Abstract
Lysosome is a key subcellular organelle in the execution of the autophagic process and at present little is known whether lysosomal function is controlled in the process of autophagy. In this study, we first found that suppression of mammalian target of rapamycin (mTOR) activity by starvation or two mTOR catalytic inhibitors (PP242 and Torin1), but not by an allosteric inhibitor (rapamycin), leads to activation of lysosomal function. Second, we provided evidence that activation of lysosomal function is associated with the suppression of mTOR complex 1 (mTORC1), but not mTORC2, and the mTORC1 localization to lysosomes is not directly correlated to its regulatory role in lysosomal function. Third, we examined the involvement of transcription factor EB (TFEB) and demonstrated that TFEB activation following mTORC1 suppression is necessary but not sufficient for lysosomal activation. Finally, Atg5 or Atg7 deletion or blockage of the autophagosome-lysosome fusion process effectively diminished lysosomal activation, suggesting that lysosomal activation occurring in the course of autophagy is dependent on autophagosome-lysosome fusion. Taken together, this study demonstrates that in the course of autophagy, lysosomal function is upregulated via a dual mechanism involving mTORC1 suppression and autophagosome-lysosome fusion.
Keywords: autophagy, lysosome, mTORC1, autophagosome
Introduction
Macroautophagy (referred as autophagy hereafter) is a cellular catabolic process in response to starvation or other stress conditions whereby cellular components and organelles are engulfed into autophagosomes and eventually delivered to lysosomes for degradation1,2. One critical stage of autophagy is the maturation and degradation of autophagosome, which involves fusion with endosome-lysosome to form autolysosome and degradation of the inner membrane together with its luminal contents3,4. At present, biogenesis of autophagosome in the early phase of autophagy is relatively well studied5, whereas the molecular mechanisms controlling the late stage of autophagy, including the autophagosome-lysosome fusion and the regulation of lysosomal function, remain poorly understood.
Lysosome is a subcellular organelle found in all animal cells that digests cellular debris, damaged organelles and invaded microorganisms6,7. There are more than 50 soluble acid hydrolases that perform the digestive function and over 120 lysosomal membrane proteins that maintain the integrity of lysosomes and regulate lysosomal trafficking, fusion and intralysosomal pH8,9. One important breakthrough in the study of lysosome is the discovery of the CLEAR (Coordinated Lysosomal Expression and Regulation) gene network and transcription factor EB (TFEB) as the master regulator of lysosome biogenesis. It is known that TFEB controls the transcription of target genes closely related to lysosomal structure and function, including hydrolases, lysosomal membrane proteins and the V-ATPase complex10,11.
One unique feature of lysosome is its highly acidic environment (pH 4.5-5.0) that provides an optimal condition for its hydrolytic enzymes to perform their catalytic function9. In mammalian cells, the size of lysosome ranges from 0.1 to 1.0 μm, which is considered to be smaller than autophagosome (0.5 to 1.5 μm)12. Therefore, the autolysosome generated from the fusion of a smaller acidic lysosome with a larger autophagosome (with a neutral interior pH) would need to have certain form of functional adjustment to maintain optimal acidic pH for performing the hydrolytic function to digest the luminal contents. In fact, there is evidence suggesting that the autolysosome possesses even lower pH than lysosome upon fusion with autophagosome. For instance, LysoTracker staining has been commonly used for examining starvation-induced autophagy in the Drosophila fat body and the increased staining indicates the reduced pH in the lysosome-autolysosome13,14. In yeast, glucose starvation was able to enhance the antimicrobial activity of lysosome15. However, at present, the functional changes of lysosome in the course of autophagy remain largely unknown. In this study, we provide evidence demonstrating the functional activation of lysosome achieved via a dual mechanism involving mTORC1 suppression and autophagosome-lysosome fusion.
Results
Induction of autophagy by starvation and mTOR inhibitors
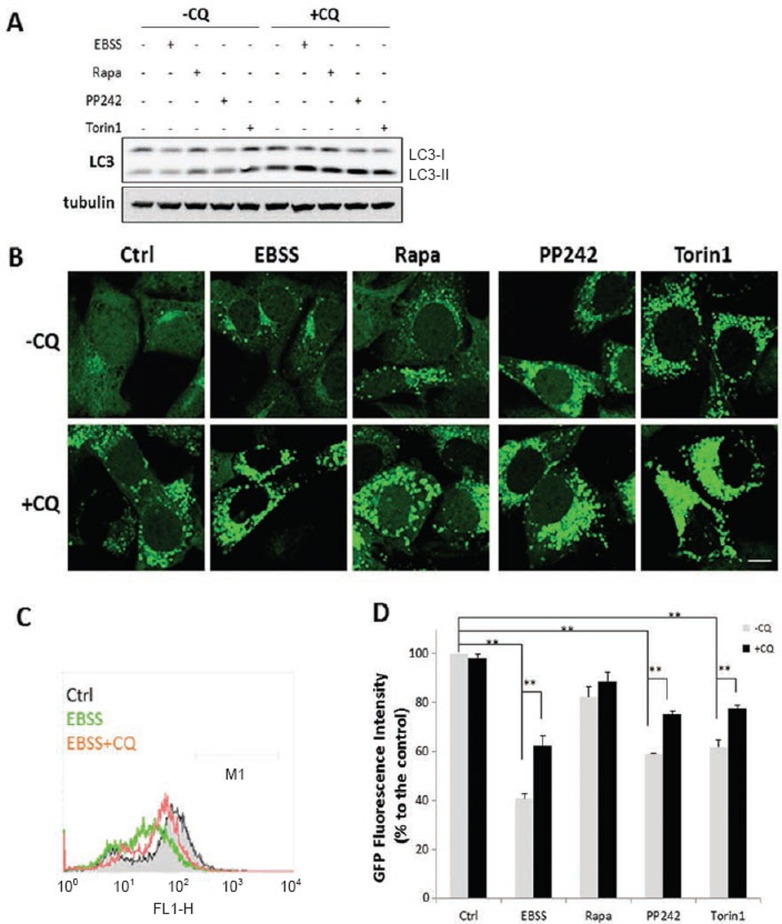
We first examined the autophagy induced by starvation (by culturing cells in Earle's Balanced Salt Solution (EBSS)) and three different mTOR inhibitors, rapamycin, PP242 and Torin1. Rapamycin is an allosteric inhibitor of mTOR and only suppresses part of mTORC1 function, whereas both PP242 and Torin1 are catalytic inhibitors that are able to completely suppress both mTORC1 and mTORC2 via binding to ATP-binding sites16,17. All treatments led to increased LC3-II protein level (Figure 1A) and number of the GFP-LC3 puncta (Figure 1B), and both are markedly enhanced by chloroquine (CQ), a lysosomotropic agent widely used to neutralize lysosomal pH and block lysosomal degradation18. Notably, in the presence of CQ, the LC3-II level or the number of the GFP-LC3 puncta among all treatments were rather similar. Next, we quantified the GFP fluorescence intensity in MEFs with stable expression of GFP-LC3 using flow cytometry, a method that has been established for measuring the autophagic flux/turnover19. Starvation, PP242 and Torin1, but not rapamycin, markedly reduced the total GFP intensity (Figure 1C and 1D); and the reduction of GFP intensity was significantly reversed by CQ. We also conducted the same experiments in HeLa cells with stable expression of GFP-LC3 and observed the same trends for LC3-II protein level, GFP-LC3 puncta and levels of the GFP fluorescence intensity (Supplementary information, Figure S1A-S1D). Our data are consistent with the earlier findings that rapamycin is a relatively weaker autophagy inducer, in comparison to catalytic mTOR inhibitors16,20.
Figure 1.
Induction of autophagy by starvation and mTOR inhibitors in MEFs. (A) MEFs with stable expression of GFP-LC3 were treated with EBSS, rapamycin, PP242 or Torin1 (all at 1 μM) with or without CQ (50 μM) for 3 h. At the end of treatment, cell lysate was collected and subject to immunoblotting. (B) MEFs with stable expression of GFP-LC3 were treated as described in panel (A). Scale bar, 10 μm. (C and D) MEFs with stable expression of GFP-LC3 were treated as indicated in Panel (A), and total GFP intensity were measured by flow cytometry. Typical histograms were shown in panel (C) and the quantification data in panel (D). Data are presented as mean ± SD from two independent experiments (each in duplicate) (**P< 0.01, Student's t-test).
