Abstract
In a recent paper published in Nature, Carrieri et al. have identified in mouse a neuron-specific antisense lncRNA transcribed in the opposite strand of Uchl1 mRNA. Antisense Uchl1 specifically promotes the translation of UCHL1 under rapamycin treatment. To do so, the lncRNA only requires a SINEB2 repeat and a small region with sequence complementarity to the regulated mRNA.
Long non-coding RNAs (lncRNAs) represent a surprisingly important portion of the mammalian transcriptome, as conservative estimations have determined that at least 75% of the genome is transcribed into noncoding RNAs1. The growing body of work shows that lncRNAs are involved in virtually every biological process, acting as modulators of gene expression. However only a handful of lncRNAs have been studied in some depth, showing that many lncRNAs regulate transcription either by serving as scaffolds that hold and guide chromatin complexes, as transcriptional enhancers, or by interfering with the transcription machinery. Additionally, some lncRNAs maintain the structure of nuclear speckles, while other lncRNAs work as posttranscriptional regulators of splicing (reviewed in2,3,4). But lncRNAs are probably present at every level of gene regulation. Indeed, recent reports have shown that lncRNAs may also act as molecular decoys for microRNAs5, inhibit protein translation6 or control mRNA stability7. All the available evidence confirms that the world of lncRNAs is enormously heterogeneous, and the lack of identifiable common features for their functional classification challenges investigators.
An elegant work recently published in Nature by Carrieri et al.8 has shed some light into the complex universe of functional lncRNAs. This paper describes the identification of an antisense lncRNA transcribed in the opposite strand of the mouse Ubiquitin carboxy-terminal hydrolase L1 (Uchl1) gene, that can specifically induce the translation of Uchl1 under certain stress conditions (Figure 1).
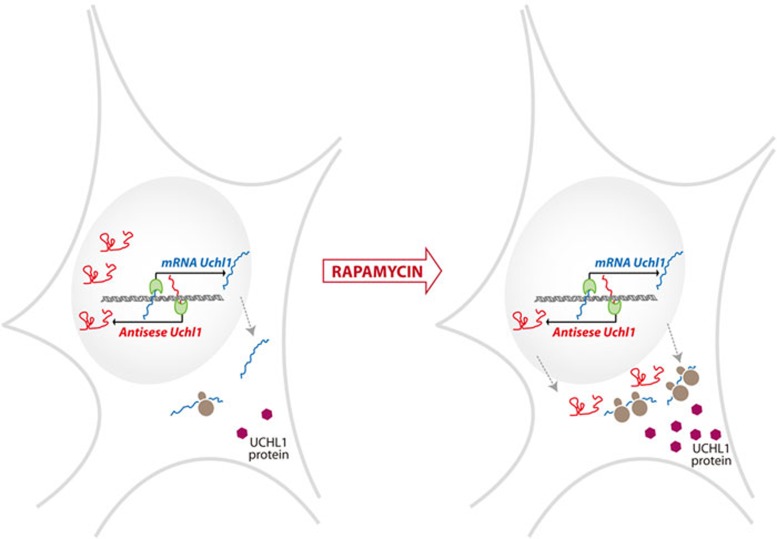
Figure 1.
Effect of antisense Uchl1 on UCHL1 protein expression. Under basal conditions (left), antisense Uchl1 is mostly retained in the nucleus of mouse dopaminergic neurons, while Uchl1 mRNA is exported to the cytoplasm. Under rapamycin treatment (right), antisense Uchl1 is exported to the cytoplasm, Uchl1 mRNA is relocated to polyribosomes and UCHL1 protein levels increase.
Many ncRNAs are transcribed antisense of protein-coding genes, and in several cases it has been reported that they regulate the expression of their counterpart mRNA either transcriptionally or posttranscriptionally9,10. Even though it is suspected that sense-antisense sequence homology is critical for this activity, so far there was little evidence for that. Carrieri and colleagues show that overexpression of mouse antisense Uchl1 induces UCHL1 protein levels, while the mRNA levels remain constant. Interestingly, for this activity the lncRNA only requires the partial sequence overlap between the two transcripts (73 nt including the AUG codon), as well as a SINEB2 repeat of the B3 subclass in the correct orientation. Furthermore, this mechanism can be applied to other sense-antisense pairs. In fact, an artificial construct containing a 72-nucleotide-long sequence antisense to the AUG-containing region of GFP, and the 3′ end of antisense Uchl1 containing the SINEB2 repeat, is able to induce an increase of GFP protein levels when cotransfected with a GFP plasmid.
Antisense Uchl1 lncRNA presents similar expression patterns to Uchl1 mRNA, which encodes for a neuron-specific protein that functions as deubiquitinating enzyme, ubiquitin ligase and monoubiquitin stabilizer, and is possibly involved in Parkinson's and Alzheimer's diseases. Unlike Uchl1 mRNA, the antisense Uchl1 is enriched in the nucleus of cells, but relocalizes to the cytoplasm when cells are treated with the mTOR inhibitor rapamycin, a general inhibitor of translation of capped mRNAs. Interestingly, this drug treatment results in the localization of Uchl1 mRNA to polyribosomes and the induction of UCHL1 protein levels (Figure 1). The increase of UCHL1 protein levels requires the presence of Uchl1 antisense lncRNA, and can be inhibited by the overexpression of dominant-negative forms of antisense Uchl1 lacking either of its functional elements (the 5′ region complementary to Uchl1 mRNA or the SINEB2 repeat). The translational enhancement promoted by endogenous antisense Uchl1 has only been observed under this particular drug treatment that interferes with cap-dependent translation. It remains unclear what the physiological role of the lncRNA, primarily localized to the nucleus of dopaminergic neurons under basal conditions might be. It is possible that other stimuli or stresses different from rapamycin induce the export of the antisense ncRNA to the cytoplasm to exert UCHL1 protein induction, although basal cytoplasmic levels may already fine-tune UCHL1 protein expression.
