Abstract
Protein homeostasis in higher eukaryotes is balanced by a dynamic network of adaptive mechanisms, including the unfolded protein response (UPR) and autophagy. In a paper recently published in Cell Research, Zhu and co-workers uncover a novel biological function of the unspliced form of the UPR transcription factor XBP1 in the modulation of autophagy through the control of FoxO1 turnover.
Disruption of protein homeostasis underlies the etiology of several human diseases, including cancer, diabetes and neurodegenerative diseases, among other pathologies. The endoplasmic reticulum (ER) is the main organelle involved in protein folding and secretion, where a complex signaling network known as the unfolded protein response (UPR) operates to overcome alterations in the function of the ER1. In doing so, the UPR controls several cellular processes to decrease the unfolded protein load, highlighting the ER-associated degradation pathway and autophagy.
The UPR signals through the activation of several ER stress sensors including IRE1α. Upon activation IRE1α catalyzes the splicing of the mRNA encoding the transcription factor XBP1, leading to the expression of a potent transcription factor termed XBP1s. The non-processed mRNA encodes a protein that is highly unstable with no transcriptional activity, known as XBP1u (for unspliced XBP1)2. IRE1α signaling mediates in part the connection between the UPR and autophagy through the binding of TRAF2 to its cytosolic domain, and the activation of JNK3,4. Besides, XBP1 deficiency in the nervous system enhances autophagy levels, providing protection against amyotrophic lateral sclerosis5 and Huntington's disease6. A recent study described a negative regulation of the transcription factor FoxO1 by XBP1s in pancreatic β cells, mediated by a physical interaction7. FoxO1 has key functions during aging and in the regulation of autophagy in neurons8. In agreement with this, an accumulation of FoxO1 was observed in the brain of XBP1-deficient mice, correlating with the enhancement of autophagy6. However, the mechanisms linking XBP1 with the modulation of FoxO1 are not fully understood.
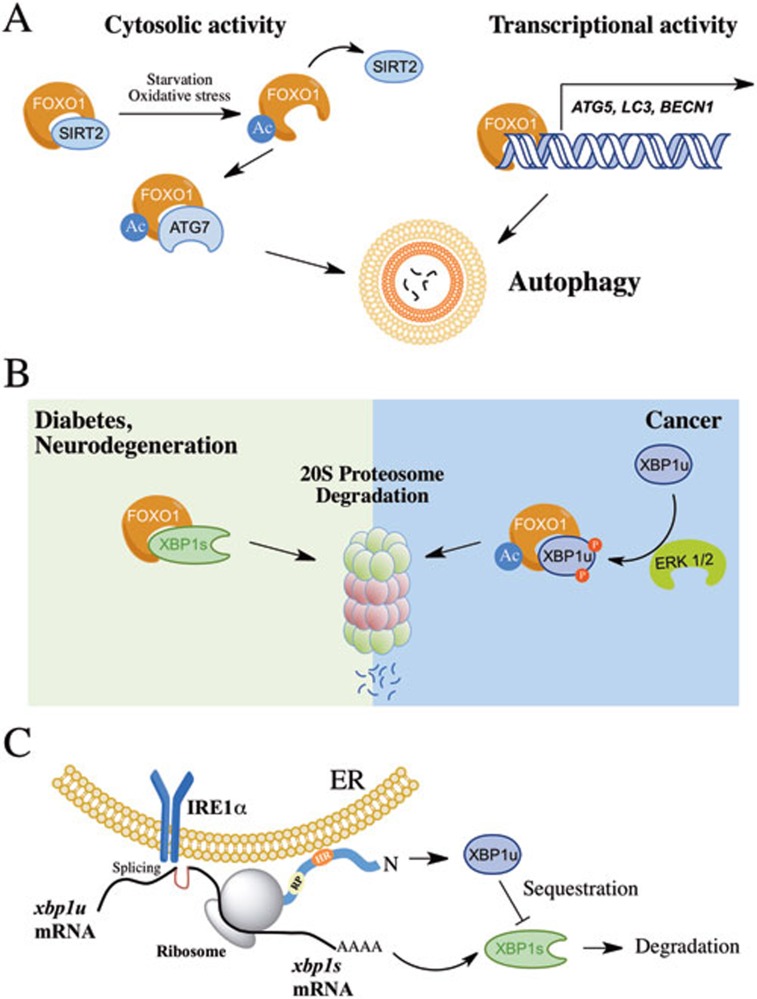
FoxO1 regulates autophagy by two distinct mechanisms: it enhances the transcription of a variety of autophagy-related genes (i.e., atg5, lc3, beclin-1)8; and it is able to induce autophagy in the cytosol through its acetylation and direct binding to ATG79 (Figure 1A). In a paper recently published in Cell Research, Zhao and colleagues uncover a new role of XBP1u in the control of FoxO1 levels and autophagy10. A physical interaction between FoxO1 and XBP1u was observed, which was dependent on the phosphorylation of XBP1u by ERK, leading to the degradation of FoxO1 through the 20S proteasome (Figure 1B). Remarkably, an inverse correlation between FoxO1 and XBP1u levels was discovered in a panel of cancer samples, consistent with previous finding showing enhanced XBP1u levels in breast cancer11.