Activation of lysosomal function in cells under starvation or treated with Torin1 and PP242, but not in cells treated with rapamycin
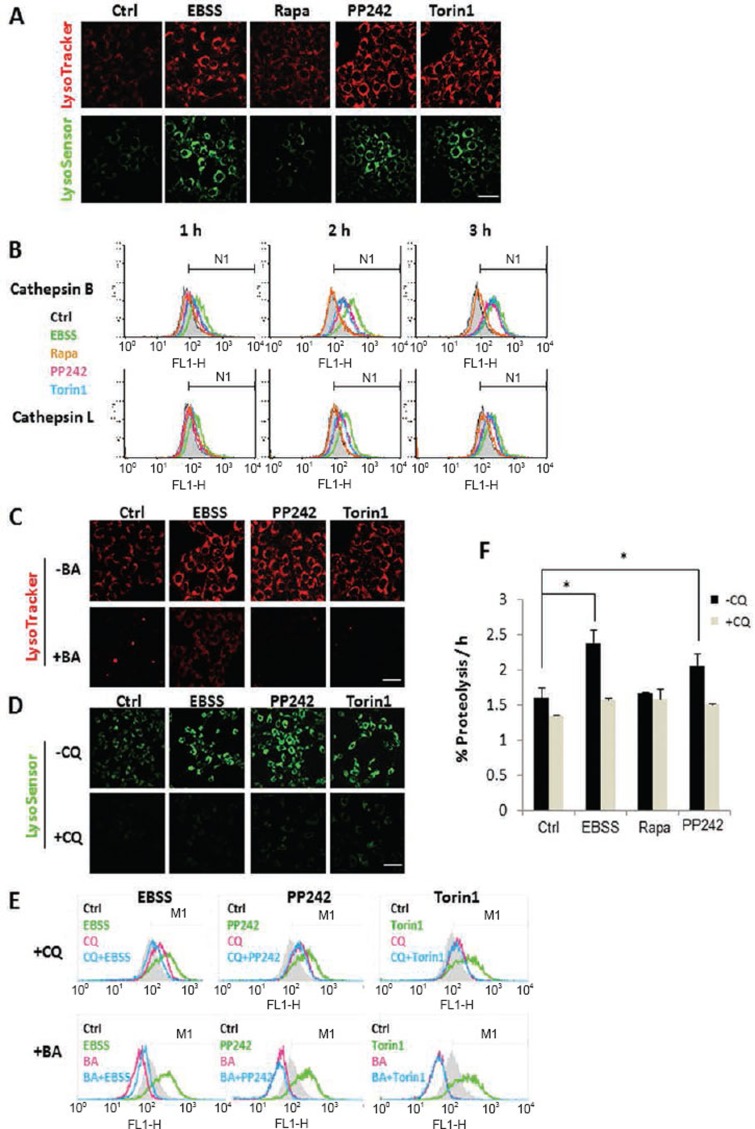
Here, we used an array of techniques to test the changes of lysosomal activity in the course of autophagy. First, significant increase of both LysoTracker and LysoSensor staining was observed in cells treated with EBSS, PP242 and Torin1, but not in cells with rapamycin for 3 h (Figure 2A), indicating enhanced acidification of lysosome (reduced pH). Second, the enzyme activities of lysosomal cathepsin B and L were measured. Both time- (Figure 2B) and dose-dependent increases (Supplementary information, Figure S2A) were found in cells treated with EBSS, PP242 and Torin1, but not rapamycin. To support the above findings, we also utilized a cell lysate-based cathepsin enzyme activity assay and generally we observed a similar trend: treatment with EBSS and PP242, but not rapamycin, enhanced cathepsin B and L enzyme activities (Supplementary information, Figure S2B). In addition, we applied the same treatments to HeLa cells and observed the similar patterns for LysoTracker staining and cathepsin B activation (Supplementary information, Figure S3A and S3B). Bafilomycin A1 (BA) is a specific V-ATPase inhibitor and is thus able to abolish lysosomal acidification21. Cotreatment with BA or CQ abolished the LysoTracker or LysoSensor staining under various treatments (Figure 2C and 2D). As expected, BA and CQ also effectively prevented the cathepsin B activation induced by EBSS, PP242 or Torin1 (Figure 2E). Treatment with BA even markedly reduced the basal level of cathepsin B enzyme activity (Figure 2E). Finally, we measured the proteolysis rate of the long-lived protein, considered as a gold standard for lysosomal function22. Treatment with EBSS and PP242, but not rapamycin, markedly enhanced proteolysis, which was completely blocked by CQ (Figure 2F). The relative weak effect of rapamycin on lysosomal acidification (Figure 2A), cathepsin activity (Figure 2B) and long-lived protein degradation (Figure 2F) are consistent with the lower autophagic flux/turnover level shown earlier (Figure 1D, Supplementary information, Figure S1D).
Figure 2.
Activation of lysosomal function in cells under starvation or treated with Torin1 and PP242, but not in cells treated with rapamycin. (A) MEFs were treated with EBSS, rapamycin, PP242 or Torin1 (all at 1 μM) for 3 h. Cells were then stained with LysoTracker Red DND-99 (50 nM) or LysoSensor Yellow/Blue DND-160 (5 μM) for 15 min. Scale bar, 50 μm. (B) MEFs were treated with EBSS, rapamycin, PP242 or Torin1 (all at 1 μM) as indicated. Cells were then loaded with Magic Red Cathepsin B or L reagent for 15 min. Fluorescence intensity of 10 000 cells per sample was measured by flow cytometry. (C and D) MEFs were treated with EBSS, PP242 (1 μM) or Torin1 (1 μM) with or without BA (50 nM) or CQ (50 μM) for 3 h. The fluorescence intensity of LysoTracker and LysoSensor was checked as described in panel (A). Scale bar, 50 μm. (E) MEFs were treated as indicated in panel (C-D), and cells were then loaded with Magic Red Cathepsin B reagent and determined as described in panel (B). (F) Rate of degradation of long-lived proteins in MEFs following the same treatment as in panel (A). Data are presented as mean ± SD from three independent experiments (*P< 0.05, Student's t test).
Activation of lysosomal function is correlated to suppression of mTORC1
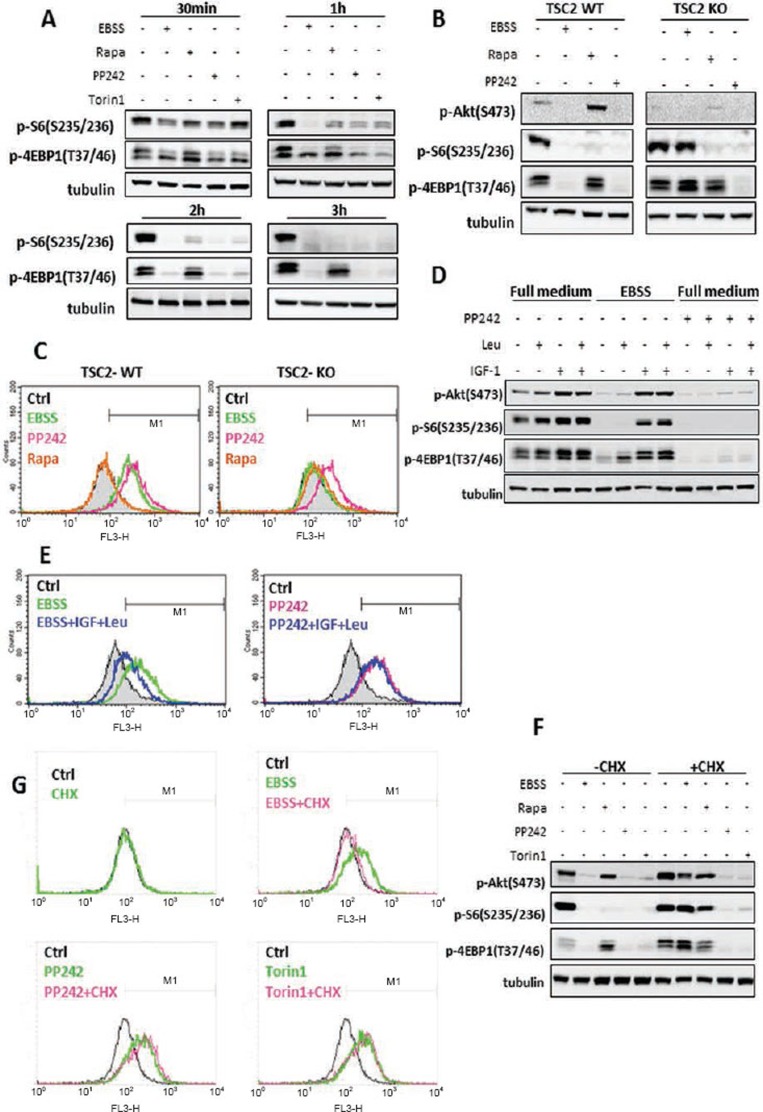
Here, we aimed to test the correlation between mTOR activity and lysosomal function. To do so, we first compared the temporal pattern of the inhibitory effects of starvation and the three mTOR inhibitors on mTORC1. Starvation and two catalytic mTOR inhibitors (PP242 and Torin1) began to suppress mTORC1 from 30 min, and completely abolished mTORC1 activity at 3 h (Figure 3A). EBSS appeared to exert its inhibitory effect on mTORC1 faster than PP242 and Torin1, especially on p-S6. Rapamycin was largely ineffective on p-4EBP1. Such observations are consistent with the current understanding that rapamycin is an allosteric inhibitor of mTOR and only suppresses part of mTORC1 function, whereas both PP242 and Torin1 are catalytic inhibitors that are able to fully suppress mTORC116.