The genomic localization and expression pattern of mouse antisense Uchl1 are conserved in other mammals, including humans, where there is an antisense ncRNA whose 5′ sequence overlaps with the 5′ end of Uchl1 mRNA. However, the human antisense Uchl1 lacks the SINEB2 repeat, so it is possible that the regulatory function over Uchl1 mRNA is not conserved in other mammals different from mouse. Still, it cannot be excluded that the function — induction of UCHL1 protein expression, but not the molecular mechanism is conserved, although this end remains to be determined.
The SINEB2 family of repeats constitute approximately 0.7% of total mouse genomic DNA and is also found at low abundance in humans, suggesting that the mechanism described for antisense Uchl1 could be common to other antisense lncRNAs. Indeed, Carrieri and colleagues have searched for other lncRNA-mRNA pairs that show the same structural features, i.e., overlap of 5′ sequences and presence of a SINEB2 repeat in the correct orientation. They found 31 of these sense-antisense pairs, and showed that at least one of them, corresponding to Ubiquitously Expressed Transcript (Uxt) gene, presents the same type of posttranscriptional regulation mediated by its antisense transcript. This mechanism could therefore be common to many lncRNAs, representing a novel functional class of regulatory RNAs.
Beyond the biological implications, the expression of artificial antisense RNAs could be used in molecular biology or even as a therapeutic tool to specifically boost protein expression in mammalian systems. However, in order to make the best use of it, some unsolved aspects of the mechanism should be better understood. For instance, although sequence complementarity between sense and antisense transcripts is required, it is unclear whether a physical interaction exists between both RNAs. The antisense Uchl1 is mainly localized to the nucleus of the cell, and exported to the cytoplasm upon rapamycin treatment. Is the lncRNA localized to polysomes together with Uchl1 mRNA under these conditions? Is the relocalization of antisense Uchl1 necessary for the translational induction? What are the proteins involved in the export of antisense Uchl1 and/or translational induction of Uchl1 mRNA?
Besides the overlap between sense and antisense sequences, the SINEB2 repeat is required in a given orientation. Interestingly, it has been shown that SINEB2 RNAs are transcribed by RNA Pol III upon heat shock and repress transcription by disrupting the binding of RNA polymerase II to certain gene promoters11,12. In the case of the spliced antisense Uchl1 lncRNA, which contains SINEB2 and SINEB1 repeats, an effect on Uchl1 mRNA levels was not observed. However, it remains to be determined whether antisense Uchl1 acts on other genes different from Uchl1, either transcriptionally or posttranscriptionally. The work of Carrieri and colleagues suggests that while the overlapping region between the antisense RNA and the mRNA confers the specificity, the functional element mediating translational induction is the SINEB2 repeat. It would be interesting to know the structural characteristics of this RNA element, as well as the interactions established with proteins, which may be determinant for the functional outcome.
This work involves antisense lncRNAs in a so far unknown mechanism of gene regulation possibly common to dozens of sense-antisense pairs. It also invites to look at genomic repeats as modular elements that confer particular functional features to RNA molecules. Furthermore, it likely represents the evolutionary plasticity of the noncoding genome, which can adopt species-specific ways to fine-tune the complex brain function.
References
- Derrien T, Johnson R, Bussotti G, et al. Genome Res. 2012. pp. 1775–1789. [DOI] [PMC free article] [PubMed]
- Guttman M, Rinn JL. Nature. 2012. pp. 339–346. [DOI] [PMC free article] [PubMed]
- Rinn JL, Chang HY. Annu Rev Biochem. 2012. pp. 145–166. [DOI] [PMC free article] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Nat Rev Genet. 2009. pp. 155–159. [DOI] [PubMed]
- Tay Y, Kats L, Salmena L, et al. Cell. 2011. pp. 344–357. [DOI] [PMC free article] [PubMed]
- Yoon JH, Abdelmohsen K, Srikantan S, et al. Mol Cell. 2012. pp. 648–655. [DOI] [PMC free article] [PubMed]
- Liu X, Li D, Zhang W, et al. EMBO J. 2012. pp. 4415–4427. [DOI] [PMC free article] [PubMed]
- Carrieri C, Cimatti L, Biagioli M, et al. Nature. 2012. pp. 454–457. [DOI] [PubMed]
- Turner AM, Morris KV. Biotechniques. 2010. pp. ix–xvi. [DOI] [PubMed]
- Beltran M, Puig I, Pena C, et al. Genes Dev. 2008. pp. 756–769. [DOI] [PMC free article] [PubMed]
- Nishihara H, Smit AF, Okada N. Genome Res. 2006. pp. 864–874. [DOI] [PMC free article] [PubMed]
- Yakovchuk P, Goodrich JA, Kugel JF. Proc Natl Acad Sci USA. 2009. pp. 5569–5574. [DOI] [PMC free article] [PubMed]