Figure 1.
Novel function of XBP1u in the control of FoxO1 and autophagy. (A) FoxO1 regulates autophagy through two mechanisms involving the transcriptional control of autophagy-related genes and the activation of ATG7 in the cytosol associated with the acetylation (Ac) of FoxO1 by the dissociation from SIRT2. (B) XBP1u regulates FoxO1 levels mediated by its phosphorylation by ERK and the degradation of FoxO1 by the 20S proteasome. This model has been associated with cancer progression. In addition, XBP1s also regulates FoxO1 levels through a physical interaction and proteasome-mediated degradation, as observed in models of neurodegeneration and diabetes. (C) Summary of the different biological activities of XBP1u in the modulation of UPR signaling. Upon translation of XBP1u, a hydrophobic region (HR) attaches the protein to the ER membrane placing the XBP1 mRNA close to IRE1α for further processing. In addition, a translational pausing (TP) motif stops translation, allowing the splicing process to occur. Besides, XBP1u may interact with and sequester XBP1s in the cytosol, mediating its degradation by the proteasome.
In addition to providing a molecular link between the UPR and autophagy, the current study resolves an important question in the ER stress field: what is the biological function of XBP1u? Recent studies suggest that XBP1u may have a modulatory role in the UPR (Figure 1C). Although XBP1u has an extremely short haft-life12, it contains two conserved domains with relevant functions. Through a membrane-interacting domain, the nascent polypeptide is attached to the ER membrane, placing the XBP1 mRNA in close proximity to IRE1α as a substrate13. This process is optimized due to the presence of a translational pausing region in the XBP1u sequence14. Besides, XBP1u operates in the termination of UPR responses in cells undergoing prolonged ER stress where it directly interacts with XBP1s, sequestering it from the nucleus to enhance its degradation15.
The novel function of XBP1u in controlling FoxO1 may help explaining the upregulation of autophagy in XBP1-deficient cells in vivo. This is particularly relevant because XBP1 mRNA splicing levels are extremely low in the brain at basal conditions. We speculate that this phenomenon may be due to the lack of XBP1u expression. As the phenotypes of IRE1α- and XBP1-deficient animals do not perfectly match1, the current study by Zhao and coworkers adds more complexity to the UPR network by identifying a new biological function of XBP1u. Finally, this report gives clues about an important conundrum: why XBP1u is expressed in mammalian cells? XBP1u may operate through integrating and modulating different homeostatic networks in multicellular organisms, mediating the interconnection between autophagy, the ER stress pathway, and possibly energy metabolism, through the FoxO1 axis.
References
- Hetz C. Nat Rev Mol Cell Biol. 2012. pp. 89–102. [DOI] [PubMed]
- Calfon M, Zeng H, Urano F, et al. Nature. 2002. pp. 92–96. [DOI] [PubMed]
- Ogata M, Hino S, Saito A, et al. Mol Cell Biol. 2006. pp. 9220–9231. [DOI] [PMC free article] [PubMed]
- Castillo K, Rojas-Rivera D, Lisbona F, et al. EMBO J. 2011. pp. 4465–4478. [DOI] [PMC free article] [PubMed]
- Hetz C, Thielen P, Matus S, et al. Genes Dev. 2009. pp. 2294–2306. [DOI] [PMC free article] [PubMed]
- Vidal RL, Figueroa A, Court FA, et al. Hum Mol Genet. 2012. pp. 2245–2262. [DOI] [PMC free article] [PubMed]
- Zhou Y, Lee J, Reno CM, et al. Nat Med. 2010. pp. 356–365. [DOI] [PMC free article] [PubMed]
- Xu P, Das M, Reilly J, Davis RJ. Genes Dev. 2011. pp. 310–322. [DOI] [PMC free article] [PubMed]
- Zhao Y, Yang J, Liao W, et al. Nat Cell Biol. 2010. pp. 665–675. [DOI] [PubMed]
- Zhao Y, Li X, Cai M, et al. Cell Res 1 Jan 2013. doi: 10.1038/cr.2013.2 [DOI]
- Davies MP, Barraclough DL, Stewart C, et al. Int J Cancer. 2008. pp. 85–88. [DOI] [PubMed]
- Navon A, Gatushkin A, Zelcbuch L, et al. FEBS Lett. 2010. pp. 67–73. [DOI] [PubMed]
- Yanagitani K, Imagawa Y, Iwawaki T, et al. Mol Cell. 2009. pp. 191–200. [DOI] [PubMed]
- Yanagitani K, Kimata Y, Kadokura H, et al. Science. 2011. pp. 586–589. [DOI] [PubMed]
- Yoshida H, Oku M, Suzuki M, et al. J Cell Biol. 2006. pp. 565–575. [DOI] [PMC free article] [PubMed]