Figure 3.
Activation of lysosomal function is correlated to the suppression of mTORC1. (A) MEFs were treated with EBSS, rapamycin, PP242 or Torin1 (all at 1 μM) for the indicated times. (B) TSC2-WT and TSC2-KO MEFs were treated with EBSS, rapamycin (1 μM) or PP242 (1 μM) for 3 h. (C) TSC2-WT and TSC2-KO MEFs were treated as indicated in panel (B), and cathepsin B enzyme activity was measured as described in Figure 2B. (D) MEFs were incubated in full medium, EBSS, or full medium with PP242 (1 μM) for 2 h, followed by the addition of IGF-1 (200 nM), or Leucine (Leu, 0.2 mg/ml) or IGF1+Leu for another 2 h. (E) MEFs were treated as indicated in panel (D), and cathepsin B enzyme activity was measured as described in Figure 2B. (F) MEFs were treated with EBSS, rapamycin, PP242 or Torin1 (all at 1 μM) in the absence or presence of CHX (10 μM) for 3 h. (G) MEFs were treated as described in panel (F) and cathepsin B enzyme activity was measured as described in Figure 2B. Cell lysate was collected and subject to immunoblotting at the end of above treatment.
To further establish the role of mTORC1 in regulating lysosomal function, we utilized the TSC2 knockout (KO) MEFs in which the mTORC1 is constitutively active23. Starvation failed to suppress mTORC1 in the TSC2-KO MEFs (no reduction of both p-S6 and p-4EBP1), whereas PP242 had the same inhibitory function in both TSC2 WT and KO MEFs (Figure 3B). Interestingly, EBSS failed to increase cathepsin B enzyme activity in TSC2 KO MEFs, whereas PP242 was found to be equally effective in TSC2 WT and KO MEFs (Figure 3C).
PP242 and Torin1 are known to block both mTORC1 and mTORC2 function16 and mTORC2 is also defective in TSC2 KO MEFs24, as suggested by the reduced p-Akt (S473) level (Figure 3B), we next tested the possible involvement of mTORC2 in lysosomal activation. Here, we used the MEFs with deletion of Sin1, an essential component of mTORC2 as described previously25. The effects of EBSS, rapamycin and PP242 on mTORC1 activity were basically similar between the Sin1 WT and KO MEFs (Supplementary information, Figure S4A). Consistently, those treatments had the same impact on lysosomal cathepsin B activation on these two cell types (Supplementary information, Figure S4B), suggesting that the mTORC2 is unlikely to have an important role in lysosomal function.
To further establish the causative relationship between mTORC1 activity and lysosomal function, several additional experiments were performed. First, we examined whether reactivation of mTORC1 would lead to the suppression of lysosomal enzyme activity. Supplement of leucine and insulin-like growth factor-1 (IGF-1) was able to reactivate mTORC1 in starved cells, but not in cells treated with PP242 (Figure 3D). Consistently, addition of leucine and IGF-1 significantly reduced cathepsin B activity in starved cells, but had no effect on cells treated with PP242 (Figure 3E). Second, we used cycloheximide (CHX) to activate mTORC1 via suppression of de novo protein synthesis and the consequent increase of intracellular pool of amino acids (AA)26. The presence of CHX restored the mTORC1 activity as evidenced by both the p-S6 and p-4EBP1 in starved cells, but not in cells treated with PP242 and Torin1 (Figure 3F). Consistently, CHX was able to block EBSS-induced cathepsin B activation, whereas it failed to do so in cells treated with PP242 or Torin1 (Figure 3G). Third, we tested the changes of lysosomal function in so-called mTOR-independent autophagy. Trehalose is a natural α-linked disaccharide that was shown to induce mTOR-independent autophagy27. As expected, trehalose induced autophagy without suppression of mTORC1 (Supplementary information, Figure S5A-S5C). Consistently, Trehalose failed to enhance LysoTracker staining (Supplementary information, Figure S5D) or cathepsin B activity (Supplementary information, Figure S5E).
mTORC1 activity, rather than its lysosomal localization, is critical for its inhibitory effect on lysosome
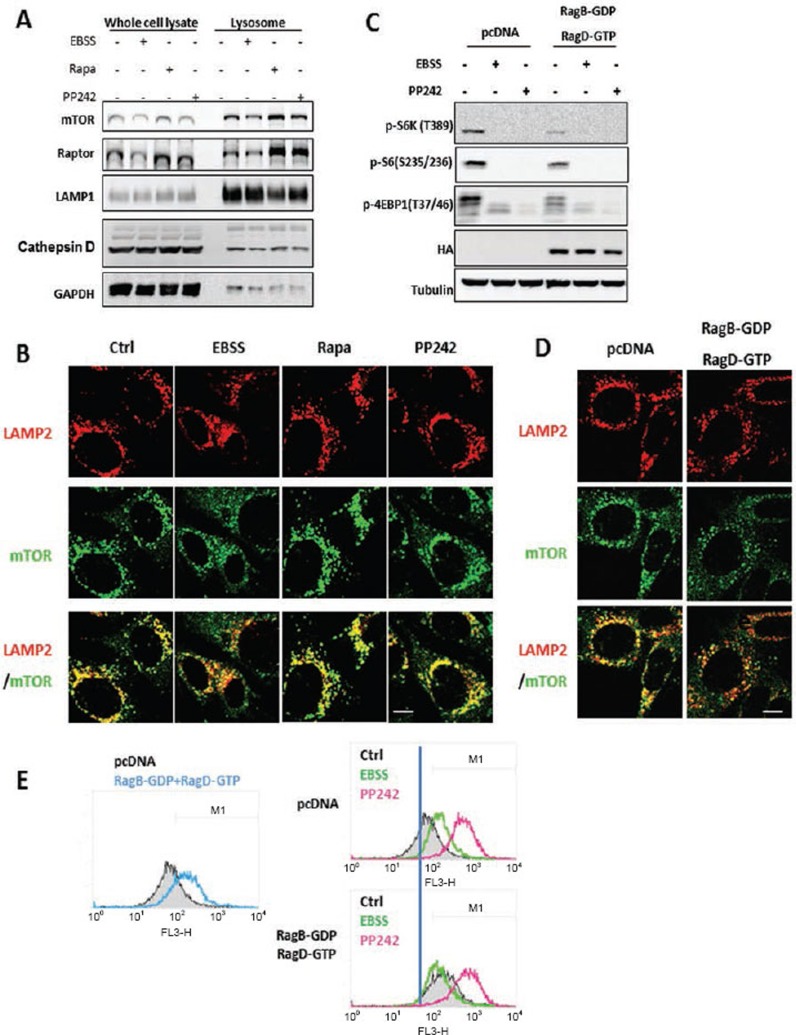
As mTORC1 activation requires its translocation to lysosome in response to AA stimulation28, we examined whether mTORC1 lysosomal localization is associated with its inhibitory effect on lysosomal function. Starvation was able to reduce the amount of mTORC1 (both mTOR and Raptor) in the lysosomal fraction (Figure 4A), as well as the colocalization of mTOR with LAMP2 (Figure 4B). In contrast, treatment with both rapamycin and PP242 markedly enhanced the mTORC1 localization to lysosome (Figure 4A and 4B). Such data are indeed in agreement with earlier reports in which mTOR inhibitors (rapamycin or Torin1) increased mTORC1 localization to lysosome29,30. Second, we utilized the dominant negative (DN) form of Rag (RagB-GDP and RagD-GTP) to block mTORC1 lysosomal translocation28 and observed the following: (i) overexpression of DN-Rag alone was able to enhance cathepsin B enzymatic activity (Figure 4E, left panel), consistent with the decreased level of mTORC1 activity in those cells (shown in Figure 4C); (ii) starvation was unable to further increase cathepsin B activity in cells expressing DN-Rag (Figure 4E, right panel), due to the fact that starvation and DN-Rag have overlapping effect on mTORC1 lysosomal translocation and activity; and (iii) PP242-induced cathepsin B activation was not affected by overexpression of DN-Rag (Figure 4E, right panel), suggesting that PP242 is able to block all mTORC1 regardless of its location (either at lysosome or in cytosol). Our data thus indicate that it is the mTORC1 activity, not the mTORC1 lysosomal localization that regulates the lysosomal function. Moreover, our data suggest that the increased lysosomal function is unlikely caused by enhanced lysosomal number or via promotion of protein trafficking to lysosome, since the LAMP1 protein level (Figure 4A) and LAMP2 immunostaining (Figure 4B), or the procathepsin D protein level (Figure 4A) in the lysosomal fraction remain consistent among different treatment groups.
