciferase SSA reporter, driven by the CMV promoter, is composed of two truncated luciferase fragments. Each fragment has a 870 bp homologous arm and is separated by a stop codon and a gRNA target site. When DSB generated by Cas9/gRNA was repaired via HDR, the luciferase activity could be recovered. The co-transfection of Cas9 expression plasmid, gRNA (or empty vector as control), SSA reporter plasmid (30 ng) and Renilla plasmid (5 ng) into HEK293T cells was carried out in 48-well plates. Each experiment was repeated three times. After 1-2 days, the cell lysate was obtained with luciferase cell lysis buffer (Promega). The relative luciferase activity was measured using a dual-luciferase assay system (Promega) and detected by a Synergy H1 hybrid multimode microplate reader (Biotek). The sequence of the SSA luciferase reporter is listed in Supplementary information, Table S5.
Cas9/gRNA mutation screens by Sanger sequencing and SURVEYOR
Genomic DNA of HEK293T cells or zebrafish embryos was extracted at 50 hpf using a genome DNA extraction kit (Tiangen) following the manufacturer's protocol. In brief, cells or embryos were resuspended in cell lysis solution and incubated at 65 °C for 15 min and 95 °C for 10 min. The genomic region surrounding the CRISPR target site for each gene was PCR amplified and cloned into the pEASY vector (Transgen), which was used for Sanger sequencing (Biomed, Beijing). A total of 200-400 ng PCR products (TakaRa) were subjected to a re-annealing process to enable heteroduplex formation: 95 °C for 10 min, 95 °C to 85 °C ramping at 2 °C/s, 85 °C to 25 °C at 0.3 °C/s and holding at 25 °C for 1 min. After re-annealing, products were treated with SURVEYOR nuclease and SURVEYOR enhancer S (Transgenomic), following the manufacturer's recommended protocol, and analyzed on 10% polyacrylamide gels. Gels were stained with 0.5 μg/ml EtBr in 1× TBE for 20 min, washed in water for 20 min and imaged with a gel-imaging system (Tanon). Quantification was based on relative band intensity.
Statistical analysis
For statistical analysis, two-sided unpaired Student's t-test was processed with Excel. P-values < 0.05 were considered statistically significant. Error bars represent the SD of at least three independent experiments, unless otherwise indicated.
Acknowledgments
We thank Dr IC Bruce (ZheJiang University) for reading the manuscript. We acknowledge funding support from the National Basic Research Program of China (2011CB809106, 2010CB529503, 2012CB944501 and 2012AA020103) and the National Natural Science Foundation of China (31271549, 31221002, 81030040 and 30600142).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Humanized Cas9 cDNA sequence
The human U6 promoter and gRNA scaffold sequence
gRNA target sequences
Primers and etsrp mloxP donor sequences
SSA luciferase reporter sequence
mut eGFP sequence
References
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2012;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Chen F, Pruett-Miller SM, Huang Y, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T, Lawson ND, Mugford JW, et al. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2006;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, et al. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system Nat Biotechnol 2013. Jan 29. doi: 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J, Yeh J, Maeder M, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS ONE. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross LM, Cook MA, Lin S, Chen JN, Rubinstein AL. Rapid analysis of angiogenesis drugs in live fluorescent zebrafish assay. Arterioscler Thromb Vasc Biol. 2003;23:911–912. doi: 10.1161/01.ATV.0000068685.72914.7E. [DOI] [PubMed] [Google Scholar]
- Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN. FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol. 2007;304:735–744. doi: 10.1016/j.ydbio.2007.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Humanized Cas9 cDNA sequence
The human U6 promoter and gRNA scaffold sequence
gRNA target sequences
Primers and etsrp mloxP donor sequences
SSA luciferase reporter sequence
mut eGFP sequence