Figure 4.
mTORC1 activity, rather than its lysosomal localization, is critical for its inhibitory effect on lysosomes. (A) MEFs were treated with EBSS, rapamycin (1 μM) or PP242 (1 μM) for 3 h, then lysosome fractions were collected as described in Materials and Methods. LAMP1 was used as a marker for the lysosome fraction. (B) MEFs were treated with EBSS, rapamycin (1 μM) or PP242 (1 μM) for 3 h, and cells were coimmunostained for LAMP2 (red) and mTOR (green). Scale bar, 10 μm. (C) HET293T cells were transfected with pcDNA or the dominant negative RagBGDP-RagDGTP heterodimer for 48 h, then exposed to EBSS or PP242 (1 μM) for 3 h. (D) HET293T cells with the same transfection as described in panel (C), and then coimmunostained for LAMP2 (red) and mTOR (green). Scale bar, 10 μm. (E) HET293T cells with the same transfection and treatment as described in panel (C), and cathepsin B enzyme activity was measured as described in Figure 2B. Cell lysate was collected and subject to immunoblotting at the end of above treatment.
Lysosomal activation induced by mTORC1 suppression is Atg or autophagosome dependent
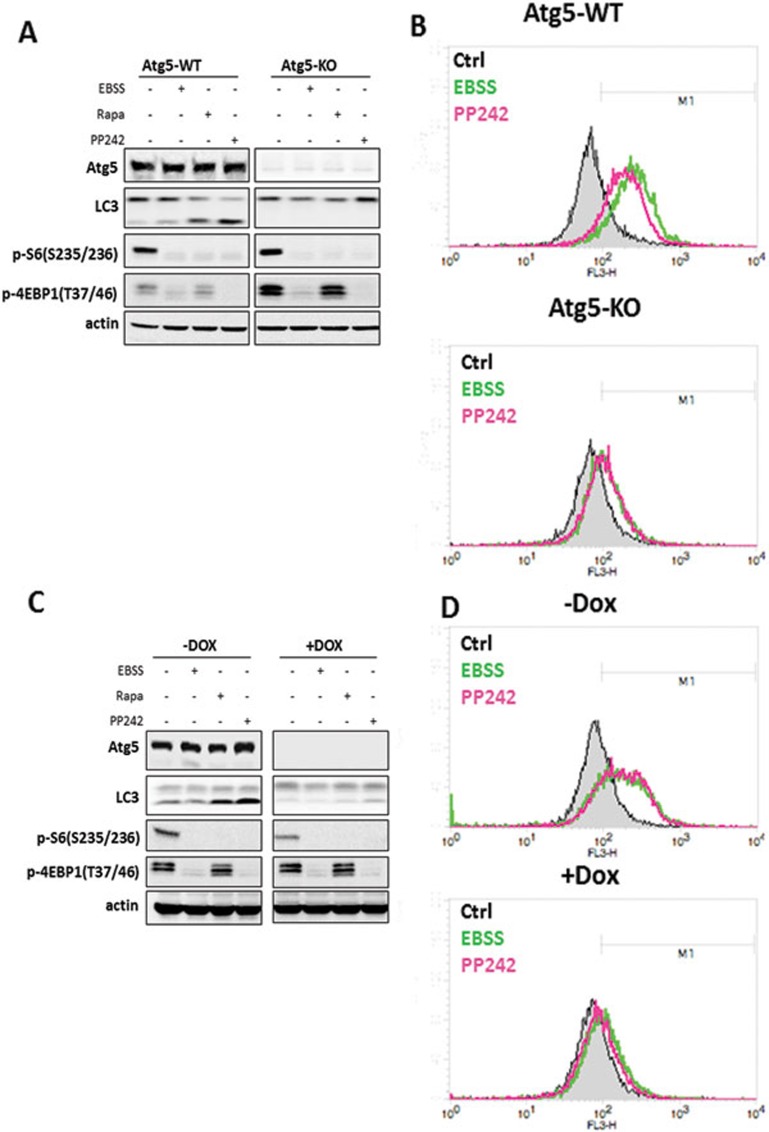
It has been well established that cells deficient for either Atg5 or Atg7 are defective in autophagosome formation31,32. To understand the relationship between the autophagy machinery and lysosomal activation under mTORC1 suppression, we first compared the lysosomal changes induced by starvation and mTOR inhibitors between Atg5-WT and Atg5-KO MEFs. It is indeed striking that no significant cathepsin B and L activation was observed in Atg5 KO MEFs treated with EBSS or PP242 (Figure 5B). To confirm our findings, we tested the effect of EBSS and PP242 on the inducible Atg5 deletion MEFs (m5-7 cells) as reported previously33. Consistently, EBSS or PP242 failed to enhance cathepsin enzyme activity in the absence of Atg5 (after incubation with DOX) (Figure 5D). Similarly, starvation and PP242 failed to enhance LysoTracker staining in Atg5-deficient cells in both systems tested (Supplementary information, Figure S6A-S6B). We also observed the same trend in primary Atg5-WT and Atg5-KO MEFs (Supplementary information, Figure S7C-S7D). Next, we tested MEFs deficient for Atg7 and found the same pattern of responses (Supplementary information, Figure S7C-S7D). Notably, starvation and mTOR inhibitors had similar effects on mTORC1 in cells with or without Atg5 or Atg7 (Figure 5A, 5C, Supplementary information, Figure S7A, S7C). Thus, our data present clear evidence that lysosome activation induced by mTOR inhibition depends on some of the key Atgs, including Atg5 and Atg7, suggesting that lysosomal activation requires autophagosome formation.
Figure 5.
Lysosomal activation induced by starvation or mTOR inhibitors is Atg5 or autophagosome dependent. (A) Atg5-WT and Atg5-KO MEFs were treated with EBSS, rapamycin (1 μM) or PP242 (1 μM) for 3 h. (B) Atg5-WT and Atg5-KO cells were treated as in panel (A), and the cathepsin B enzyme activity was measured as described in Figure 2B. (C) Atg5 Tet-off inducible MEF cells (m5-7) were pretreated with or without doxycycline (DOX) for 4 days, then treated as indicated in panel (A). (D) Cells as described in panel (C) were treated with EBSS or PP242 (1 μM) for 3 h, and the cathepsin B enzyme activity was then measured as described in Figure 2B. Cell lysate was collected and subject to immunoblotting at the end of above treatment.
TFEB activation is necessary but not sufficient for upregulation of lysosomal function following mTORC1 suppression
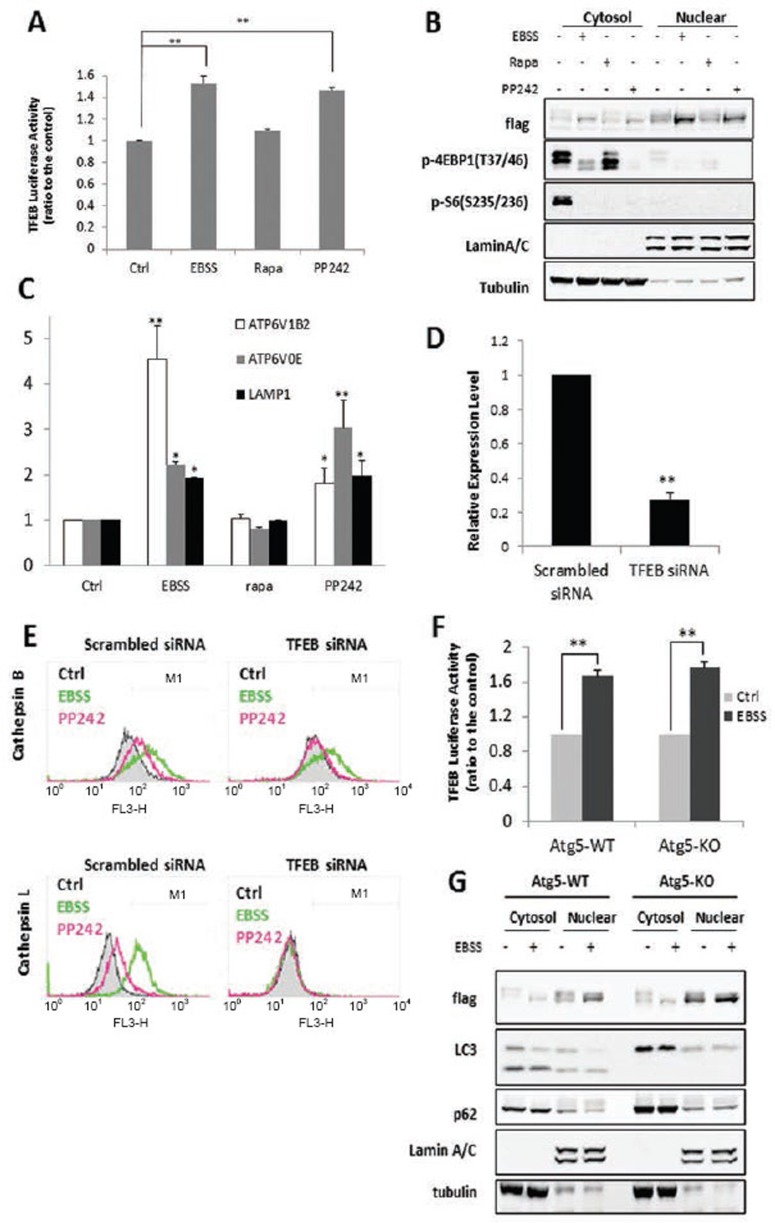
It has been reported recently that mTORC1 is a key upstream kinase that directly phosphorylates TFEB and inhibits its activity30,34. However, the changes of lysosomal function upon TFEB activation have not been examined. Here, starvation and PP242, but not rapamycin, were able to increase the TFEB transcriptional activity as measured by the TFEB luciferase assay (Figure 6A). Consistently, there was significant amount of nuclear translocation of Flag-tagged TFEB in cells treated with EBSS and PP242, but not rapamycin (Figure 6B). Measurement of the mRNA level of some known TFEB target genes, such as the V-ATPase subunits and LAMP1, showed the same pattern (Figure 6C). Next, to test the regulatory role of TFEB in lysosomal function, we carried out transient knockdown (KD) of TFEB. As shown in Figure 6D-6E, TFEB KD blocked the increase of cathepsin B and L activities induced by EBSS and PP242. Our data thus suggest that TFEB activation has an important role in lysosomal activation following the suppression of mTORC1 function. Next, to understand whether the autophagy machinery is involved in TFEB activation, we compared the effect of starvation on TFEB in Atg5 WT and KO MEFs. To our surprise, we did not observe any difference between these two cell types in terms of TFEB luciferase activity and TFEB nuclear translocation in response to starvation (Figure 6F-6G). Such observations thus suggest that (i) TFEB activation is independent of Atg or autophagosome formation, and (ii) the activated TFEB in Atg5 KO MEFs is unable to activate lysosomal function. Therefore, we conclude that TFEB activation following mTORC1 suppression is necessary but not sufficient for the upregulation of lysosomal function.
Figure 6.
TFEB activation is necessary but not sufficient for upregulation of lysosomal function following mTORC1 suppression. (A) MEFs were first transiently transfected with a TFEB luciferase reporter vector together with Renilla luciferase vector for 48 h. Cells were then treated with EBSS or PP242 (1 μM) for 4 h. Data were presented as mean ± SD from two independent experiments (each in triplicates) (**P< 0.01 comparing to the control, Student's t test). (B) MEFs were first transiently transfected with the Flag-tagged TFEB for 48 h, and then treated with EBSS, rapamycin (1 μM) or PP242 (1 μM) for 2 h. At the end of treatment, cell lysate was collected and subject to immunoblotting. (C) MEFs were treated with EBSS, rapamycin (1 μM) or PP242 (1 μM) for 4 h, and the mRNA levels were measured by RT-qPCR. Values are expressed as fold increase compared to the control group. Data are presented as mean ± SD from two independent experiments (each in triplicates) (*P< 0.05, **P < 0.01 comparing to their respective group in the control cells, Student's t test). (D) TFEB mRNA levels measured by qPCR. Data were presented as mean ± SD from two independent experiments (each with duplicates) (**P< 0.01, Student's t test). (E) After transfection with TFEB siRNA or scrambled siRNA for 48 h, cells were treated with EBSS or PP242 (1 μM) for 3 h, then cathepsin B and L activities were determined as in Figure 2B. (F) Atg5-WT and Atg5-KO MEFs were transfected with TFEB luciferase reporter vector together with Renilla luciferase vector for 48 h, cells were then treated in EBSS for 4 h. The luciferase activity was determined and presented as in panel (A). (G) Atg5-WT and Atg5-KO MEFs were first transiently transfected with the Flag-tagged TFEB for 48 h, and then treated with EBSS for 2 h. Cell lysates were collected and subjected to cytoplasm and nucleus subfraction, as described in panel (B).
Activation of lysosome function depends on autophagosome-lysosome fusion
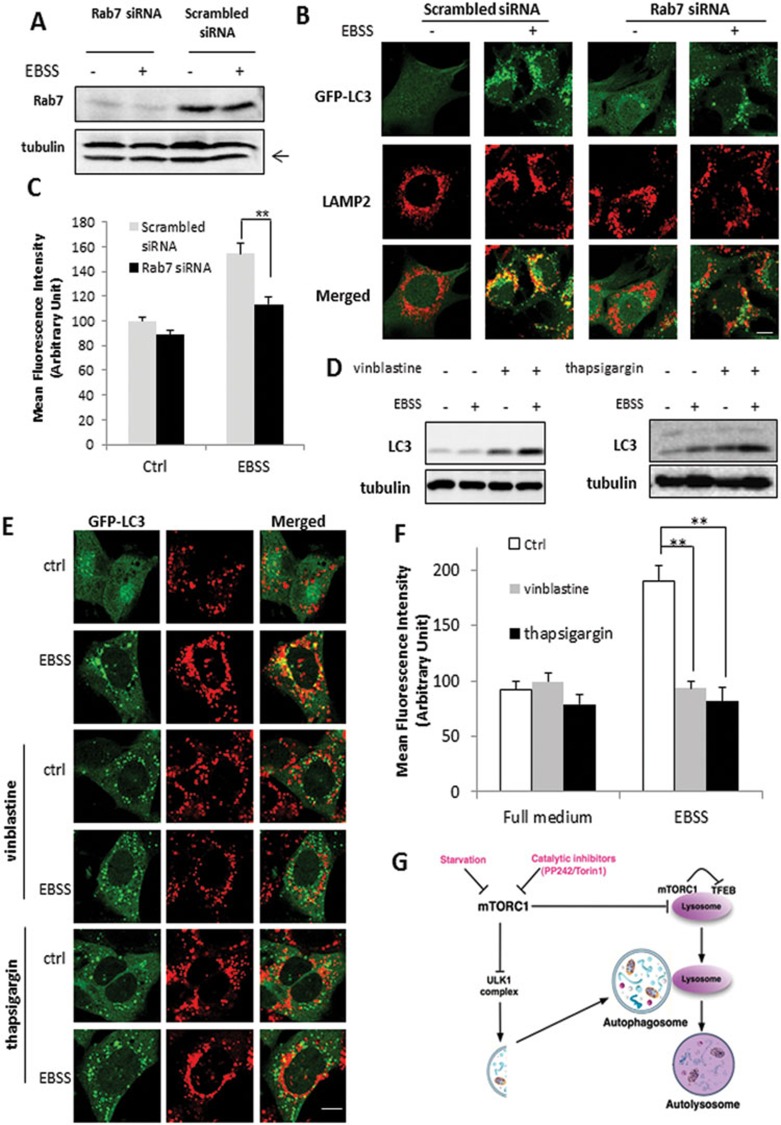
As lysosomal activation is Atg5/7 dependent, we speculate that following autophagosome formation, it is the autophagosome-lysosome fusion process that has a critical role for lysosomal activation. Rab7 is a small GTP-binding protein, which is known to have an important role in lysosome biogenesis and autophagosome-lysosome fusion35,36. Rab7 KD led to the marked reduction of Rab7 protein level in MEFs with stable expression of GFP-LC3 (Figure 7A) and significant reduction of colocalization of LAMP2 and GFP-LC3 induced by starvation, indicating the blockage of autophagosome-lysosome fusion (Figure 7B). As expected, Rab7 KD reduced, although not completely prevented, the increase of cathepsin B activity (Figure 7C) in EBSS-treated cells. Next, we used two chemical inhibitors, vinblastine and thapsigargin to block the autophagosome-lysosome fusion37,38. Vinblastine (left panel) or thapsigargin (right panel) markedly increased the accumulation of LC3-II in the cells under starvation (EBSS) (Figure 7D), and significantly reduced the colocalization of LAMP2 and GFP-LC3 (Figure 7E). Consistently, vinblastine or thapsigargin effectively abolished the increase of cathepsin B activity (Figure 7F). The above data thus support our hypothesis that lysosome activation in cells under starvation and mTOR suppression requires autophagosome-lysosome fusion.
Figure 7.
Activation of lysosome function depends on autophagosome-lysosome fusion. (A) MEFs with stable expression of GFP-LC3 were transfected with scrambled or Rab7 siRNA for 48 h. (B) MEFs with or without Rab7 KD were treated with EBSS for 3 h, then processed for LAMP-2 immunostaining (red) to observe the colocalization with GFP-LC3 (green). Scale bar, 10 μm. (C) Following the same treatments as in panel (B), cathepsin B activity was measured as described in Figure 2B. (D) MEFs were pre-treated with vinblastine (20 μM) or thapsigargin (3 μM) for 2 h, followed by EBSS for another 3 h. (E) MEFs with stable expression of GFP-LC3 were subjected to the same treatment as in panel (D) and then processed for LAMP-2 immunostaining (red) and its colocalization with GFP-LC3 (green) was examined. Scale bar, 10 μm. (F) MEFs were treated as indicated in panel (D), and cathepsin B activity was determined as described in Figure 2B. Data are presented as mean ± SD from two independent experiments (**P< 0.01, Student's t-test). (G) Illustration showing the mechanisms mediating the activation of lysosomal function in autophagy involving the mTORC1-TFEB signaling axis and autophagsome-lysosome fusion. Cell lysate was collected and subject to immunoblotting at the end of above treatment.
Discussion
In this study, we report a novel finding that lysosomal function is upregulated in autophagy and such a process depends on two critical factors: suppression of mTORC1 activity and occurrence of autophagosome-lysosome fusion (as illustrated in Figure 7G). In search of the molecular mechanisms underlying lysosomal activation in autophagy, we first establish the role of mTORC1 as a negative regulator of lysosomal function. Via multiple lines of evidence, we conclude that (i) it is the mTORC1, but not mTORC2, that executes a negative role in the regulation of lysosomal function; (ii) it is the enzymatic activity of mTORC1, not its lysosomal localization, that is directly relevant to its inhibitory effects on lysosome and (iii) following mTORC1 suppression, TFEB activation is necessary but not sufficient for upregulation of lysosomal function in the course of autophagy. At present, there are some clues linking the function of mTOR with lysosomes. First, lysosomes have a critical role in mediating mTORC1 activation in response to AA via the control of mTORC1 lysosomal localization and intralysosomal AA sensing28,39. Second, in cells under starvation, changes of lysosomal positioning also regulate mTOR activity40. Third, mTOR is required for the reformation of lysosomes at the end of autophagy41. Lastly, a very recent report provided the first evidence that mTORC1 on lysosomal membrane directly phosphorylates TFEB and suppresses its nuclear translocation30. On the other hand, mTORC1 has been well established as the key negative regulator of autophagy, via suppression of the ULK1 complex at the initiation stage of autophagy42. Therefore, it appears that mTORC1 not only exerts its inhibitory effect on the initiation of autophagy, but also inhibits the maturation and degradation of autophagy via suppression of lysosomal function (Figure 7G).
Another important mechanism underlying lysosomal activation in autophagy is autophagosome-lysosome fusion. It is known that autophagosome-lysosome fusion is a complex process involving multiple molecular machineries, including endosomal sorting complex required for transport (ESCRT)43, microtubules44, Rab735,36, soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs)45 and the lysosomal V-ATPase46. In this study, blockage of autophagosome-lysosome fusion (by Rab7 KD, vinblastine and thapsigargin) was able to abolish lysosomal activation, thus suggesting that the increased acidification and cathepsin enzyme activity were likely originated from autolysosome, not lysosome per se. On the other hand, the above-mentioned mechanisms are also known to be involved in other fusion processes, including late endosome-lysosome fusion (for the formation of endolysosome) and autophagosome-late endosome fusion (for the formation of amphisome)7,47. Therefore, the contribution of other hybrid organelles such as amphisome or endolysosome could not be excluded. Nevertheless, lysosome and autolysosome are believed to have a much stronger degradative capacity than endosome and amphisome6,12.
Understanding the activation of lysosomal function in the course of autophagy and establishing the role of mTORC1 in this process have important implications in biology and medicine. First, the inhibitory function of mTORC1 on lysosome couples the anabolism and catabolism, and places these two opposite processes into one regulatory network. It has been known for long that suppression of de novo protein synthesis (such as by CHX) is able to inhibit lysosomal proteolysis function48, without knowing the exact underlying mechanisms. In our study, treatment with CHX was able to reactivate mTORC1 in starved cells (Figure 3F), most probably via increased intracellular pool of AA26 and then abolished starvation-induced lysosomal activation (Figure 3G). Thus, our findings could well explain the fact that blockage of protein synthesis leads to the inhibition of lysosomal proteolysis via activation of mTORC1. Second, rapamycin and its analogs (temsirolimus (CCI-779), everolimus (RAD001) and deforolimus (AP23573)) are under clinical trials as cancer therapeutic agents with limited success49,50. Now a new generation of catalytic mTOR inhibitors have been developed and are in clinical trials, and some preliminary evidence has shown that these new inhibitors lead to more promising clinical outcomes51,52. Moreover, cancer-related changes in lysosomal composition and function have been increasingly appreciated53. Although still speculative, it would be of importance to evaluate whether the activation of lysosomal function is implicated in the better therapeutic efficacy of those new agents under testing. Third, our findings that the catalytic inhibitors of mTOR are able to activate lysosomal function suggest that such inhibitors might have potential therapeutic value for treatment of diseases with impaired lysosomal function such as the lysosomal storage disorder and certainly more studies are needed in this direction.
In summary, data from this study demonstrate the functional activation of lysosome in the course of autophagy via suppression of mTORC1 and autophagosome-lysosome fusion (Figure 7G). It appears that mTORC1 has a dual inhibitory effect on autophagy by imposing a double lock: first blocking the initiation of autophagy via suppression of the ULK1 complex and then inhibiting the maturation and degradation of autophagy via suppression of lysosomal function. We believe that understanding the functional activation of lysosome and the underlying mechanisms, especially the negative regulatory effect of mTORC1 on lysosomal function, integrates the anabolic and catabolic process of the cell, expands the scope of mTORC1 function in autophagy, and offers clues for developing novel interventional strategies in diseases via modulation of lysosomal function. Future studies will be focusing on the identification of possible molecular targets of mTORC1 on lysosome.
Materials and Methods
Reagents and antibodies
The chemicals used in our experiments were: chloroquine diphosphate (CQ, Sigma, C6628), bafilomycin A1 (BA, Sigma, B1793), vinblastine (Sigma, V1377), Earles Balanced Salt Solution (EBSS) (Sigma, E2888), ATP (Sigma, A1852), Valinomycin (Sigma, V0627), bis-tris-propane (Sigma, B4679), PP242 (Sigma, P0037), rapamycin (Sigma, R8781), Torin1 (initially provided by Dr DM Sabatini and subsequently purchased from Tocris Bioscience, 4247), trehalose (Katayama Chemicals, #1489), cycloheximide (Sigma, C7698), IGF-1 (Sigma, I8779), leucine (Invitrogen, L8912), LysoTracker Red DND-99 (Invitrogen, L7528), LysoSensor Yellow/Blue DND-160 (Invitrogen, L7545), Magic Red cathepsin B reagent (Immunochemistry Technologies, LLC, #938) and Magic Red cathepsin L reagent (Immunochemistry Technologies, LLC, #942). Cathepsin substrates Z-RR-AMC (#219392) for cathepsin B or Ac-HRYR-ACC (#219497) for cathepsin L were from Calbiochem. The antibodies used in our experiments included: microtubule-associated protein 1 light chain 3 (LC3) (Sigma, L7543), Atg7 (ProScience 3617), Atg5 (Nanotools, #0262), lysosome-associated membrane protein 1 (LAMP-1) (Developmental Studies Hybridoma Bank, 1D4B), lysosome-associated membrane protein 2 (LAMP-2) (Developmental Studies Hybridoma Bank, ADL-95), Rab7 (Santa Cruz, SC10767), Cathepsin D (C-20, SC-6486), tubulin (Sigma, T6199), Flag (Sigma, F3165), β-actin (Sigma, A5441), Lamin A/C (Cell Signaling Technology (CST), #2032), p-S6K (T389) (CST, #9205), p-S6(S235/236) (CST, #2211), p-4EBP1(T37/46) (CST, #2855), p-Akt (S473) (CST, #9271), GAPDH (CST, #2118X), mTOR (CST, #2983) and Sin1 (Bethyl Laboratories, #300-910A), HA (Sigma, H3663).
Cell culture
HeLa cells with stable expression of GFP-LC3, Atg5-WT and Atg5-KO MEFs and Atg5 Tet-off inducible MEFs (m5-7) stably expressing GFP-LC3 were all kindly provided by Dr N Mizushima33,54. Atg7-WT and Atg7-KO MEFs were kindly provided Dr M Komatsu32. TSC2-WT and TSC2-KO MEFs were obtained from Dr DJ Kwiatkowski55. Sin1-WT and Sin1-KO MEFs were provided by Dr B Su25. All cell lines were maintained in DMEM (Sigma, D1152) containing 10% fetal bovine serum (HyClone, SV30160.03) in a 5% CO2 atmosphere at 37 °C.
Estimation of intralysosomal pH using LysoSensor and LysoTracker
The intralysosomal pH was estimated using LysoSenor and LysoTracker, following manufacturer's instructions. The fluorescence intensity was observed under a confocal microscope (Olympus Fluoview FV1000) and representative cells were selected and photographed.
Cathepsin B and L activity assay
The cathepsin B and L enzymatic activity was measured using two different methods. First, following an earlier report56, MEFs and HeLa cells were cultured in 24-well plates, after designated treatment, cells were further loaded with Magic Red cathepsin B or cathepsin L reagents for 15 min. Fluorescence intensities of 10 000 cells per sample were measured by flow cytometry using the FACS cytometer (BD Biosciences). Second, we used a cell lysate-based assay that has already been established in our laboratory57,58. Briefly, cells were lysed in M2 buffer and the lysate and then incubated with 50 μM of the fluorogenic cathepsin B/L substrate (Z-RR-AMC or Ac-HRYR-ACC, respectively) in 100 μl cell-free system buffer (10 mM HEPES-NaOH, pH 7.4, 220 mM mannitol, 68 mM sucrose, 2 mM NaCl, 2.5 mM KH2PO4, 0.5 mM EGTA, 2 mM MgCl2, 5 mM pyruvate, 0.1 mM PMSF and 1 mM dithiothreitol) in a 96-well plate for 1 h at 37 °C. The fluorescence intensity was monitored by a fluorometer (Tecan SpectraFluor Plus) at an excitation wavelength of 380 nm and an emission wavelength of 460 nm. Data are expressed as percentage of fluorescence intensity compared with the control group.
Measurement of GFP-LC3 intensity using flow cytometry
The experiment was carried out based on an earlier report with modifications19. MEFs and HeLa cells stably expressing GFP-LC3 were cultured in 24-well plate overnight. After the designated treatments, the cells were then collected and the total fluorescence intensity of GFP-LC3 of 10 000 cells was measured by flow cytometry using the FACS cytometer (BD Biosciences).
Western blotting
At the end of the designated treatments, cells were lysed in Laemmli SDS buffer (62.5 mM Tris, pH 6.8, 25% glycerol, 2% SDS, phosphatase inhibitor and proteinase inhibitor cocktail). An equal amount of protein was resolved by SDS-PAGE and transferred onto PVDF membrane. After blocking with 5% non-fat milk, the membrane was probed with designated primary and secondary antibodies, developed with the enhanced chemiluminescence method and visualized with the Kodak Image Station 4000R (Kodak).
Cell fractions
Lysosome isolation was carried out according to the manufacturer's protocol from the Lysosome Enrichment Kit for Tissue and Cultured Cells (Thermo Scientific, 89839). 5 × 150 mm2 culture dishes were used for each designated treatments before being collected and pooled together for lysosome isolation. The isolated lysosome fractions were then subjected to western blotting analysis to check the purity of the isolated lysosome and mTOR translocation. pCMV-TFEB-3X Flag plasmid was provided by Dr A Ballabio10. MEFs were transfected with pCMV-TFEB-3× Flag plasmid using Lipofectmine™ 2000 (Invitrogen, 11668019) for 48 h. Nuclear and cytosolic extracts were then prepared by NE-PER nuclear and cytoplasmic extraction reagents (Pierce, 78833) according to the manufacturer's protocol.
Immunofluorescence staining and confocal microscopy
GFP-LC3-expressing stable MEFs or normal MEFs were seeded to a coverglass slide chamber (Lab-Tek, NUNC, 155411), and after the designated treatments, cells were washed with PBS, then fixed with 4% paraformaldehyde in PBS for 15 min at room temperature, and permeabilized with 0.1% saponin in PBS for 15 min. Cells were washed with PBS and blocked with 1% BSA in PBS for 30 min, then incubated with primary antibodies and corresponding secondary antibodies. The cells were examined and recorded using a confocal microscope (Olympus Fluoview FV1000) and representative cells were selected and photographed.
Luciferase assays
TFEB luciferase vector was provided by Dr A Ballabio11. The transient transfection of TFEB luciferase vector was done in MEFs using Lipofectmine™ 2000 transfection reagent according to the manufacturer's protocols. Renilla luciferase vector was used as a transfection control. The luciferase activity was measured 48 h after transfection using a Dual-Luciferase Reporter Assay System (Promega, E1960) based on the protocol provided by the manufacturer.
Small interfering RNA (siRNA) and transient transfection
The scrambled RNAi oligonucleotides and siRNAs targeting TFEB (Dharmacon, 21425) or Rab7 (Dharmacon, 19349) were transfected into MEFs using the DharmaFECT 4 Transfection Reagent (Dharmacon, T-2001-02) according to the manufacturer's protocol. For plasmid transfection, HEK-293T or MEFs were transiently transfected with pcDNA, pRK5-HAGST RagB 54L, pRK5-HAGST RagB 99L, pRK5-HA GST RagD 77L, pRK5-HA GST RagD 121L or pCMV-TFEB-3X Flag plasmids using Lipofectamine™ 2000 according to the protocol from manufacturer.
Proteolysis of long-lived proteins
This assay was performed based on the established method22. Briefly, MEFs were incubated for 24 h at 37 °C with 0.2 μCi/ml L-[14C] valine. Cells were then incubated in complete medium supplemented with 10 mM cold valine for 1 h, the medium was then replaced with fresh medium (MEM plus 10% of bovine serum albumin and 10 mM cold valine), and the incubation was continued for an additional 4 h. Cells then underwent designated treatments, and cells together with the radiolabeling proteins from the 4 h chase medium were precipitated in trichloroacetic acid at a final concentration of 10% (v/v) at 4 °C. Radioactivity was determined by liquid scintillation counting. Protein degradation was calculated by dividing the acid-soluble radioactivity recovered from both cells and medium by the radioactivity contained in precipitated proteins from both cells and medium.
Reverse transcription and quantitative real-time PCR
RNA was extracted with the RNeasy kit (Qiagen, 217004). A reverse transcription reaction was performed using 1 μg of total RNA with High Capacity cDNA Reverse Transcription kit (Applied Biosystems, 4368814). The mRNA expression levels were determined by real-time PCR using SsoFast EvaGreen Supermix (Bio-Rad, 172-5201) and CFX96 Touch Real-time PCR Detection System (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control of RNA integrity. Real-time PCR was performed in triplicate. The primers used for GAPDH, TFEB, ATP6V1B2, ATP6V0E and LAMP1 were based on the previous report10.
Statistical analysis
All western blot and image data presented are representatives from at least three independent experiments. The numeric data are presented as means ± SD from 2-3 independent experiments (each in duplicates or triplicates) and analyzed using Student's t-test.
Acknowledgments
We thank Dr N Mizushima (Tokyo Medical and Dental University, Japan) for providing the Atg5−/− MEFs and the Tet-off Atg5 MEFs with stable expression of GFP-LC3, as well as the HeLa cells with stable expression of GFP-LC3, Dr A Ballabio (Telethon Institute of Genetics and Medicine, Italy) for providing the Flag-TFEB expression vector and the TFEB-luciferase construct, Dr DJ Kwiatkowski (Harvard University, USA) for providing the pair of TSC2 WT and KO MEFs, and Dr DM Sabatini (Massachusetts Institute of Technology, USA) for providing Torin1. We also thank Dr N Mizushima and Dr WX Ding for the insightful discussions. SHT, NDY and JZ were supported by research scholarships from NUS. This study was supported in part by research grants from Singapore National Medical Research Council (NMRC/1260/2010), Singapore Biomedical Research Council (BMRC /08/1/21/19/554) to HMS and by INSERM grants to PC.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Induction of autophagy by starvation and mTOR inhibitors in HeLa cells.
PP242 and Torin1, but not rapamycin enhance cathepsin activity.
Effects of Starvation, PP242, Torin1 and rapamycin on lysosomal function in HeLa cells.
Effects of starvation, PP242 and rapamycin on lysosome in Sin1-WT and KO MEFs.
Effects of Trehalose on mTORC1 activity and lysosomal function.
Changes of LysoTracker staining in Atg5-deficient cells.
Lysosomal activation induced by starvation or mTOR inhibitors in primary Atg5 WT/KO MEFs and Atg7 WT/KO MEFs.
References
- Mehrpour M, Esclatine A, Beau I, Codogno P. Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am J Physiol Cell Physiol. 2010;298:C776–C785. doi: 10.1152/ajpcell.00507.2009. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- Lubke T, Lobel P, Sleat DE. Proteomics of the lysosome. Biochim Biophys Acta. 2009;1793:625–635. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA. Lysosomal acidification mechanisms. Annu Rev Physiol. 2012;74:69–86. doi: 10.1146/annurev-physiol-012110-142317. [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells. Cell Struct Funct. 2002;27:421–429. doi: 10.1247/csf.27.421. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Yoon J, Chang ST, Park JS, Kim YH, Min J. Functional characterization of starvation-induced lysosomal activity in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2010;88:283–289. doi: 10.1007/s00253-010-2755-4. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- Las G, Serada SB, Wikstrom JD, Twig G, Shirihai OS. Fatty acids suppress autophagic turnover in beta-cells. J Biol Chem. 2011;286:42534–42544. doi: 10.1074/jbc.M111.242412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyfeler B, Bergman P, Triantafellow E, et al. Relieving autophagy and 4EBP1 from rapamycin resistance. Mol Cell Biol. 2011;31:2867–2876. doi: 10.1128/MCB.05430-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Bauvy C, Meijer AJ, Codogno P. Assaying of autophagic protein degradation. Methods Enzymol. 2009;452:47–61. doi: 10.1016/S0076-6879(08)03604-5. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ. Rhebbing up mTOR: new insights on TSC1 and TSC2, and the pathogenesis of tuberous sclerosis. Cancer Biol Ther. 2003;2:471–476. doi: 10.4161/cbt.2.5.446. [DOI] [PubMed] [Google Scholar]
- Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Price DJ, Nemenoff RA, Avruch J. Purification of a hepatic S6 kinase from cycloheximide-treated rats. J Biol Chem. 1989;264:13825–13833. [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Suzuki M, Shinohara Y, Fujimoto T. Lysosomal accumulation of mTOR is enhanced by rapamycin. Histochem Cell Biol. 2010;134:537–544. doi: 10.1007/s00418-010-0759-x. [DOI] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. Embo J. 2012. [DOI] [PMC free article] [PubMed]
- Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–145. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011. [DOI] [PMC free article] [PubMed]
- Yu L, McPhee CK, Zheng L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- Fraldi A, Annunziata F, Lombardi A, et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–3620. doi: 10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Williamson WR, Wang D, Haberman AS, Hiesinger PR. A dual function of V0-ATPase a1 provides an endolysosomal degradation mechanism in Drosophila melanogaster photoreceptors. J Cell Biol. 2010;189:885–899. doi: 10.1083/jcb.201003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- Thoene JG, Lemons R, Boskovich S, Borysko K. Inhibitors of protein synthesis also inhibit lysosomal proteolysis. Studies using cystinotic fibroblasts. J Clin Invest. 1985;75:370–376. doi: 10.1172/JCI111709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancey JE, Curiel R, Purvis J. Evaluating temsirolimus activity in multiple tumors: a review of clinical trials. Semin Oncol. 2009;36 Suppl 3:S46–S58. doi: 10.1053/j.seminoncol.2009.10.010. [DOI] [PubMed] [Google Scholar]
- Konings IR, Verweij J, Wiemer EA, Sleijfer S. The applicability of mTOR inhibition in solid tumors. Curr Cancer Drug Targets. 2009;9:439–450. doi: 10.2174/156800909788166556. [DOI] [PubMed] [Google Scholar]
- Zask A, Verheijen JC, Richard DJ. Recent advances in the discovery of small-molecule ATP competitive mTOR inhibitors: a patent review. Expert Opin Ther Pat. 2011;21:1109–1127. doi: 10.1517/13543776.2011.584871. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Duan Y, Zheng XF. Targeting the mTOR kinase domain: the second generation of mTOR inhibitors. Drug Discov Today. 2011;16:325–331. doi: 10.1016/j.drudis.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T, Olsen OD, Jaattela M.Cancer-associated lysosomal changes: friends or foes Oncogene 2012. Jul 9. doi: 10.1038/onc.2012.292 [DOI] [PubMed]
- Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cicchetti G, Onda H, et al. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest. 2003;112:1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni HM, Bockus A, Wozniak AL, et al. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Huang Q, et al. Autophagy plays a protective role during zVAD-induced necrotic cell death. Autophagy. 2008;4:457–466. doi: 10.4161/auto.5662. [DOI] [PubMed] [Google Scholar]
- Zhou J, Hu SE, Tan SH, et al. Andrographolide sensitizes cisplatin-induced apoptosis via suppression of autophagosome-lysosome fusion in human cancer cells. Autophagy. 2012;8:338–349. doi: 10.4161/auto.18721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Induction of autophagy by starvation and mTOR inhibitors in HeLa cells.
PP242 and Torin1, but not rapamycin enhance cathepsin activity.
Effects of Starvation, PP242, Torin1 and rapamycin on lysosomal function in HeLa cells.
Effects of starvation, PP242 and rapamycin on lysosome in Sin1-WT and KO MEFs.
Effects of Trehalose on mTORC1 activity and lysosomal function.
Changes of LysoTracker staining in Atg5-deficient cells.
Lysosomal activation induced by starvation or mTOR inhibitors in primary Atg5 WT/KO MEFs and Atg7 WT/KO MEFs.